
A prognostic predictive model constituted with gene mutations of APC, BRCA2, CDH1, SMO, and TSC2 in colorectal cancer
Introduction
Colorectal cancer (CRC) is one of the most common malignant tumors and one of the main causes of cancer-related deaths worldwide (1). According to the International Agency for Research on Cancer (IARC), there were approximately 1.8 million new cases of CRC (10% of all cancers), and more than 860,000 CRC-related deaths (9% of cancer-related deaths) in the world in 2018 (2,3). In China, CRC is one of the most common cancers and its incidence rate is increasing over time (4). CRC is often preventable and curable, with a slow progression, and the main treatment is surgery combined with postoperative chemotherapy (5). However, because CRC symptoms are usually only observed at the late stage of the disease, the lack of early diagnosis and metastasis are the two main causes of CRC-related death (6). In addition, due to the prevalence of COVID-19, the isolation measures have been taken in various regions, the normal diagnosis and treatment process of patients with malignant tumors are seriously affected, including treatment suspension, delay or seeking alternative solutions, which has a huge impact on the prognosis and mental state of patients (7). Therefore, early detection of CRC through active population screening has a significant impact on reducing its mortality (8). In addition, it is increasingly recognized that the treatment of common cancers can be tailored to the patient’s expected prognosis or response to treatment (9). In CRC, with the development of understanding the interaction between molecular drivers, significant progress in drug development of targeted drugs reveals the clinical value of tumor profiling. The response of metastatic CRC to current targeted drugs and immunotherapy is highly dependent on driver genes. Recently, much more clinical studies showed that biomarkers are associated with treatment stratification, treatment decision based on molecular subtypes has developed significantly. Using next-generation sequencing (NGS) technology to detect the changes of multiple genes over time has great potential in cancers, which can promote the drug development of molecular subtypes (10). Thus, screening for CRC-related risk factors is of great significance in evaluating prognosis.
Cancer is the accumulation of mutations in oncogenes and tumor donor genes, which are involved in cell cycle control, DNA repair, or apoptosis. Powerful reactive biomarkers can be used in some new molecular therapies, which usually contain mutations targeting specific proteins (11). Prognostic biomarkers still have considerable potential clinical importance, and gene mutations can be used as an index to predict the prognosis of CRC (12). Such markers can guide more or less aggressive treatment regimens and enable clinicians to balance the expected outcomes with the toxicity of early and late treatment. However, due to individual differences and the cost of gene testing, truly useful mutant genes are very limited (9). Hence, it is necessary and meaningful to find more oncogene mutations and their roles in predicting prognosis.
In this study, we identified the mutated genes in clinical patients, and comprehensively evaluated the mutation status, clinical, pathological, and other factors in the TCGA cohort to determine a prognostic prediction model for CRC. We present the following article in accordance with the TRIPOD reporting checklist (available at http://dx.doi.org/10.21037/atm-21-1010).
Methods
Sample and data collection
From October 2017 to September 2020, thirty-four patients who were diagnosed with CRC in Tianjin Medical University Cancer Institute and Hospital (Tianjin, China) were enrolled in this study. Cancer tissues were collected from the patients and 1,000-cancer gene panel targeted NGS was performed, with paracancerous tissues used as the control. In addition, clinical information including sex, age, history of smoking or drinking, past medical history and complications, pathological type, stage, maximum diameter of tumors, treatment, recurrence and metastasis, and survival were collected. All procedures performed in studies involving human participants were in accordance with the ethical standards of the institutional and national research committee and with the Helsinki declaration (as revised in 2013). This study was approved by Tianjin Medical University Cancer Institute and Hospital and written informed consent was obtained from all patients.
The mutation results of 536 CRC patients analyzed by MuTect software (GATK, Broad Institute, USA) were downloaded from The Cancer Genome Atlas (TCGA) database (https://portal.gdc.cancer.gov/), including 399 cases of colon cancer data and 137 cases of rectal cancer data. Patients with nil or incomplete clinical data were excluded, and a final 526 samples were reserved for follow-up analysis.
