
Global alternative splicing landscape of skeletal muscle atrophy induced by hindlimb unloading
Introduction
After entering the space orbit, the spacecraft will be in a state of weightlessness. Extended periods of weightlessness or in a microgravity environment restrict astronauts’ movement, resulting in skeletal muscle atrophy. It has been found that after bed rest, physical restriction, or space flight for about 5 days, skeletal muscles will atrophy significantly (1,2). The specific characteristics of skeletal muscle atrophy are as follows: decreased muscle mass, muscle fiber cross-sectional area, and muscle contraction capacity. Further studies have shown that soleus muscle atrophy is the most severe type of atrophic muscle, accompanied by the transition from slow-switch fiber to fast-switch fiber (3,4). The molecular mechanism of skeletal muscle atrophy is complex and still unclear. Therefore, there are no effective drugs or methods to prevent skeletal muscle atrophy caused by weightlessness.
Although the causes of skeletal muscle atrophy are diverse, such as denervation, physical damage, cancer, aging, and disuse, they have similar pathological characteristics and some common activation pathways (5-7). The cause of the atrophic phenotype is the imbalance of protein metabolism, that is, protein synthesis is inhibited and protein degradation is activated (8). Three major protein degradation pathways are activated during muscle atrophy as follows: the ubiquitin-proteasome pathway, autophagy-lysosome pathway, and calpain pathway (9-11). Two studies using proteomics in denervated muscle atrophy model found that a large number of protein expression changes were accompanied by muscle atrophy, ubiquitin proteasome pathway was widely activated (12-14). Our study showed that the ubiquitin-proteasome pathway is activated in the middle and late stages of skeletal muscle atrophy, suggesting that this pathway does not trigger muscle atrophy. In the early stage of skeletal muscle atrophy, oxidative stress, and inflammation-related pathways are relatively active, which may be an important trigger of skeletal muscle atrophy (15,16). Also, numerous studies have shown that a large number of non-coding RNAs are differentially expressed during muscle atrophy. They form a complex competing endogenous RNA network with target genes and participate in the process of muscle atrophy (17-20). Therefore, skeletal muscle atrophy is regulated by diverse events.
However, as an indispensable part of gene expression regulation, the role of alternative splicing in the process of muscle atrophy has attracted less attention. A single gene produces multiple transcripts through alternative splicing, which greatly expands the proteome’s complexity and the diversity of protein functions (21). Alternative splicing can also change mRNA’s stability and the production of non-coding RNA and regulate the expression of a large number of genes (22-24). It has been found that 95% of human multiple exon genes are alternative spliced, and they are specifically regulated under different physiological or pathological conditions (25). There are extensive alternative splicing phenomena in skeletal muscle, which are involved in various physiological processes of muscle cells, such as contraction, aging, development, and regeneration (21,26-30).
Moreover, the imbalance of RNA splicing can also cause pathological changes in skeletal muscle (31), and RNA binding protein (RBP) deletion can directly lead to the loss of skeletal muscle mass (32-34). This suggests that alternative splicing may be a new regulatory mechanism of muscle atrophy. However, there are few studies on alternative splicing events during muscle atrophy.
Hind limbs suspension can simulate weightlessness, and hanging for extended periods can cause a phenotype similar to the clinical muscle atrophy caused by disuse. In the present study, we established a rat hindlimb unloading model. RNA sequencing (RNA-seq) was performed on the soleus muscle, which was severely atrophic during unloading (12 h, 24 h, 36 h, 3 days, and 7 days). We analyzed the dynamic changes and biological functions of the genes that underwent alternative splicing during skeletal muscle atrophy caused by hindlimb unloading and found several genes involved in the ubiquitin-proteasome pathway have alternative splicing phenomenon. We also analyzed the expression of RBPs, which drove the changes of alternative splicing. This can enrich the molecular mechanism of skeletal muscle atrophy and provide a scientific basis for the targeted therapy of muscle atrophy.
We present the following article following the ARRIVE reporting checklist (available at http://dx.doi.org/10.21037/atm-20-5388).
Methods
Establishment of rat models of hindlimb unloading-induced atrophy
Eighteen 8-week-old male Sprague-Dawley rats were used in the present study. The experiments were done following the recommendations of and approved by the Institutional Animal Care and Use Committee of Nantong University, China (No. S20200312-003). The rats’ hindlimbs were just off the ground, and the tilt angle between the body and the ground was approximately 30 degrees, following the internationally recognized tail unloading method of rodents (35). At this time, the rats could move freely with access to food in the cage. The animals were raised in individual cages at 23±2 °C, and with 12 h of light per day. According to the randomization principle, the rats were killed by cervical dislocation at 6 time points (0 h, 12 h, 24 h, 36 h, 3 days, 7 days), and the soleus muscles were taken. After cutting off excess fat and fascia, the samples were quickly immersed in liquid nitrogen for storage. Normal (0 h) rat soleus muscle was used as the control group, and samples at each time point were obtained from 3 independent repeated experiments (15 suspended and 3 control animals in total).
