
The G199X and V157fs mutations in the TP53 gene promote malignancy in serous ovarian cancer: an analysis using whole-exome sequencing
Introduction
Ovarian cancer represents the deadliest gynecologic malignancy (1). The World Health Organization (WHO) estimates that approximately 225,500 new ovarian cancer cases and 140,200 related deaths occur each year (2). High-grade serous ovarian cancer (HGSOC) is often detected in women at stage III or IV, with 5-year survival rates in the range of 26–42% (3). New treatments such as targeted therapy have emerged, with various targets being explored. Since the discovery of BRCA1 and BRCA2 mutations, HGSOC patients with these two mutations have been shown to benefit significantly from Olaparib compared to non-carriers (4). With the development of clinical genetics, novel candidates are urgently needed for improving ovarian cancer diagnosis and treatment.
Recently, multi-gene panels have been used to assess gene mutations, with a large amount of unknown but significant variants identified. While exons only represent 1.5% of all genomic DNA, they are implicated in most human diseases. Whole exome sequencing only assesses exons or protein coding regions, however, it has already been applied in prenatal diagnosis (5). Next generation sequencing (NGS) technologies provide rapid replication of susceptible genes in large sample sizes. In addition, it can be used to examine the coding regions in the human genome that may contribute to disease predisposition. This current investigation aimed to identify candidate genes with uncommon mutations associated with serous ovarian cancer using whole-exome sequencing and targeted sequencing. In vitro experiments were conducted to detect the functions and impacts of these mutations on oncogenesis.
We present the following article in accordance with the MDAR reporting checklist (available at http://dx.doi.org/10.21037/atm-21-583).
Methods
Patients and specimens
A total of 88 serous ovarian tumor and para-tumor tissue specimens were obtained from 76 individuals treated in the Department of Gynecology and Obstetrics, Sichuan Academy of Medical Sciences & Sichuan Provincial People’s Hospital, People’s Republic of China from 2015 to 2020. For whole-exome sequencing, 24 samples were obtained from 12 patients at the time of diagnosis (tumor and para-tumor tissues). Targeted deep sequencing was conducted on another 64 ovarian cancer tissue specimens obtained from pathology slides. Peripheral blood samples were collected from 156 healthy women as control samples for the targeted deep sequencing studies. All procedures performed in this study involving human participants were in accordance with the Declaration of Helsinki (as revised in 2013). The study was approved by the Ethics Committee of Sichuan Provincial People’s Hospital and signed informed consents were obtained from all participants.
DNA isolation and whole-exome sequencing
DNA was extraction from primary ovarian cancers and matched para-tumor tissue specimens from the same individuals. The TruSeq DNA Sample Preparation Kit (Illumina) was used for preparation of the DNA library. Exomes were enriched in-solution with Agilent SureSelectXT Human All Exon Kit V6, and DNA sequencing was performed with a SureSelectXT Reagent kit on a Hiseq2000 Sequencing System (Illumina).
Targeted gene sequencing
The candidate gene was screened to detect mutations in 220 samples (including 64 cancer tissues and 156 normal blood samples) using Illumina cBot Cluster Station/Illumina Hiseq. Nextera Rapid Capture Exome Kit (Illumina) was utilized for library construction and exome enrichment. NGS results were primarily assessed by generating FASTQ files and assessing data quality. The obtained reads underwent alignment with the human reference genome hg19.
Copy number (CN) variation detection
Approximately 250 ng of DNA/specimen was used for hybridization using CytoScan 750K Array (Affymetrix) according to the manufacturer’s instructions. Data analysis was performed using the Picard Software (https://broadinstitute.github.io/picard/), and the SNP-FASST2 segmentation algorithm was applied for normalization. After normalization, Nexus v7.5 was used to visualize probe intensity and allele ratio. All primers and probes were provided by Applied Biosystems.
Patient data
Patient data were obtained from the medical records system and follow-up records. Collated data included drugs administered, physical examinations, and laboratory results. Follow-up duration was the time from surgical treatment to disease progression/relapse, death, final hospital visit, or phone call. Overall survival (OS) was assessed from the date of surgical treatment to death. The cut-off time for survival analysis was February 2021.