Targeted NGS sequencing
DNA in tissue samples was extracted using a genomic DNA extraction kit (Qiagen, Valencia, CA) according to the manufacturer’s instructions. After interruption by ultrasound, the DNA fragments were purified and size selected with Agencourt AMPure XP beads (Agencourt Biosciences, Beverly, MA), then PCR amplification was performed. SeqCap EZ MedExome Enrichment kits (Roche, Basel, CH) were used to capture the target sequences, and Roche’s customized 1,000-gene probes (Roche, Basel, CH) were used to capture and elute the hybridization library. The obtained libraries were subjected to targeted sequencing using an Illumina HiSeq Xten sequencer (San Diego, CA, USA).
Mutation identification and functional enrichment analysis
The Burrows-Wheeler Aligner (BWA) software was used to map the cancer tissue sequencing data to the corresponding paracancerous sequencing data for tumor-specific somatic mutation detection. MuTect (version 1.1.4) was used to identify mutation types, the mutation frequency was counted by a self-developed Python script to search for high-frequency mutation genes, and the GenVisR package (version 1.18.1) in R (version 3.6.3) was used to draw the mutation landscape map.
Afterward, the clusterProfiler package (version 3.14.3) was used to enrich the selected high-frequency mutation genes with Gene Ontology (GO) terms and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathways, using the Benjamini and Hochberg (BH) P value correction method, where only P<0.05 and q<0.05 were retained.
Construction of the prognostic model based on TCGA data
In TCGA, a Cox proportional hazards model univariate analysis was performed using the survival package (version 3.2-3) and survminer package (version 0.4.8) in R (version 3.6.3) to analyze the correlation between clinical parameters or mutation status of the high-frequency genes and overall survival (OS), recurrence, and metastasis. Afterward, the TCGA samples were randomly divided into a model group and a validation group at a ratio of 7:3, and a Cox proportional hazards model multivariate analysis was performed to screen the risk factors affecting the OS of patients, and the model was obtained and optimized by a successive sweep method.
The ROC curves of the model and validation groups were drawn by the survivaloc package (version 1.16.2) in R (version 3.6.3). The bootstrap method was used to repeat sampling 1000 times, the calibration curves in the model and validation groups were drawn, and the consistency test was conducted. The R package ggplot2 (version 3.3.2) and gridextra (version 2.3) were used to draw the risk value map and survival scatter diagram. The Survminer package (version 0.4.8) was used to draw the survival curve.
Statistical analysis
Statistical analysis was carried out using R (Windows version 3.6.3), and a P value <0.05 was considered statistically significant. Unpaired t-tests were used for normally distributed data, and the Mann-Whitney U test was used for non-normal distributions of data. The Chi-square test or Fisher exact probability method was used for categorical variables, and the Wilcoxon rank-sum test was used to analyze the continuous variables.
Results
Mutation analysis
A total of 423 mutations in 75 genes were detected from 34 clinical patients. By analyzing the mutation frequency of each gene, we finally selected 26 genes with mutation frequencies greater than 15% in all clinical patients as the high-frequency mutation genes for the follow-up analysis. Figure 1A presents the mutation distribution of 26 genes, of which TP53 has the highest mutation frequency (47.06%), followed by APC and KRAS (both 32.35%).
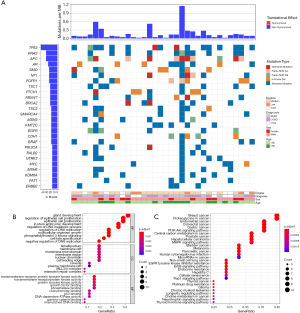
In addition, the functional terms and pathways of these 26 high-frequency mutant genes were enriched. A total of 751 GO terms (including 700 BPs, 28 CCs, and 23 MFs) and 87 KEGG pathways were enriched. Figure 1B and C show the top 10 BPs, CCs, MFs, and top 30 pathways according to the P value. The GO terms of gland development, regulation of epithelial cell proliferation, and epithelial cell proliferation, and the KEGG pathways of breast cancer and proteoglycans in cancer, enriched the most number of genes.