RNA extraction
Total RNA was extracted using TRIzol (Vazyme, Nanjing, Jiangsu, China). Briefly, the muscle tissue was soaked in 1 mL TRIzol and completely crushed by a homogenizer. Using 1/5 volume of chloroform, RNA was extracted at 12,000 rpm at 4 °C. An equal volume of isopropanol was used to precipitate the RNA, and then the precipitate was washed with 75% ethanol.
cDNA synthesis and library construction
In total, 4 µg RNA was purified and enriched by magnetic beads with oligo(dT). The mRNA was broken into short fragments by adding interruption of reagent. Using interrupted mRNA as a template, the first-strand cDNA was synthesized by using 6-base random primers. Double-stranded cDNA was synthesized and purified. Purified double-stranded cDNA was then subjected to end repair. Poly(A) tailing was added and connected to a sequencing adapter. Finally, polymerase chain reaction amplification was performed. After the Agilent 2100 Bioanalyzer qualified the constructed library, it was sequenced using the Illumina HiSeq X Ten sequencer to generate 125 or 150 bp double-ended data.
Filtering and mapping of sequencing data
The raw data (raw reads) generated by high-throughput sequencing in FASTQ format. To obtain high-quality reads that could be used for subsequent analyses, further quality filtering of raw reads was required. First, Trimmomatic software was used to control quality. Low-quality bases and N bases were filtered out, and finally, high-quality clean reads were obtained. The clean reads were compared using HISAT2 as the reference genome of the species. The software parameters were the default parameters, and the genome mapping rate evaluated samples.
Identification of alternative splicing events
To analyze the differences in alternative splicing events between different samples, rMATS software was used to classify alternative splicing events based on RNA-seq data. Alternative splicing events were divided into the following 5 categories using rMATS: skipped exon (SE), alternative 5' splice site (A5SS), alternative 3' splice site (A3SS), mutually exclusive exon (MEX), and retained intron (RI). In the analysis, junction counts were used for the alternative splicing event detection. The number of reads across the inclusion junction site and across the skipping junction site was calculated. The value of the inclusion level (IncLevel) was used to define the degree of alternative splicing. The IncLevel was equal to the number of reads across the inclusion junction site/(number of reads across the inclusion junction site + number of reads across the skipping junction site). The difference in the value of the IncLevel difference between the experimental and control groups was >0.01, and the false discovery rate (FDR) was not >0.05 as the criterion for screening differential alternative splicing events. The ΔIncLevel was equal to IncLevel2-IncLevel1.
Gene function analysis
We performed Gene Ontology (GO) enrichment analysis on differentially alternative spliced genes to describe gene function. The method for the enrichment analysis of the GO function was as follows. All protein coding genes/transcripts were used as the background list. A list of differential protein encoding genes/transcripts was utilized as a candidate list screened from the background list. The hypergeometric distribution test was used to calculate the P value, representing whether the GO function library was significantly enriched in the differential protein coding genes/transcripts list. The P value was corrected by Benjamini-Hochberg multiple tests to obtain FDR. For the Kyoto Encyclopedia of Genes and Genomes (KEGG) analysis (http://www.genome.jp/kegg/), a hypergeometric distribution test was also used to calculate the significance of differential gene enrichment in each pathway entry. The calculated results could return a significant P value, and a low P value indicated that the differential genes were enriched in the pathway. The STRING database (https://string-db.org/) was used to analyze the interaction network between RBPs.
Statistical analysis
The values of gene expression and the IncLevel alternative splicing events in the present study were calculated from the average of 3 repeated experiments. The relative change represented the difference between each experimental group and the control group, and the negative binomial distribution test was used to test the significance (P value) of the difference between the 2 groups of data.
Results
Differential alternative splicing events during muscle atrophy caused by hindlimb unloading
To identify the alternative splicing events during muscle atrophy caused by hindlimb unloading, we acquired the soleus muscle at 12 h, 24 h, 36 h, 3 days, and 7 days and performed RNA-seq of the poly(A) method to construct the library. We obtained the original base number >7.4 G/sample, the effective base percentage >93.46%, and Q30 >94.2%. At least 97.15% of reads matched the rat genome, suggesting higher reliability of sequencing (Table S1).
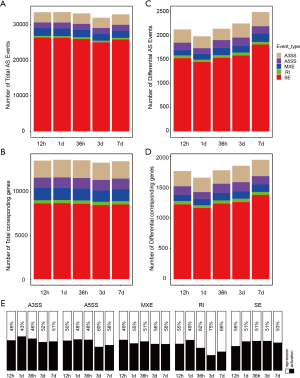
Mammals contained the following 5 common alternative splicing modes: SE, A5SS, A3SS, MEX, and RI. Using rMATS software for the data analysis (36,37), we identified a total of 33,505, 33,547, 33,179, 31,966, and 32,906 alternative splicing events at 12 h, 24 h, 36 h, 3 days, and 7 days after hindlimb unloading, which matched the 13,616, 13,707, 13,650, 13,374, and 13,543 genes, respectively (Figure 1A, B). Compared with the control group, according to the absolute value of the IncLevel difference >0.01 and FDR ≤0.05, we identified 2,118, 1,975, 2,132, 2,242, and 2,478 differential alternative splicing events at 5 time points, which matched the 1,762, 1,655, 1,775, 1,849, and 1,946 genes, respectively (Figure 1C, D). The number of differentially spliced genes was almost the same at every time point. This suggests that the responses were early and constant and did not change much over the time course of 7 days. Of these, SE was the main alternative splicing mode, regardless of the event or gene, and accounted for >70%. We then analyzed the changing trends of these differential splicing events at 5 time points. Compared with the control group, the IncLevel difference >0 was defined as splicing activation, representing a longer transcript, and the IncLevel difference <0 was defined as splicing inhibition. The results showed that for the alternative splicing events in the MEX, A5SS, and A3SS modes, the proportions of activation and inhibition were not significantly different. The inhibited alternative splicing events accounted for a relatively large number in the late stage of atrophy for the RI model, reaching 75% (3 days) and 69% (7 days). As the most common alternative splicing mode, SE increased the number of alternative splicing events activated during muscle atrophy (Figure 1E).