Cell culture and transfection
Ovarian cancer A2780 cells were purchased from BNCC (BeNa Culture Collection, Beijing, China) and cultured in complete Roswell Park Memorial Institute (RPMI)-1640 medium. A2780 cells lack endogenous TP53 expression (6). For transfection, plasmids were purchased from YouBio (Changsha, China). TP53 bearing the G199X (7578254:C>A) and V157fs (7578459-7578459:ins A) mutations were introduced into the pcDNA3.1 plasmid using the Q5 Site-Directed Mutagenesis Kit (New England Biolabs, USA). The following primers were used: CCGAGTGGAATGAAATTTGCG (forward) and ATAAGATGCTGAGGAGGG (reverse) for G199X, and TCCGCGCCATGGCCATCTACAAGCAGTCAC (forward) and ACGCGGGTGCCGGGCGGG (reverse) for V157fs. The resulting constructs were confirmed by Sanger sequencing and were transfection into A2780 cells with Lipofectamine 3000 (Invitrogen, USA) as per the manufacturer’s protocols. Meanwhile, wild-type TP53 and empty vector plasmids were transfected as controls.
Western blot analysis
After 48 hours of transfection, A2780 cells were collected and lysed. The lysates underwent fractionation, electroblotting, and overnight incubation at 4 °C with primary antibodies targeting P53 (1:200; mouse monoclonal) and glyceraldehyde 3-phosphate dehydrogenase (GAPDH 1:1,000). Subsequently, goat anti-mouse secondary antibodies (1:1,000) were probed at 25 °C for 1 hour, and the membranes were exposed to films. Immunoreactivity was determined using Fusion FX7 Spectra (Vilber, France). All antibodies were purchased from Cell Signaling Technology (USA).
Cell Counting Kit-8 (CCK-8) assay
The CCK-8 assay was performed to assess cell proliferation. Briefly, cells were plated into 96-well plates at 1×104 cells per well in triplicates, and transfected with plasmids bearing the G199X or V157fs mutation, or the empty vector. After 48 hours, 10 µL CCK-8 reagent (Boster, USA) was added per well, followed by incubation at 37 °C for 1 hour. Optical density at 450 nm was obtained after 12, 24, 36, and 48 hours on a Model 680 Microplate Reader (Bio-Rad, USA).
Scratch wound-healing assay
The scratch wound-healing assay was conducted to assess cell migration. Cells in 6-well plates were grown to 80% confluency, and a scratch was created with a 10-µL pipette tip on the cell monolayer to obtain a cell-free zone. Cells were then incubated in serum-free culture medium. Cells migrating into the cell-free zone were monitored after 24 hours under an OLYMPUS IX71 inverted microscope (Olympus, Japan), and the percentage of wound closure was analyzed with ImageJ software.
Transwell assay
The transwell assay was performed to evaluate cell invasion ability. Briefly, after Matrigel (Corning, USA) coating, 2×105 cells were suspended in serum-free culture and added to the upper chambers. After a 48-hour incubation at 37 °C, the cells remaining in the upper chambers were gently swabbed. Cells that had migrated through the membrane underwent methanol fixation and crystal violet (0.1%) staining. Ten randomly selected high-power fields (×400) per specimen were analyzed under an OLYMPUS IX71 inverted microscope (Olympus).
Statistical analysis
The SPSS 22.0 software was used for data analysis. Results are expressed as mean ± standard error of the mean (SEM). The association of mutations with clinical features was examined by the χ2 test. OS was analyzed using the Kaplan-Meier test. Student’s t-test was performed for comparisons. Results were considered statistically significant when P<0.05.
Results
Whole-exome sequencing detected mutated genes in ovarian cancer samples
Whole-exome sequencing was performed to examine 12 pairs of serous ovarian cancer tissue specimens and case-matched normal tissue samples to profile somatic mutations. In the present study, 52,464 somatic single nucleotide variants (SNVs), including 27,130 nonsynonymous and 25,334 synonymous SNVs, were identified in ovarian cancer exomes. Of these, 484 were non-frameshift deletions, 347 were stop-gain mutations, 270 were frameshift deletions, 327 were non-frameshift insertions, 172 were frameshift insertions, and 27 were stop-loss mutations. The C>T substitution had the highest frequency among substitution types in serous ovarian cancer. Moreover, C>T mutations at the TCN sites dominated in the trinucleotide signature, particularly in TCA containing sequences (Figure 1).