Establishment and assessment of the predictive model
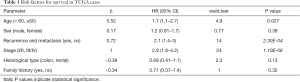
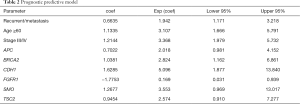
In TCGA samples, after Cox univariate analysis, the clinical parameters of age (<60, ≥60), stage (I/II, III/IV), and recurrence and metastasis were significantly correlated with the OS of patients (P<0.05, Table 1). Therefore, age, stage, metastasis and recurrence, and the 26 high-frequency mutation genes were included in the multivariate analysis. In the model group, recurrence, age ≥60, stage III/IV, mutation status of APC, BRCA2, CDH1, SMO, and TSC2 were risk factors for prognosis, and FGFR1 mutation was a protective factor for CRC, so these factors were used to construct the predictive model (Table 2).
Full table
Full table
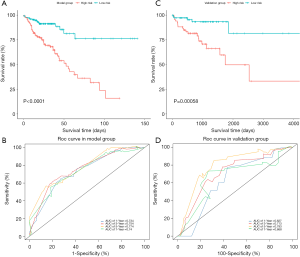
According to the model, the samples can be divided into two groups based on the median risk score (Figure S1A,B). Figure 2A shows the survival of high-risk and low-risk patients in the model group, where the OS of high-risk patients is significantly shorter than that of low-risk patients (P<0.001, Figure 2A). The receiver operating characteristic (ROC) curve indicated that the model had a good discriminative ability (Figure 2B), with the area under the curve (AUC) values of 0.734, 0.754, 0.774, and 0.74 for 1-, 3-, 5- and 7-year survival, respectively. The large AUC represents high resolution. The consistency test of the 1-, 3-, and 5-year predicted OS (Figure S1C,D,E) showed that the point line (red line) was close to the dotted line (gray line), indicating that our model had good consistency.
Validation of the predictive model
In the validation group of TCGA, both the high-risk and low-risk patients were predicted by the model (Figure S1F,G). Consistent with the model group, the OS of high-risk patients was significantly shorter than that of low-risk patients in the validation group (P<0.001, Figure 2C). The AUC values of 1-, 3-, 5- and 7-year survival were 0.627, 0.713, 0.793, and 0.668, respectively (Figure 2D). The calibration curve showed good calibration (Figure S1H,I,J), indicating that our model had sufficient credibility.
Preliminary molecular mechanism of CRC progression
To explore the molecular mechanism of the progression of CRC, we first analyzed the significance between the mutation status of the 26 high-frequency mutation genes and the clinical parameters. In TCGA cohort, mutations of BRAF and KMT2C were significantly associated with more clinical parameters, and the tumor stage was statistically related to a greater proportion of mutated genes. Patients with recurrence and metastasis had a higher proportion of KMT2C and KRAS mutations (P<0.05, Figure 3A).
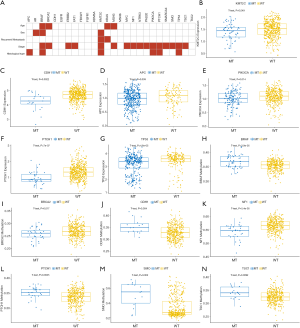
Then, we analyzed the correlation between OS and the mutation frequency, expression, and methylation levels of the 26 high-frequency mutant genes in TCGA cohort. A total of seven genes with a significant effect on OS were screened out and are listed in Table 3. Afterward, to further investigate the effect of mutation on gene expression and methylation, we analyzed the differences in expression and methylation levels between mutated and non-mutated cases. The mutations of KMT2C, CDH1, APC, PIK3CA, PTCH1, and TP53 significantly affected the gene expression level (Figure 3B,C,D,E,F,G), while BRAF, BRCA2, CDH1, NF1, PTCH1, SMO, and TSC1 gene mutations affected methylation (Figure 3H,I,J,K,L,M,N).