Distribution of differential alternative splicing events on genes and their influence on open reading frames
Alternative splicing occurred in the process of pre-mRNA excision of introns to form mature mRNA. The mRNA contained 3 regions, 5' untranslated region (5'UTR), coding sequence (CDS), and 3' untranslated region (3'UTR). Alternative splicing in different regions may affect gene expression through different mechanisms, such as transcriptional activation, protein-coding, and mRNA stability. Therefore, we analyzed the location of SE form alternative splicing events on the mRNA at 5 time points. The results showed that most alternative splicing occurred in the CDS region. Taking 7 days as an example, the alternative splicing events that occurred in the 5'UTR, 5'UTR-CDS, CDS, CDS-3'UTR, and 3'UTR regions were 183, 233, 985, 170, and 148, respectively, of which the CDS region accounted for 57.3% (Figure 2A).
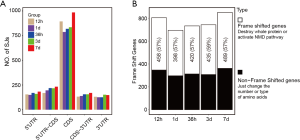
If the alternative exon's base number was not a multiple of 3, alternative splicing could cause protein frameshift or activate the nonsense-mediated mRNA decay pathway to degrade mRNA (38). The impact on the protein was devastating. If the base number of the alternative exon was a multiple of 3, it would only cause changes in the number and type of several amino acids. In most cases, the impact on protein function was relatively minimal. The number and proportion of genes containing frameshift alternative splicing events in the CDS region are shown in Figure 2A. The findings indicated that the proportion of frameshift alternative splicing genes was higher than that of non-frameshift alternative splicing genes. At the 5 time points, we identified 458, 398, 420, 435, and 489 frameshift alternative splicing genes, respectively (Figure 2B).
Functional analysis of differential alternative splicing genes
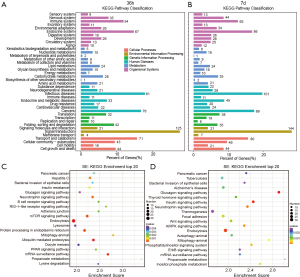
To obtain a functional view of differential alternative splicing genes, we selected 2 representative time points as follows: the intermediate time point of 36 h and the late time point of atrophy of 7 days. Because SE is the main alternative splicing mode, we performed a KEGG analysis of genes with alternative differential splicing in the SE model. According to the different biological functions, these differential alternative splicing genes were classified into 6 processes as follows: cellular processes, environmental information processing, genetic information processing, human diseases, metabolism, and organismal systems. The results showed that the number of genes enriched in each pathway at 7 days was generally greater than those at 36 h, suggesting that skeletal muscle had more active molecular changes at 7 days. The following 4 pathways had the most significant increase in the number of genes: endocrine system, infectious diseases, endocrine, and metabolic diseases, and signal transduction (Figure 3A,B). We analyzed the Top 20 most significant KEGG pathways, and the findings indicated that 9 pathways were significantly activated at 36 h and 7 days as follows: pancreatic cancer, bacterial invasion of epithelial cells, glucagon signaling pathway, insulin signaling pathway, neurotropin signaling pathway, endocytosis, mitophagy-animal, mRNA surveillance pathway, and propanoate metabolism. Of these, the insulin signaling pathway, endocytosis pathway, and mitophagy-animal pathway have been reported to be associated with muscle atrophy (Figure 3C,D) (39,40). It is worth noting that the mammalian target of rapamycin (mTOR) signaling pathway was enriched to Top20 at 36 h instead of 7 days (by comparing the P value, it is 0.0003 in 36 h and 0.0072 in 7 days, by comparing the enrichment score, it is 2.2257 in 36 h and 1.7689 in 7 days), suggesting that alternative splicing may affect protein synthesis at the late stage of atrophy (41).
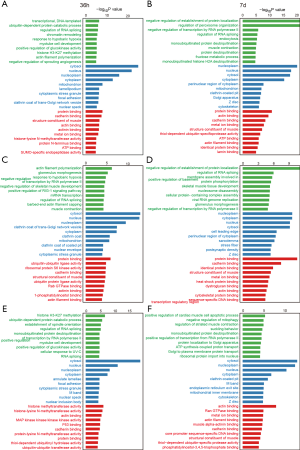
Next, we analyzed the GO of the alternative splicing genes with SE mode. The most significant GO terms of Top30 were divided into the following 3 aspects: biological process, molecular function, and cellular component. We first compared the functions of differentially alternative splicing genes at 36 h and 7 days. Overall, the degree of activation at 7 days was generally stronger than at 36 h, which was mainly reflected in the negative regulation of the establishment of the protein localization, regulation of peroxisome organization, protein binding, actin binding, and cadherin binding pathways (Figure 4A,B). It is worth noting that 1 ubiquitin-related pathway was enriched at 3 days, and 4 ubiquitin-related pathways were enriched at 7 days, suggesting that changes in alternative splicing events may promote significant activation of the ubiquitin-proteasome pathway at 7 days (Figure 4A, B). Next, we analyzed the function of SE mode splicing activation gene and splicing repression gene; the results are shown in Figure 4C (36-h activation), Figure 4D (7-day activation), Figure 4E (36-h repression), and Figure 4F (7-day repression). After analysis, we found that the ubiquitin-proteasome pathway was mainly represented by splicing repressing genes, rather than splicing activating genes. Interestingly, the regulation of RNA splicing and the regulation of transcription by RNA polymerase II were widely activated, suggesting that alternative splicing may cause expression changes in a large number of downstream genes (Figure 4C,D,E,F).