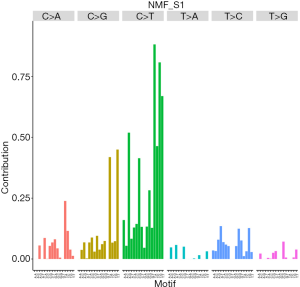
In total, 25 non-silent mutations were most frequently detected in ovarian cancer samples, including in the mucin 4 (MUC4; 66.7%), titin (TTN; 50.0%), TP53 (41.7%), and serine protease 3 (PRSS3; 41.7%) genes (Figure 2). Since TP53 mutations have been reported to occur in more than 50% of human tumors (7), TP53 was selected as a candidate gene for subsequent analyses. A total of 5 point mutations in the TP53 gene was identified in 5 cases, including p.R123W (7577094:G>A), p.R273H (7577120:C>T), p.G199X (7578254:C>A), p.H154R (7578271:T>C), and V157fs (7578459-7578459:ins A). Among them, 3 mutations were synonymous or hotspots, while the remaining 2, named G199X and V157fs, were newly discovered. G199X is a stop-gain mutation while V157fs is a frameshift insertion. Although TP53 mutations have been shown to be related to ovarian cancer, G199X (7578254:C>A) and V157fs (7578459-7578459:ins A) have not been previously reported.
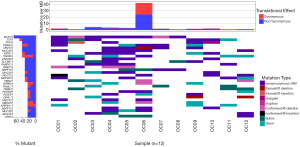
Next-generation sequencing (NGS) detected TP53 mutations in clinical samples
NGS was used to sequence TP53 in 220 samples, including another 64 cases confirmed with high-grade (III–IV serous ovarian cancer by pathology after surgery, and 156 blood samples from healthy women. Combining the whole-exome data and the targeted sequencing data, a total of 76 specimens were assessed (12 samples in the whole-exome analysis and 64 samples in targeted gene sequencing). A total of 45 samples showed TP53 mutations (5 in the whole-exome analysis and 40 in the targeted gene sequencing analysis). TP53 mutations were identified in 59.2% (45 of 76) of the cancer cases. In the control group including 156 healthy women, the TP53 mutation rate in blood samples was approximately 9.0%, which was much lower than that of the ovarian cancer group.
TP53 mutations and clinical outcomes
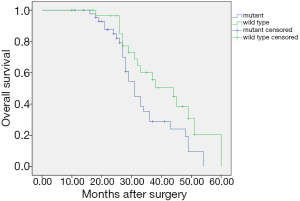
The 76 ovarian cancer patients were followed up to assess their clinical data. Patients with TP53 mutations (n=45) had significantly poorer prognosis compared to patients with wild type TP53 (n=31), with Kaplan-Meier analysis showing a median survival of 34 and 41 months, respectively (χ2=4.176, P=0.041; Figure 3). The 5-year OS rate was significantly lower (approximately 33.3%) in the TP53 mutation group compared with the TP53 wild type group (45.2%, P<0.05).
TP53 G199X and V157fs mutants inhibited P53 expression
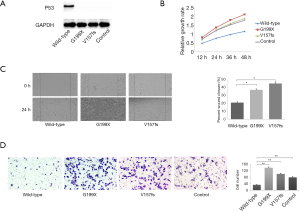
To verify the tumorigenicity of TP53 mutations, the TP53 mutants were overexpressed in the P53-defective human serous ovarian cancer cell line A2780. Cells were transfected with plasmids harboring the G199X (7578254:C>A) mutant or the V157fs (7578459-7578459:ins A) mutant. Western blot analysis showed that transfection with wild typeTP53 resulted in increasing P53 protein expression in A2780 cells compared with control cells (empty vector). Overexpression of the G199X or V157fs mutant resulted in no detectable P53 protein expression (Figure 4A). These results demonstrated that the TP53 G199X and V157fs mutants inhibited P53 expression in A2780 cells.
TP53 G199X and V157fs mutants promoted A2780 cell proliferation
To assess the effect of TP53 mutations on cell proliferation, the CCK-8 assay was performed in A2780 cells. Overexpression of the G199X and V157fs mutants resulted in increased A2780 cell proliferation compared with control cells (P<0.05; Figure 4B), while overexpression of wild type TP53 suppressed cell proliferation compared with control cells. These findings suggested that the TP53 G199X and V157fs mutants increased the cell proliferation ability of A2780 cells.