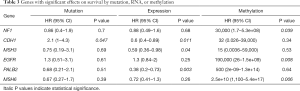
Full table
Discussion
CRC is one of the most common malignant tumors in the world and has a high incidence and mortality rate. Whole-exome sequencing (WES) technology focuses on the coding regions (exons) of the genome to find common or rare variants related to diseases or phenotypes, which reduces cost and time compared with whole genome sequencing (13). The most common method of WES is to “capture” the targeted DNA fragment using oligonucleotide probe hybridization. In CRC patients, WES is widely used for detecting mutations of driver genes and therapeutic targets to direct treatment (14,15). In this study, we screened high-frequency mutated genes in CRC patients by 1,000-gene targeted NGS sequencing, which captured a total of 1.1 MB from 1,000 known cancer-related genes and enriched some key pathways. Combined with clinical parameters, a series of comprehensive analyses were carried out that resulted in 6 key mutation genes and 3 clinical parameters being identified, which were closely related to the prognosis of CRC patients.
Early diagnosis and an accurate prognostic evaluation are two key strategies to improve the survival rate of cancer patients. Tumor biomarkers provide an effective tool for early cancer detection and an evaluation of prognosis. The combined application of multiple markers can improve the accuracy of cancer screening and diagnosis, and the combination of clinical indicators and marker gene targets can reduce the probability of false positives or false negatives, thus improving the diagnostic efficiency of gene detection (16). In this study, multiple mutated genes and clinical indicators were used to predict the prognosis of CRC patients. The Cox proportional hazard regression model takes survival outcome and survival time as dependent variables and is mainly used for the correlation analysis of survival data (17). We used this model to analyze the prognostic risk of TCGA patients, and the clinical parameters of recurrence/metastasis, age ≥60, stage III/IV, and mutation genes of APC, BRCA2, CDH1, FGFR1, SMO, and TSC2 were identified as risk factors for poor outcome, which were verified in the validation group (Figure 2). Age is one of the key factors related to the survival of CRC patients. Study of Leong et al. (18) showed that the 5-year survival rate of older patients (≥70) was the worst compared with other ages. However, in some studies, there was no significant difference in mortality among different age groups of CRC patients (19). The reasons may be that: (I) with the improvement of medical level, the survival rate of elderly patients with CRC has improved; (II) the cancer of young patients is more aggressive and less responsive to treatment; and (III) ethnicity is an important predictor for survival of CRC patients (18). Therefore, further study is necessary to explore the relationship between age and survival in Chinese CRC patients.
To further explore the mechanism of genes in the occurrence and development of CRC, we analyzed the relationship between mutation status and expression level, methylation level, high-risk clinical parameters, and OS in TCGA cohort. Among the high-frequency mutation genes, mutations in KMT2C, CDH1, APC, PIK3CA, PTCH1, and TP53 significantly affected gene expression levels (Figure 3B,C,D,E,F,G), while BRAF, BRCA2, CDH1, NF1, PTCH1, SMO, and TSC1 gene mutations affected methylation level (Figure 3H,I,J,K,L,M,N). Among the risk factors for OS, almost all mutated genes are included in genes that affect methylation (BRCA2, CDH1, SMO, TSC1), but the methylation level of these genes is not statistically significant with OS. We speculated that mutations in these genes affect the methylation level, which in turn affects their downstream target genes, cytokines, or pathways related to CRC occurrence and development, and ultimately affects the survival of patients. Researchers have found that inactivating mutations of APC gene led to hyperactivation of the WNT signaling pathway, which is a hallmark of CRC (20,21). Besides, there were some studies proved that BRCA2 mutations were associated with CRC, but studies on the determination of the excess incidence of CRC in BRCA mutation carriers have conflicting results (22). For example Gay-Bellile et al. (23) found that BRCA2 variants could be implicated in familial CRC inheritance, but another meta-analysis showed that BRCA1 and/or BRCA2 mutation carriers were not at a higher risk of CRC (24). Therefore, our results are necessary to be confirmed in large scale studies in the future.