Alternative splicing continues to affect the ubiquitin–proteasome pathway during muscle atrophy caused by hindlimb unloading
Muscle atrophy is a gradual process and may require certain molecules to continue to function. Therefore, we took the intersection of the differential alternative splicing genes at different time points, and 201 continuously changing genes were identified (Figure 5A). GO analysis of these genes showed that the biological processes of cell-cell adhesion, transcription, DNA-templated, protein deubiquitination, skeletal muscle contraction, skeletal muscle tissue development, actin filament organization, cytoskeleton organization, microtubule cytoskeleton organization, regulation of transcription, DNA-templated, and regulation of muscle contraction were enriched (Figure 5B). Molecular functions of poly(A) binding, protein binding, actin-binding, structural constituent of muscle, cadherin binding involved in cell-cell adhesion, N6-methyladenosine-containing RNA binding, thiol-dependent ubiquitinyl hydrolase activity, tropomyosin binding, lamin binding, and actin filament binding were enriched (Figure 5C). As the ubiquitin-proteasome pathway plays an important role in muscle atrophy, we focused on the splicing changes of 6 genes in protein deubiquitination and thiol-dependent ubiquitinyl hydrolase activity pathway and found that Brca1 associated protein 1 (Bap1), ubiquitin specific peptidase 15 (Usp15), and ubiquitin specific peptidase 54 (Usp54) splicing was continuously suppressed; ubiquitin specific peptidase 2 (Usp2) splicing was continuously activated; ubiquitin specific peptidase 36 (Usp36) and ubiquitin specific peptidase 9x (Usp9x) splicing showed fluctuating changes (Figure 5D). Our data suggest that AS changes may represent a new ubiquitin-proteasome activation mechanism associated with skeletal muscle atrophy.

Identification of differentially expressed RBPs during skeletal muscle atrophy
To explore the mechanism of alternative splicing changes during skeletal muscle atrophy, we analyzed the expression of RBPs, which are considered splicing regulators. In the sequencing results, we identified a total of 15,781 genes that corresponded with 2,269 identified rat RBPs (42), and 1,639 RBP genes were obtained (Figure 6A). We identified the differentially expressed RBPs at 5 time points (P<0.01) and calculated the proportion of their numbers in 1,639 genes. The results showed that ~20% of RBP expression changed during skeletal muscle atrophy (Figure 6B). We took the intersection of RBPs that were differentially expressed at 5 time points to obtain the following 6 RBP genes: exportin 4 (Xpo4), eukaryotic translation initiation factor 4E family member 2 (Eif4e2), prolyl 4-hydroxylase subunit alpha 1 (P4ha1), LRR binding FLII interacting protein 1 (Lrrfip1), zinc finger CCCH type containing 14 (Zc3h14), and EMG1 N1-specific pseudouridine methyltransferase (Emg1). Analysis of the dynamic changes of these 6 genes during atrophy demonstrated continuous change; only Zc3h14 showed an upregulated trend, and the rest were all downregulated. Of these genes, Xpo4 demonstrated a continuous downward trend (Figure 6C). It has been reported that alternative splicing plays an important role in skeletal muscle biology, which may be strongly associated with the role of certain RBP family proteins (Hnrnp, Rbfox, Celf, Mbnl, Pabpc, and Pabpn) (21,43). Our results confirmed that Hnrnp h1, and Mbnl2 were significantly downregulated during skeletal muscle atrophy caused by unloading, and Rbfox1 and Mbnl1 were significantly upregulated during skeletal muscle atrophy caused by unloading (Figure 6D). The interaction network between these RBPs is shown in Figure 6E. The findings indicated that RBPs have complex interactions and connections, making the molecular mechanism of muscle atrophy more complex.

Discussion
In the present study, we conducted an alternative splicing analysis of the process of rat skeletal muscle atrophy caused by hindlimb unloading. We found that many differential splicing events occurred at 12 h, 24 h, 36 h, 3 days, and 7 days, and their main locations were the CDS region of the gene. Most of these alternative splicing events occurred in the CDS region, resulting in the gene’s frameshift. We found that alternative splicing activated the insulin signaling, endocytosis, mitophagy-animal, and ubiquitin-proteasome pathways at 36 h and 7 days through functional analysis. The alternative splicing of several USP-related genes continues to be altered during muscle atrophy. In terms of molecular mechanism, we found that 6 RBP genes’ expressions continued to change during atrophy, and 4 common RBPs showed significantly differential expressions. Our results emphasize that alternative splicing is a new focus of muscle atrophy, and in the present study, we preliminarily explored the molecular mechanism of alternative splicing changes.