TP53 G199X and V157fs mutants promoted A2780 cell migration
To determine the effects of the TP53 G199X and V157fs mutants on cell migration, a scratch wound-healing assay was carried out. After 24 hours, migration was higher in cells expressing G199X and V157fs compared with cells transfected with wild type TP53 (Figure 4C). The cells in the V157fs and G199X groups had migrated to filled half of their respective scratched areas, while little change was observed in the wild-type group. This suggested that the TP53 G199X and V157fs mutations enhanced the migratory ability of ovarian cancer cells.
TP53 G199X and V157fs mutants increased A2780 cell invasion
The effects of the TP53 G199X and V157fs mutants on cell invasion were determined using Matrigel™ in the transwell assays. After transfection, cells expressing TP53 G199X and V157fs invaded significantly more compared to the wild-type and control counterparts (Figure 4D). These results suggested that the TP53 G199X and V157fs mutations increased the invasive properties of ovarian cancer cells.
Discussion
The 5-year OS rate for HGSOC patients is approximately 45%. However, vastly different outcomes are associated with the various stages of the disease. While stage I and II HGSOC patients have a 5-year OS rate exceeding 70%, the OS for individuals with late stage (III and IV) disease is only 26–42% (3). Novel treatments such as gene therapy have emerged in recent years. The United States Food and Drug Administration (FDA) has approved Olaparib, a poly-ADP ribose polymerase (PARP) suppressor, as third-line or later-line treatment for germline BRCA mutant (gBRCAmt) ovarian cancer (8). Olaparib has been shown to prolong the progression-free survival (PFS) of patients with gBRCAmt or somatic BRCA1/2 mutant (sBRCAmt) cells. A methylated derivative and structural analogue ofPRIMA-1, i.e., APR-246, and a third generation thiosemicarbazone developed by Critical Outcome Technologies Inc. (COTI-2) COTI-2 were reported to reactive mutant p53 protein by converting it to a form exhibiting wildtype properties. A phase I/IIa clinical trial to advanced serous ovarian cancer with APR-246 reported no major adverse effect (9). Multiple additional genes harboring low-penetrance mutations, including ATM (ataxia telangiectasia-mutated gene), BARD1 (BRCA1-associated RING domain protein 1), BRIP1 (BRCA1 interacting protein C-terminal helicase 1), CHEK2 (checkpoint kinase 2), PALB2 (partner and localizer of BRCA2), RAD51C (RAD51paralog C), and RAD51D (RAD51paralog D), have been linked to a greater risk of ovarian cancer (2). Gene mutations have been tested concurrently by multiple assays to identify unknown significant variants for disease diagnosis and treatment.
The present study identified two point mutations in the TP53 gene in ovarian cancer tissues using whole-exome sequencing combined with NGS. Plasmids carrying TP53 wild-type, or the TP53 mutants G199X and V157fs, were transfected into A2780 cells. The TP53 mutations G199X and V157fs both inhibited P53 protein expression. In addition, these mutations promoted cell migration, invasion, and proliferation in ovarian cancer cells. Furthermore, the TP53 mutation status was correlated to the OS in ovarian cancer patients and may represent a predictive indicator.
In this study cohort, the TP53 mutation rate in patients with serous ovarian cancer was 59.2%. The Cancer Genome Project revealed high-grade serous ovarian adenocarcinoma featured TP53 mutations in 96% of the cases studied (10). However, Song et al. (11) reported relatively lower TP53 mutation rates in Chinese HGSOC patients with a rate of 52.3%. The latter is similar to the mutation rate observed in our study. This variance has been reported with other cancers. Gouas et al. (12) reported that the rate ofTP53 mutation was 75% in hepatocellular carcinoma patients in high-incidence areas of China or East Africa. However, incidences of TP53 mutation are rarer in hepatocellular carcinoma in Europe and the United States, and this may be due to site-specific mutagenesis by aflatoxin. The differences in the mutation rate of TP53 may be due to different mutation sites caused by variations in diet, living habits, and even geographical differences, and this warrants further research.