Besides, among the above genes, the mutation of CDH1 not only reduces the expression level and increases the methylation level but also is an independent influencing factor of patient survival. CDH1 is a tumor suppressor gene located on chromosome 16q22.1. The mutation of the CDH1 gene and the loss of its related protein, E-cadherin, leads to an epithelial-mesenchymal transition (EMT) process, which in turn causes the loss of cell-cell adhesion (25). The genetic, or epigenetic, changes of E-cadherin result in epithelial cell adhesion and cell structure changes, abnormal matrix interactions, and altered cell migration and signal transduction, thereby promoting tumor occurrence (26-29). CDH1 gene exon mutations are one of the most important factors that cause multiple tumors such as gastric, colorectal, and breast cancers. Screening the mutation can be used to guide clinical diagnosis and the treatment of tumors. Aitchison et al. (30) discovered CDH1 germline mutations in some patients with signet ring cell carcinoma of the rectum, and the expression of E-cadherin was decreased in these tumors, which supports the previous discovery of the role of a CDH1 mutation in the development of CRC. Also, for these patients, the detection of germline mutations in family members may be helpful for tumor screening and early detection. Other changes in the CDH1 gene, such as intron mutation, gene methylation, and single nucleotide polymorphism may also affect its expression, but the relationship between these changes and tumor diagnosis requires further exploration.
In addition, we also found that the NF1 methylation level was significantly decreased in patients with the NF1 gene mutation, and there was a statistical correlation between NF1 methylation and prognosis in TCGA. NF1 is a tumor suppressor gene located on chromosome 17q11.2, and its hereditary mutation is associated with neurofibromatosis type 1 (NF1) (31). Acquired somatic mutations of NF1 have also been detected in many non-NF1-related malignant tumors; for example, studies have found that NF1 mutations occur in a variety of malignant tumors, including melanoma, breast cancer, pancreatic cancer, gastric cancer, lung cancer, and CRC (32). In 2012, genome-scale analysis of 212 CRC cases in TCGA revealed that the NF1 mutation rate was about 5.6% (11/212) (12), and subsequent studies confirmed that NF1 mutation occurred in 5.6% (4/72) and 5.8% (39/619) of cases, respectively (33,34). NF1 is the main regulator of the Ras-MAPK pathway, therefore mutation of the NF1 gene can lead to Ras-MAPK pathway disorder (35,36), which is consistent with our enrichment analysis results indicating that NF1 is enriched in the MAPK and Ras signaling pathways. However, the mechanism of NF1 mutation in the occurrence and development of CRC has not been elucidated, and further investigation is warranted to verify and advance this finding.
This research is mainly limited in the following aspects: Most of the patients were newly diagnosed, and because the follow-up time was short, it was insufficient to analyze the correlation between mutations or clinical indicators and prognosis in the clinical cohort. In the future, we will continue to follow up to get enough survival data to further verify the results. In addition, this was a preliminary study, the results are exploratory and should be further addressed; the number of patients in this study is small, and a large sample study should be added for verification. Moreover, the mechanisms of these mutated genes in the progression and metastasis of CRC are still unclear, which are necessary to be explored through investigations at molecular and cellular level in the future.
In conclusion, we screened the prognostic risk factors of CRC based on clinical parameters and mutated genes, constructed a prognostic predictive model, and preliminarily explored the molecular mechanism affecting the prognosis. This provides new insight into the search for novel biomarkers of CRC. Further in vitro and in vivo experiments and large-scale clinical trials are necessary to verify these results.
Acknowledgments
We thank all patients who participated in this study.
Funding: This study was supported by the National Natural Science Foundation of China (82073010) and the Science and Technology Development Fund of Tianjin Education Commission for Higher Education (2020KJ133).