The splicing of mammalian precursor mRNA is the process of intron removal and exon interconnection and is an important link during gene expression. In a specific time and space (such as different developmental stages or pathological conditions), cells usually produce proteins with different or even opposite functions by alternative splicing to adapt to environmental changes (44). Although alternative splicing is strongly associated with skeletal muscle physiology (26,45), the role of alternative splicing during skeletal muscle atrophy caused by hindlimb unloading has not been studied at the whole-gene level. In the present study, we found that a large number of differential alternative splicing events (genes) occurred during skeletal muscle atrophy caused by hindlimb unloading, which expanded our understanding of the process of skeletal muscle atrophy. It has been found that splicing disorders of some genes may cause skeletal muscle lesions, leading to muscle atrophy (46,47). Antisense nucleic acid technology is a potential therapy to correct and splice neuromuscular diseases. At present, antisense nucleic acid drugs for spinal muscular atrophy and Duchenne muscular dystrophy have been approved by the American Food and Drug Administration (48). The present study provides several new alternative splicing targets for the development of anti-atrophic drugs.
In the present study, we identified the differential alternative splicing events (genes) during muscle atrophy and analyzed their location on the gene and their impact on the open reading frame. The findings indicated that 59% of these differential alternative splicing events occurred in the CDS region and caused protein frameshifts. In addition to changing the reading frame, frameshifted transcripts were also one of the conditions for the activation of nonsense-mediated mRNA decay (49), and the transcripts that activated nonsense-mediated mRNA decay would eventually be degraded. Several recent studies have found that skeletal muscle atrophy was regulated by miRNAs, such as miR-29b (50), miR-125b-5p (18), and miR-351 (51). Although our results show that most AS occurs in the CDS region, a considerable portion still occurs in the UTR area. Alternative splicing events at 3'UTR will increase or decrease the binding site of miRNA.
Moreover, alternative splicing that occurs in 3'UTR can reduce the 3'UTR length to avoid miRNA or RBP-mediated regulation, it is also called alternative cleavage and polyadenylation (APA). APA also greatly contributes to the proteome’s selectivity and determines the ultimate fate of mRNA (52). Therefore, our results suggest that skeletal muscle atrophy is an extremely complex pathological process, and there are synergistic effects of multiple mechanisms.
The activation of the ubiquitin–proteasome pathway is one of the characteristics of skeletal muscle atrophy. In addition to the 5 classic molecules MAFbx, MuRF1, Traf6, UBR5, TRIM62, many other E3 ubiquitin ligases have been shown to regulate skeletal muscle atrophy (53-56). In the present study, 6 genes were found to be involved in the ubiquitin-proteasome pathway, including Usp15 and Usp2, and were found to have splicing differences. These splicing differences are widespread in muscle atrophy and provide a new perspective for the activation of ubiquitin-proteasome. Seven Usp2 gene alternative splicing products have been reported, 5 of which can encode proteins (57). The functions of the 3 main Usp2 alternative splicing products, Usp2-201, Usp2-202, and Usp2-204, have been extensively studied (51). Usp2-201 is involved in recycling epidermal growth factor receptor and plays a vital role in the cell cycle, Usp2-202 can activate the apoptosis signaling pathway and participate in apoptosis, and Usp2-204 can reduce the antiviral response of cells (58). Kotani et al. confirmed that the changes in inclusion (activation) or skipping (inhibition) of exon 7 of Usp15 could affect its substrate specificity and interactome (59). In our results, Usp15 splicing was inhibited during muscle atrophy, and Usp2 splicing was activated, implying that these changes may have a broad impact on substrate selection and downstream signaling pathways in the ubiquitin response.
RBPs are direct regulators of alternative splicing. The present study’s findings indicated that approximately 20% of RBPs were differentially expressed during skeletal muscle atrophy caused by hindlimb unloading, which could explain why a large number of alternative splicing events occur during atrophy caused by hindlimb unloading. We found that 6 RBP genes continued to change during muscle atrophy. Zc3h14 is generally upregulated during muscle atrophy and is involved in the formation of the poly(A) tail of mRNA, which affects the stability of mRNA (60); Xpo4 continues to be downregulated during muscle atrophy and encodes a nuclear transport protein that participates in the nuclear output of eIF5A and Smad3 (61). Although there is currently no evidence that Xpo4 are involved in alternative splicing regulation, they may still play an important role in muscle atrophy.
Multiple families of RBPs have been reported to participate in the physiological process of skeletal muscle (43), they may contain key alternative splicing regulators that we focus on. Singh et al. found that Rbfox1/2 is necessary to maintain skeletal muscle quality and proteostasis. Rbfox1/2 knockout in skeletal muscle has been found to cause changes in alternative splicing of many genes (32). Our results showed that the expression of Rbfox1 was disordered, suggesting that it may play an important role in skeletal muscle atrophy by regulating alternative splicing. Mbnl1 and Mbnl2 are also RBPs with large changes in expression during muscle atrophy. It has been reported that Mbnl1/2 is a skeletal muscle-specific RBP and participates in the tissue-specific alternative splicing regulation of skeletal muscle. Mbnl expression’s dysregulation will affect the severity of hereditary muscle atrophy, such as myotonic dystrophy (62). Interestingly, Rbfox1 and Mbnl1 have been reported to bind the same cis-acting element competitively and coordinately regulate alternative splicing (63), suggesting that the differential alternative splicing event at each time point may be the result of the interaction of many differentially expressed RBPs at that time point.