The whole-exome sequencing data for ovarian cancer samples detected five point mutations in the TP53 gene in five cases, including p.R123W (7577094:G>A), p.R273H (7577120:C>T), p.G199X (7578254:C>A), p.H154R (7578271:T>C), and p.V157fs (7578459-7578459:ins A). Among them, 3 mutations were synonymous or hotspots, while the remaining 2 mutations, namely G199X and V157fs, had not been previously discovered. Prior evidence suggested that theTP53 R273H, R248Q, R175H, and R248W point mutations were the four most frequent mutations (or “hotspots”) in many cancers, including bladder, lung, liver, breast, larynx, and skin cancers (13). In ovarian cancer, TP53 mutations includingp.E298X, p.R248Q, p.Y163C, c.997delG, and c.833delC have been reported (14,15). Other studies have shown that approximately 75% of TP53 mutations were of the missense type. TP53 point mutations in most cancers are most often detected in the amino acid codons of this gene. Approximately 80% are clustered between exons 5 and 8, which encodes the highly conserved DNA binding domain of the P53 protein. In our study, G199X in exon 6 and V157fs in exon 5 were stop-gain mutations and frame-shift insertions, respectively. The mutations generated 3 different phenotypes according to their impacts on P53 protein function, including loss of function, dominant negative, and gain of function (16). In 10,522 cancer genomes from The Cancer Genome Atlas, more than 91% of TP53-mutant cancers exhibit second allele loss by mutation, chromosomal deletion, or copy-neutral loss of heterozygosity. TP53 mutations are associated with enhanced chromosomal instability, including increased amplification of oncogenes and deep deletion of tumor suppressor genes. Tumors with TP53 mutations differ from their non-mutated counterparts in RNA, miRNA, and protein expression patterns, with mutant TP53 tumors displaying enhanced expression of cell cycle progression genes and proteins (17).
In this study, patients with the TP53 G199X and V157fs mutations showed reduced OS, suggested that TP53 mutations induced gene signatures with poor prognosis in ovarian cancer, consistent with the current observations. Between 65–90% of studies in the literature have reported that TP53 mutations are linked to poor prognosis in cancers such as breast, head and neck, bladder, colorectal, and hematological malignancies. In a large-scale study of 264 HGSOC cases, Brachova et al. (18) concluded that individuals harboring oncomorphicTP53 mutations had markedly reduced PFS. Previous investigations have also demonstrated that TP53 mutations can promote drug resistance in a growing array of clinical malignancies (13). However, many studies still reported a lack of such associations, and few reported a contrary trend. For instance, in brain, lung, and ovarian cancers, 50% of reports detected no association between TP53 mutations and patient outcomes. In fact, some studies have even shown thatTP53 mutations may be associated with good prognosis (13). Kang et al. (19) found no marked differences in OS and PFS between gain of function (GOF) by missense mutations in exons 4–9 and no evidence GOF (NE-GOF) mutp53 patients. One possible reason was that the TP53 mutation resided in a different exon or functional domain, even in the non-specific residue (7,20). According to prognostic related genes, 1915, 1194, 822, 423 and 291 were identified good prognostic genes for the HGSCs and LGSCs. Additionally, 2075, 2274, 724, 535 and 303 genes were identified bad prognostic (21). Nakayama (22) revealed global gene upregulation by mutant p53using RNA sequencing analyses and concluded that mutant p53 inducing colorectal cancer progression was not only by a cell intrinsic mechanism, but also by the generation or activation of the microenvironment.
Tumor microenvironment (TME) including immune cells and immunoregulatory molecules are involved both in anti-tumor immune responses of the host and in immunosuppressive mechanisms promoting cancerprogression.TP53 mutation was enriched in samples with higher aneuploidy, which was anti-correlated with expression of immune signaling genes (23). Tumor-infiltrating lymphocytes (TILs) infiltrate to the tumor epithelium (intraepithelial, i.e., TILs) or locate at the peritumoral stroma (stromal, sTILs) (24). Tumor-associated macrophages (TAMs) are further divided into M1 and M2subtypes, of which M1 TAMs contribute to elimination of cancer cells, whereas anti-inflammatory M2 type is involved with tissue repair, angiogenesis and cancer progression (25). TP53 acts as a physiological break forM2 macrophages polarization (26). CD8+ T cells play a critical role in controlling the metastasis and prognosis of cancer (27,28), such as in ovarian cancer (29). Jäntti et al. (29) detected that combined high intraepithelial CD8 and granzyme B expression correlated with prolonged PFS of high-grade serous ovarian cancer patients. Yang et al. (30) reported that the patients with higher CD274, CD8A, GZMA, HLA-A, HLA-B, HLA-C, IRF1, and PRF1 expression levels had significantly longer overall survival in the high-grade serous carcinoma. Ovarian Serous Cystadenocarcinoma (OV) patients with mutant TP53 had significantly higher macrophages infiltration than those with wild-type TP53 (P value <0.05) and poor prognosis associated (21).