Footnote
Reporting Checklist: The authors have completed the TRIPOD reporting checklist. Available at http://dx.doi.org/10.21037/atm-21-1010
Data Sharing Statement: Available at http://dx.doi.org/10.21037/atm-21-1010
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/atm-21-1010). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. All procedures performed in studies involving human participants were in accordance with the ethical standards of the institutional and national research committee and with the Helsinki declaration (as revised in 2013). This study was approved by Tianjin Medical University Cancer Institute and Hospital and written informed consent was obtained from all patients.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Binefa G, Rodriguez-Moranta F, Teule A, et al. Colorectal cancer: from prevention to personalized medicine. World J Gastroenterol 2014;20:6786-808. [Crossref] [PubMed]
- Jung G, Hernández-Illán E, Moreira L, et al. Epigenetics of colorectal cancer: biomarker and therapeutic potential. Nat Rev Gastroenterol Hepatol 2020;17:111-30. [Crossref] [PubMed]
- Bray F, Ferlay J, Soerjomataram I, et al. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin 2018;68:394-424. [Crossref] [PubMed]
- Chen W, Zheng R, Baade PD, et al. Cancer statistics in China, 2015. CA Cancer J Clin 2016;66:115-32. [Crossref] [PubMed]
- Bertelsen CA, Bols B, Ingeholm P, et al. Can the quality of colonic surgery be improved by standardization of surgical technique with complete mesocolic excision? Colorectal Dis 2011;13:1123-9. [Crossref] [PubMed]
- Wei J, Ge X, Tang Y, et al. An Autophagy-Related Long Noncoding RNA Signature Contributes to Poor Prognosis in Colorectal Cancer. J Oncol 2020;2020:4728947 [Crossref] [PubMed]
- Yang Y, Gao ZY, Chen YK, et al. Effects on the integrated treatment of colorectal cancer patients during COVID-19 epidemic in China: a cross-sectional study. Zhonghua Wei Chang Wai Ke Za Zhi 2020;23:795-800. [PubMed]
- Loktionov A. Biomarkers for detecting colorectal cancer non-invasively: DNA, RNA or proteins? World J Gastrointest Oncol 2020;12:124-48. [Crossref] [PubMed]
- Domingo E, Camps C, Kaisaki PJ, et al. Mutation burden and other molecular markers of prognosis in colorectal cancer treated with curative intent: results from the QUASAR 2 clinical trial and an Australian community-based series. Lancet Gastroenterol Hepatol 2018;3:635-43. [Crossref] [PubMed]
- Dienstmann R, Salazar R, Tabernero J. Molecular Subtypes and the Evolution of Treatment Decisions in Metastatic Colorectal Cancer. Am Soc Clin Oncol Educ Book 2018;38:231-8. [Crossref] [PubMed]
- de Fraipont F, Moro-Sibilot D, Michelland S, et al. Promoter methylation of genes in bronchial lavages: A marker for early diagnosis of primary and relapsing non-small cell lung cancer? Lung Cancer 2005;50:199-209. [Crossref] [PubMed]
- Cancer Genome Atlas Network. Comprehensive molecular characterization of human colon and rectal cancer. Nature 2012;487:330-7. [Crossref] [PubMed]
- Serratì S, De Summa S, Pilato B, et al. Next-generation sequencing: advances and applications in cancer diagnosis. Onco Targets Ther 2016;9:7355-65. [Crossref] [PubMed]
- Feng L, Hong S, Gao J, et al. Whole-Exome Sequencing Characterized the Landscape of Somatic Mutations and Pathways in Colorectal Cancer Liver Metastasis. J Oncol 2019;2019:2684075 [Crossref] [PubMed]
- Chang YS, Lee CC, Ke TW, et al. Molecular characterization of colorectal cancer using whole-exome sequencing in a Taiwanese population. Cancer Med 2019;8:3738-47. [Crossref] [PubMed]