Splicing regulation depends on the interaction of cis-acting elements and trans-acting factors. Although we found several potentially differentially expressed RBPs, it is unclear why those RBPs are differentially expressed in atrophic skeletal muscle. Therefore, it is important to determine the complex regulatory relationship between them and the differential alternative splicing events in future studies so that the molecular mechanism of muscle atrophy can be further elucidated. This can also provide a scientific basis for the prevention and treatment of muscle atrophy.
Acknowledgments
Funding: This work was supported by the National Natural Science Foundation of China (Nos. 82072160, 32000841, 31730031 and 81901933), the National Key Research and Development Program of China (No. 2017YFA0104703), the Major Natural Science Research Projects in Universities of Jiangsu Province (No. 20KJA310012), the Natural Science Foundation of Jiangsu Province (No. BK20202013, BK20201209), the Priority Academic Program Development of Jiangsu Higher Education Institutions, and Nantong Science and Technology Innovation Program (No. JC2019028).
Footnote
Reporting Checklist: The authors have completed the ARRIVE reporting checklist Available at http://dx.doi.org/10.21037/atm-20-5388
Data Sharing Statement: Available at http://dx.doi.org/10.21037/atm-20-5388
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/atm-20-5388). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. The experiments were done following the recommendations of and approved by the Institutional Animal Care and Use Committee of Nantong University, China (No. S20200312-003).
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Fitts RH, Riley DR, Widrick JJ. Physiology of a microgravity environment invited review: microgravity and skeletal muscle. J Appl Physiol (1985) 2000;89:823-39. [Crossref] [PubMed]
- Fitts RH, Riley DR, Widrick JJ. Functional and structural adaptations of skeletal muscle to microgravity. J Exp Biol 2001;204:3201-8. [PubMed]
- Li S, Yang Z, Gao Y, et al. Ligustrazine and the contractile properties of soleus muscle in hindlimb-unloaded rats. Aviat Space Environ Med 2012;83:1049-54. [Crossref] [PubMed]
- Brooks NE, Myburgh KH. Skeletal muscle wasting with disuse atrophy is multi-dimensional: the response and interaction of myonuclei, satellite cells and signaling pathways. Front Physiol 2014;5:99. [Crossref] [PubMed]
- Powers SK, Lynch GS, Murphy KT, et al. Disease-Induced Skeletal Muscle Atrophy and Fatigue. Med Sci Sports Exerc 2016;48:2307-19. [Crossref] [PubMed]
- Lecker SH, Jagoe RT, Gilbert A, et al. Multiple types of skeletal muscle atrophy involve a common program of changes in gene expression. FASEB J 2004;18:39-51. [Crossref] [PubMed]
- Huang Z, Zhu J, Ma W, et al. Strategies and potential therapeutic agents to counter skeletal muscle atrophy. Biotarget 2018;2:8. [Crossref]
- Ma W, Xu T, Wang Y, et al. The role of inflammatory factors in skeletal muscle injury. Biotarget 2018;2:7. [Crossref]
- Ebert SM, Al-Zougbi A, Bodine SC, et al. Skeletal Muscle Atrophy: Discovery of Mechanisms and Potential Therapies. Physiology (Bethesda) 2019;34:232-9. [Crossref] [PubMed]
- Bonaldo P, Sandri M. Cellular and molecular mechanisms of muscle atrophy. Dis Model Mech 2013;6:25-39. [Crossref] [PubMed]
- Wan Q, Zhang L, Huang Z, et al. Aspirin alleviates denervation-induced muscle atrophy via regulating the Sirt1/PGC-1α axis and STAT3 signaling. Ann Transl Med 2020;8:1524. [Crossref] [PubMed]
- Lang F, Aravamudhan S, Nolte H, et al. Dynamic changes in the mouse skeletal muscle proteome during denervation-induced atrophy. Dis Model Mech 2017;10:881-96. [Crossref] [PubMed]
- Sun H, Qiu J, Chen Y, et al. Proteomic and bioinformatic analysis of differentially expressed proteins in denervated skeletal muscle. Int J Mol Med 2014;33:1586-96. [Crossref] [PubMed]
- Huang Z, Zhong L, Zhu J, et al. Inhibition of IL-6/JAK/STAT3 pathway rescues denervation-induced skeletal muscle atrophy. Annals of Translational Medicine 2020;8:1681. [Crossref] [PubMed]
- Shen Y, Zhang R, Xu L, et al. Microarray Analysis of Gene Expression Provides New Insights Into Denervation-Induced Skeletal Muscle Atrophy. Front Physiol 2019;10:1298. [Crossref] [PubMed]