In the present study, the TP53 G199X and V157fs mutations triggered malignancy by promoting cell migration, invasion, and proliferation. TP53 encodes the P53 protein and is a tumor suppressor gene. P53 regulates cell proliferation, DNA repair, programmed cell death, genomic stability, senescence, and metabolic homeostasis (31). Liu et al. (32) reported that TP53 mutations could lead to dysfunction of the tumor suppressor Rb protein (pRb), which may cause cervical cancer. The possible mechanisms by which TP53 mutations enhance cell proliferation may lie in regulation of the nuclear transcription factor Y (NF-Y) target genes [CCNA (cyclin A), CCNB2 (cyclin B2), CDK1 (cyclin-dependent kinases 1), and CDC25C (cell division cyclin 25 homolog C)] (33), MAP2K3 (mitogen-activated protein kinase 3) (34), and MAD1L1 (MAD1 mitotic arrest deficient-like 1) (35). Mutant TP53 variants were associated with higher levels of immune cells such as stromal CD3+, CD4+ and FOXP3+ cells in breast ductal carcinoma in situ and increased the growth rate of tumors (36). In addition, mutant P53 up-regulated invasiveness may be mediated through NF-κB target genes, including chemokine (C-X-C motif) ligand 1 (CXCL1), interleukin 1β and metalloproteinase 3 (MMP3) (13). As to migration and metastasis, mutant p53 in vivo could form a complex with phosphorylated Smad2 and p63, simultaneously blocking wild-type p63 transcriptional targets while activating oncogenic genes, such as SHARP1(split and hairy related protein-1), Dicer, and cyclin G2 (37). Weissmueller et al. (38) demonstrated that mutant p53 could directly bind to p73, preventing p73-mediated inactivation of platelet-derived growth factor β (PDGFβ), resulting in an invasive and metastatic phenotype in a pancreatic cancer mouse model. Our future studies will investigate whether TP53 G199X and V157fs mutations cause tumorigenesis through the above-mentioned pathways or genes.
Overall, the present study identified high rates of TP53 G199X and V157fs mutations in ovarian cancer using whole-exome sequencing. These mutations are related to tumorigenesis by promoting tumor cell migration, invasion, and proliferation. Moreover, such mutations were associations with poor prognosis. The current findings suggest that TP53 mutations may provide novel insights into ovarian cancer and may serve as potential diagnostic and therapeutic targets. However, the limited number of patients in the present study precluded the possibility of exploring this further, and future large-scale studies are warranted.
Acknowledgments
We acknowledge support from the Department of Gynecology and Obstetrics, Sichuan Provincial People’s Hospital and Sichuan Provincial Key Laboratory for Human Disease Gene Study.
Funding: This work was supported by the Science & Technology Department of Sichuan Province (grant number: 2015SZ0243) and the Science & Technology Department of Chengdu, Sichuan (grant number: 2019-YF05-00244-SN).