- Bustos García de Castro A, Ferreirós Domínguez J, Delgado Bolton R, et al. PET-CT in presurgical lymph node staging in non-small cell lung cancer: the importance of false-negative and false-positive findings. Radiologia 2017;59:147-58. [PubMed]
- Geng H, Li S, Guo Y, et al. Survival prediction for patients with lung adenocarcinoma: A prognostic risk model based on gene mutations. Cancer Biomark 2020;27:525-32. [Crossref] [PubMed]
- Leong E, Ong SK, Madli F, et al. Survival Rates and Associated Factors of Colorectal Cancer Patients in Brunei Darussalam. Asian Pac J Cancer Prev 2020;21:259-65. [Crossref] [PubMed]
- Wan Ibrahim NR, Chan HK, Soelar SA, et al. Incidence, Clinico-demographic Profiles and Survival Rates of Colorectal Cancer in Northern Malaysia: Comparing Patients Above and Below 50 Years of Age. Asian Pac J Cancer Prev 2020;21:1057-61. [Crossref] [PubMed]
- Schatoff EM, Goswami S, Zafra MP, et al. Distinct Colorectal Cancer-Associated APC Mutations Dictate Response to Tankyrase Inhibition. Cancer Discov 2019;9:1358-71. [Crossref] [PubMed]
- Aghabozorgi AS, Bahreyni A, Soleimani A, et al. Role of adenomatous polyposis coli (APC) gene mutations in the pathogenesis of colorectal cancer; current status and perspectives. Biochimie 2019;157:64-71. [Crossref] [PubMed]
- Sopik V, Phelan C, Cybulski C, et al. BRCA1 and BRCA2 mutations and the risk for colorectal cancer. Clin Genet 2015;87:411-8. [Crossref] [PubMed]
- Gay-Bellile M, Privat M, Martins A, et al. Is BRCA2 involved in early onset colorectal cancer risk? Clin Genet 2020;97:668-9. [Crossref] [PubMed]
- Cullinane CM, Creavin B, O'Connell EP, et al. Risk of colorectal cancer associated with BRCA1 and/or BRCA2 mutation carriers: systematic review and meta-analysis. Br J Surg 2020;107:951-9. [Crossref] [PubMed]
- Shenoy S. CDH1 (E-Cadherin) Mutation and Gastric Cancer: Genetics, Molecular Mechanisms and Guidelines for Management. Cancer Manag Res 2019;11:10477-86. [Crossref] [PubMed]
- Bruner HC, Derksen PWB. Loss of E-Cadherin-dependent cell-cell adhesion and the development and progression of cancer. Cold Spring Harb Perspect Biol 2018;10:a029330 [Crossref] [PubMed]
- Li D, Lo W, Rudloff U. Merging perspectives: genotype-directed molecular therapy for hereditary diffuse gastric cancer (HDGC) and E-cadherin-EGFR crosstalk. Clin Transl Med 2018;7:7. [Crossref] [PubMed]
- Figueiredo J, Melo S, Carneiro P, et al. Clinical spectrum and pleiotropic nature of CDH1 germline mutations. J Med Genet 2019;56:199-208. [Crossref] [PubMed]
- Nam S, Kim JH, Lee DH. RHOA in gastric cancer: functional roles and therapeutic potential. Front Genet 2019;10:438. [Crossref] [PubMed]
- Aitchison A, Hakkaart C, Whitehead M, et al. CDH1 gene mutation in early-onset, colorectal signet-ring cell carcinoma. Pathol Res Pract 2020;216:152912 [Crossref] [PubMed]
- Bollag G, McCormick F. Differential regulation of rasGAP and neurofibromatosis gene product activities. Nature 1991;351:576-9. [Crossref] [PubMed]
- Philpott C, Tovell H, Frayling IM, et al. The NF1 somatic mutational landscape in sporadic human cancers. Hum Genomics 2017;11:13. [Crossref] [PubMed]
- Giannakis M, Mu XJ, Shukla SA, et al. Genomic correlates of immune-cell infiltrates in colorectal carcinoma. Cell Rep 2016;15:857-65. [Crossref] [PubMed]
- Seshagiri S, Stawiski EW, Durinck S, et al. Recurrent R-spondin fusions in colon cancer. Nature 2012;488:660-4. [Crossref] [PubMed]
- Stites EC, Trampont PC, Haney LB, et al. Cooperation between noncanonical Ras network mutations. Cell Rep 2015;10:307-16. [Crossref] [PubMed]
- Rasmussen SA, Friedman JM. NF1 gene and neurofibromatosis 1. Am J Epidemiol 2000;151:33-40. [Crossref] [PubMed]
(English Language Editor: D. Fitzgerald)