- Shen Y, Zhang Q, Huang Z, et al. Isoquercitrin Delays Denervated Soleus Muscle Atrophy by Inhibiting Oxidative Stress and Inflammation. Front Physiol 2020;11:988. [Crossref] [PubMed]
- Weng J, Zhang P, Yin X, et al. The Whole Transcriptome Involved in Denervated Muscle Atrophy Following Peripheral Nerve Injury. Front Mol Neurosci 2018;11:69. [Crossref] [PubMed]
- Qiu J, Zhu J, Zhang R, et al. miR-125b-5p targeting TRAF6 relieves skeletal muscle atrophy induced by fasting or denervation. Ann Transl Med 2019;7:456. [Crossref] [PubMed]
- Hitachi K, Nakatani M, Tsuchida K. Long Non-Coding RNA Myoparr Regulates GDF5 Expression in Denervated Mouse Skeletal Muscle. Noncoding RNA 2019;5:33. [Crossref] [PubMed]
- Alessio E, Buson L, Chemello F, et al. Single cell analysis reveals the involvement of the long non-coding RNA Pvt1 in the modulation of muscle atrophy and mitochondrial network. Nucleic Acids Res 2019;47:1653-70. [Crossref] [PubMed]
- Nakka K, Ghigna C, Gabellini D, et al. Diversification of the muscle proteome through alternative splicing. Skelet Muscle 2018;8:8. [Crossref] [PubMed]
- Kelemen O, Convertini P, Zhang Z, et al. Function of alternative splicing. Gene 2013;514:1-30. [Crossref] [PubMed]
- Ohtsuka M, Tanemura M, Akamatsu H. Long noncoding RNAs regulate malignant phenotypes in colorectal cancer. Biotarget 2018;2:4. [Crossref]
- Desai N. Divergent functions of a ribosome maturation factor. Biotarget 2017;1:17. [Crossref]
- Castle JC, Zhang C, Shah JK, et al. Expression of 24,426 human alternative splicing events and predicted cis regulation in 48 tissues and cell lines. Nat Genet 2008;40:1416-25. [Crossref] [PubMed]
- Giudice J, Loehr JA, Rodney GG, et al. Alternative Splicing of Four Trafficking Genes Regulates Myofiber Structure and Skeletal Muscle Physiology. Cell Rep 2016;17:1923-33. [Crossref] [PubMed]
- Brinegar AE, Xia Z, Loehr JA, et al. Extensive alternative splicing transitions during postnatal skeletal muscle development are required for calcium handling functions. Elife 2017;6. [PubMed]
- Rodríguez SA, Grochova D, McKenna T, et al. Global genome splicing analysis reveals an increased number of alternatively spliced genes with aging. Aging Cell 2016;15:267-78. [Crossref] [PubMed]
- Bland CS, Wang ET, Vu A, et al. Global regulation of alternative splicing during myogenic differentiation. Nucleic Acids Res 2010;38:7651-64. [Crossref] [PubMed]
- Liu N, Nelson BR, Bezprozvannaya S, et al. Requirement of MEF2A, C, and D for skeletal muscle regeneration. Proc Natl Acad Sci U S A 2014;111:4109-14. [Crossref] [PubMed]
- Yu Z, Wang AM, Robins DM, et al. Altered RNA splicing contributes to skeletal muscle pathology in Kennedy disease knock-in mice. Dis Model Mech 2009;2:500-7. [Crossref] [PubMed]
- Singh RK, Kolonin AM, Fiorotto ML, et al. Rbfox-Splicing Factors Maintain Skeletal Muscle Mass by Regulating Calpain3 and Proteostasis. Cell Rep 2018;24:197-208. [Crossref] [PubMed]
- Hosokawa M, Takeuchi A, Tanihata J, et al. Loss of RNA-Binding Protein Sfpq Causes Long-Gene Transcriptopathy in Skeletal Muscle and Severe Muscle Mass Reduction with Metabolic Myopathy. iScience 2019;13:229-42. [Crossref] [PubMed]
- Anderson DM, Cannavino J, Li H, et al. Severe muscle wasting and denervation in mice lacking the RNA-binding protein ZFP106. Proc Natl Acad Sci U S A 2016;113:E4494-503. [Crossref] [PubMed]
- Marzuca-Nassr GN, Vitzel KF, Murata GM, et al. Experimental Model of HindLimb Suspension-Induced Skeletal Muscle Atrophy in Rodents. Methods Mol Biol 2019;1916:167-76. [Crossref] [PubMed]
- Shen S, Park JW, Lu ZX, et al. rMATS: robust and flexible detection of differential alternative splicing from replicate RNA-Seq data. Proc Natl Acad Sci U S A 2014;111:E5593-601. [Crossref] [PubMed]
- Shen S, Park JW, Huang J, et al. MATS: a Bayesian framework for flexible detection of differential alternative splicing from RNA-Seq data. Nucleic Acids Res 2012;40:e61 [Crossref] [PubMed]
- Nasif S, Contu L, Muhlemann O. Beyond quality control: The role of nonsense-mediated mRNA decay (NMD) in regulating gene expression. Semin Cell Dev Biol 2018;75:78-87. [Crossref] [PubMed]
- Martín AI, Priego T, Lopez-Calderon A. Hormones and Muscle Atrophy. Adv Exp Med Biol 2018;1088:207-33. [Crossref] [PubMed]