Footnote
Reporting Checklist: The authors have completed the MDAR reporting checklist. Available at http://dx.doi.org/10.21037/atm-21-583
Data Sharing Statement: Available at http://dx.doi.org/10.21037/atm-21-583
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/atm-21-583). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. All procedures performed in this study involving human participants were in accordance with the Declaration of Helsinki (as revised in 2013). The study was approved by the Ethics Committee of Sichuan Provincial People’s Hospital and signed informed consents were obtained from all participants.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Siegel RL, Miller KD, Jemal A. Cancer statistics, 2019. CA Cancer J Clin 2019;69:7-34. [Crossref] [PubMed]
- Lisio MA, Fu L, Goyeneche A, et al. High-Grade Serous Ovarian Cancer: Basic Sciences, Clinical and Therapeutic Standpoints. Int J Mol Sci 2019;20:952. [Crossref] [PubMed]
- Torre LA, Trabert B, DeSantis CE, et al. Ovarian cancer statistics, 2018. CA Cancer J Clin 2018;68:284-96. [Crossref] [PubMed]
- Domchek SM, Aghajanian C, Shapira-Frommer R, et al. Efficacy and safety of olaparib monotherapy in germline BRCA1 / 2 mutation carriers with advanced ovarian cancer and three or more lines of prior therapy. Gynecol Oncol 2016;140:199-203. [Crossref] [PubMed]
- Jelin AC, Vora N. Whole Exome Sequencing: Applications in Prenatal Genetics. Obstet Gynecol Clin North Am 2018;45:69-81. [Crossref] [PubMed]
- Beaufort CM, Helmijr JC, Piskorz AM, et al. Ovarian cancer cell line panel (OCCP): clinical importance of in vitro morphological subtypes. PLoS One 2014;9:e103988 [Crossref] [PubMed]
- Leroy B, Girard L, Hollestelle A, et al. Analysis of TP53 mutation status in human cancer cell lines: a reassessment. Hum Mutat 2014;35:756-65. [Crossref] [PubMed]
- Matsumoto K, Nishimura M, Onoe T, et al. PARP inhibitors for BRCA wild type ovarian cancer; gene alterations, homologous recombination deficiency and combination therapy. Jpn J Clin Oncol 2019;49:703-7. [Crossref] [PubMed]
- Duffy MJ, Synnott NC, Crown J. Mutant p53 as a target for cancer treatment. Eur J Cancer 2017;83:258-65. [Crossref] [PubMed]
- Cancer Genome Atlas Research Network. Integrated genomic analyses of ovarian carcinoma. Nature 2011;474:609-15. Erratum in: Nature 2012;490:298. [Crossref] [PubMed]
- Song F, Li L, Zhang B, et al. Tumor specific methylome in Chinese high-grade serous ovarian cancer characterized by gene expression profile and tumor genotype. Gynecol Oncol 2020;158:178-87. [Crossref] [PubMed]
- Gouas D, Shi H, Hainaut P. The aflatoxin-induced TP53 mutation at codon 249 (R249S): biomarker of exposure, early detection and target for therapy. Cancer Lett 2009;286:29-37. [Crossref] [PubMed]
- Brosh R, Rotter V. When mutants gain new powers: news from the mutant p53 field. Nat Rev Cancer 2009;9:701-13. [Crossref] [PubMed]
- Sokolenko AP, Bizin IV, Preobrazhenskaya EV, et al. Molecular Profiles of BRCA1-Associated Ovarian Cancer Treated by Platinum-Based Therapy: Analysis of Primary, Residual and Relapsed Tumours. Int J Cancer 2020;146:1879-88. [Crossref] [PubMed]
- Arildsen NS, Martin de la Fuente L, Masback A, et al. Detecting TP53 mutations in diagnostic and archival liquid-based Pap samples from ovarian cancer patients using an ultra-sensitive ddPCR method. Sci Rep 2019;9:15506. [Crossref] [PubMed]
- Zhang Y, Cao L, Nguyen D, et al. TP53 mutations in epithelial ovarian cancer. Transl Cancer Res 2016;5:650-63. [Crossref] [PubMed]
- Donehower LA, Soussi T, Korkut A, et al. Integrated Analysis of TP53 Gene and Pathway Alterations in The Cancer Genome Atlas. Cell Rep 2019;28:1370-1384.e5. [Crossref] [PubMed]
- Brachova P, Mueting SR, Carlson MJ, et al. TP53 oncomorphic mutations predict resistance to platinum and taxanebased standard chemotherapy in patients diagnosed with advanced serous ovarian carcinoma. Int J Oncol 2015;46:607-18. [Crossref] [PubMed]
- Kang HJ, Chun SM, Kim KR, et al. Clinical relevance of gain-of-function mutations of p53 in high-grade serous ovarian carcinoma. PLoS One 2013;8:e72609 [Crossref] [PubMed]
- Seagle BL, Eng KH, Dandapani M, et al. Survival of patients with structurally-grouped TP53 mutations in ovarian and breast cancers. Oncotarget 2015;6:18641-52. [Crossref] [PubMed]
- El-Arabey AA, Abdalla M, Abd-Allah AR. SnapShot: TP53 status and macrophages infiltration in TCGA-analyzed tumors. Int Immunopharmacol 2020;86:106758 [Crossref] [PubMed]
- Nakayama M, Sakai E, Echizen K, et al. Intestinal cancer progression by mutant p53 through the acquisition of invasiveness associated with complex glandular formation. Oncogene 2017;36:5885-96. [Crossref] [PubMed]
- Taylor AM, Shih J, Ha G, et al. Genomic and Functional Approaches to Understanding Cancer Aneuploidy. Cancer Cell 2018;33:676-689.e3. [Crossref] [PubMed]
- Santoiemma PP, Powell DJ Jr. Tumor infiltrating lymphocytes in ovarian cancer. Cancer Biol Ther 2015;16:807-20. [Crossref] [PubMed]
- Cheng H, Wang Z, Fu L, et al. Macrophage Polarization in the Development and Progression of Ovarian Cancers: An Overview. Front Oncol 2019;9:421. [Crossref] [PubMed]
- Li L, Ng DS, Mah WC, et al. A unique role for p53 in the regulation of M2 macrophage polarization. Cell Death Differ 2015;22:1081-93. [Crossref] [PubMed]
- Khairallah AS, Genestie C, Auguste A, et al. Impact of neoadjuvant chemotherapy on the immune microenvironment in advanced epithelial ovarian cancer: Prognostic and therapeutic implications. Int J Cancer 2018;143:8-15. [Crossref] [PubMed]
- Mesnage SJL, Auguste A, Genestie C, et al. Neoadjuvant chemotherapy (NACT) increases immune infiltration and programmed death-ligand 1 (PD-L1) expression in epithelial ovarian cancer (EOC). Ann Oncol 2017;28:651-7. [Crossref] [PubMed]
- Jäntti T, Luhtala S, Maenpaa J, et al. Characterization of immunoreactivity with whole-slide imaging and digital analysis in high-grade serous ovarian cancer. Tumour Biol 2020;42:1010428320971404 [Crossref] [PubMed]
- Yang L, Wang S, Zhang Q, et al. Clinical significance of the immune microenvironment in ovarian cancer patients. Mol Omics 2018;14:341-51. [Crossref] [PubMed]
- Nakamura M, Obata T, Daikoku T, et al. The Association and Significance of p53 in Gynecologic Cancers: The Potential of Targeted Therapy. Int J Mol Sci 2019;20:5482. [Crossref] [PubMed]
- Liu Z, Gersbach E, Zhang X, et al. miR-106a represses the Rb tumor suppressor p130 to regulate cellular proliferation and differentiation in high-grade serous ovarian carcinoma. Mol Cancer Res 2013;11:1314-25. [Crossref] [PubMed]
- Di Agostino S, Strano S, Emiliozzi V, et al. Gain of function of mutant p53: the mutant p53/NF-Y protein complex reveals an aberrant transcriptional mechanism of cell cycle regulation. Cancer Cell 2006;10:191-202. [Crossref] [PubMed]
- Bossi G, Marampon F, Maor-Aloni R, et al. Conditional RNA interference in vivo to study mutant p53 oncogenic gain of function on tumor malignancy. Cell Cycle 2008;7:1870-9. [Crossref] [PubMed]
- Iwanaga Y, Jeang KT. Expression of Mitotic Spindle Checkpoint Protein hsMAD1 Correlates with Cellular Proliferation and Is Activated by a Gain-of-Function p53 Mutant. Cancer Res 2002;62:2618-24. [PubMed]
- Toss MS, Abidi A, Lesche D, et al. The prognostic significance of immune microenvironment in breast ductal carcinoma in situ. Br J Cancer 2020;122:1496-506. [Crossref] [PubMed]
- Su X, Chakravarti D, Cho MS, et al. TAp63 suppresses metastasis through coordinate regulation of Dicer and miRNAs. Nature 2010;467:986-90. [Crossref] [PubMed]
- Weissmueller S, Manchado E, Saborowski M, et al. Mutant p53 drives pancreatic cancer metastasis through cell-autonomous PDGF receptor beta signaling. Cell 2014;157:382-94. [Crossref] [PubMed]
(English Language Editor: J. Teoh)