- Ma W, Zhang R, Huang Z, et al. PQQ ameliorates skeletal muscle atrophy, mitophagy and fiber type transition induced by denervation via inhibition of the inflammatory signaling pathways. Ann Transl Med 2019;7:440. [Crossref] [PubMed]
- Cohen S, Nathan JA, Goldberg AL. Muscle wasting in disease: molecular mechanisms and promising therapies. Nat Rev Drug Discov 2015;14:58-74. [Crossref] [PubMed]
- Liao JY, Yang B, Zhang YC, et al. EuRBPDB: a comprehensive resource for annotation, functional and oncological investigation of eukaryotic RNA binding proteins (RBPs). Nucleic Acids Res 2020;48:D307-13. [Crossref] [PubMed]
- Hinkle ER, Wiedner HJ, Black AJ, et al. RNA processing in skeletal muscle biology and disease. Transcription 2019;10:1-20. [Crossref] [PubMed]
- Montes M, Sanford BL, Comiskey DF, et al. RNA Splicing and Disease: Animal Models to Therapies. Trends Genet 2019;35:68-87. [Crossref] [PubMed]
- Spletter ML, Schnorrer F. Transcriptional regulation and alternative splicing cooperate in muscle fiber-type specification in flies and mammals. Exp Cell Res 2014;321:90-8. [Crossref] [PubMed]
- Renna LV, Bose F, Brigonzi E, et al. Aberrant insulin receptor expression is associated with insulin resistance and skeletal muscle atrophy in myotonic dystrophies. PLoS One 2019;14:e0214254 [Crossref] [PubMed]
- Nutter CA, Jaworski E, Verma SK, et al. Developmentally regulated alternative splicing is perturbed in type 1 diabetic skeletal muscle. Muscle Nerve 2017;56:744-9. [Crossref] [PubMed]
- Bennett CF, Krainer AR, Cleveland DW. Antisense Oligonucleotide Therapies for Neurodegenerative Diseases. Annu Rev Neurosci 2019;42:385-406. [Crossref] [PubMed]
- Lykke-Andersen S, Jensen TH. Nonsense-mediated mRNA decay: an intricate machinery that shapes transcriptomes. Nat Rev Mol Cell Biol 2015;16:665-77. [Crossref] [PubMed]
- Li J, Chan MC, Yu Y, et al. miR-29b contributes to multiple types of muscle atrophy. Nat Commun 2017;8:15201. [Crossref] [PubMed]
- He Q, Qiu J, Dai M, et al. MicroRNA-351 inhibits denervation-induced muscle atrophy by targeting TRAF6. Exp Ther Med 2016;12:4029-34. [Crossref] [PubMed]
- Gruber AJ, Zavolan M. Alternative cleavage and polyadenylation in health and disease. Nat Rev Genet 2019;20:599-614. [Crossref] [PubMed]
- Rom O, Reznick AZ. The role of E3 ubiquitin-ligases MuRF-1 and MAFbx in loss of skeletal muscle mass. Free Radic Biol Med 2016;98:218-30. [Crossref] [PubMed]
- Paul PK, Bhatnagar S, Mishra V, et al. The E3 ubiquitin ligase TRAF6 intercedes in starvation-induced skeletal muscle atrophy through multiple mechanisms. Mol Cell Biol 2012;32:1248-59. [Crossref] [PubMed]
- Seaborne RA, Hughes DC, Turner DC, et al. UBR5 is a novel E3 ubiquitin ligase involved in skeletal muscle hypertrophy and recovery from atrophy. J Physiol 2019;597:3727-49. [Crossref] [PubMed]
- Schmidt F, Kny M, Zhu X, et al. The E3 ubiquitin ligase TRIM62 and inflammation-induced skeletal muscle atrophy. Crit Care 2014;18:545. [Crossref] [PubMed]
- Gousseva N, Baker RT. Gene structure, alternate splicing, tissue distribution, cellular localization, and developmental expression pattern of mouse deubiquitinating enzyme isoforms Usp2-45 and Usp2-69. Gene Expr 2003;11:163-79. [Crossref] [PubMed]
- Zhu HQ, Gao FH. The Molecular Mechanisms of Regulation on USP2's Alternative Splicing and the Significance of Its Products. Int J Biol Sci 2017;13:1489-96. [Crossref] [PubMed]
- Kotani Y, Morito D, Sakata K, et al. Alternative exon skipping biases substrate preference of the deubiquitylase USP15 for mysterin/RNF213, the moyamoya disease susceptibility factor. Sci Rep 2017;7:44293. [Crossref] [PubMed]
- Wigington CP, Williams KR, Meers MP, et al. Poly(A) RNA-binding proteins and polyadenosine RNA: new members and novel functions. Wiley Interdiscip Rev RNA 2014;5:601-22. [Crossref] [PubMed]
- Aksu M, Trakhanov S, Gorlich D. Structure of the exportin Xpo4 in complex with RanGTP and the hypusine-containing translation factor eIF5A. Nat Commun 2016;7:11952. [Crossref] [PubMed]
- Konieczny P, Stepniak-Konieczna E, Sobczak K. MBNL proteins and their target RNAs, interaction and splicing regulation. Nucleic Acids Res 2014;42:10873-87. [Crossref] [PubMed]
- Sellier C, Cerro-Herreros E, Blatter M, et al. rbFOX1/MBNL1 competition for CCUG RNA repeats binding contributes to myotonic dystrophy type 1/type 2 differences. Nat Commun 2018;9:2009. [Crossref] [PubMed]


