
A narrative review of circular RNAs as potential biomarkers and therapeutic targets for cardiovascular diseases
Introduction
Circular RNAs (circRNAs), a novel class of non-coding RNA, were first discovered in 1970s (1,2). Initially, they were considered to be byproducts of splicing (3-6). In recent years, high-throughput sequencing and large-scale bioinformatics analysis have revealed the tremendous amounts of this novel non-coding RNA species. In 2016, by integrating deep-sequencing data, Zheng et al. annotated 14,867 circRNAs in human, and among these, 1,260 are orthologous to mouse circRNAs (7). In addition to mammals, circRNAs are also detected in other species, such as fungi, protists, and plants (8-11). The length of most human exonic circRNAs is less than 1,500 nucleotides (nt) with the median of 500 nt (12). Despite general low expression levels, less than 10% of their cognate linear transcripts (13), recent studies have showed the potential roles of circRNAs in physiological or pathological conditions.
Cardiovascular diseases (CVDs) are the leading cause of mortality and morbidity worldwide, and result in substantial health and financial burden (14). CVDs are disorders occurring in heart and blood vessels, including myocardial infarction (MI), cardiac hypertrophy, heart failure (HF), atherosclerosis (AS), cardiac senescence, hypertension (HT), cardiac fibrosis, coronary heart disease (CHD), stroke, etc. Despite current improvements in diagnostic and therapeutic approaches, survival and prognosis of CVDs patients remain relatively poor. Therefore, novel diagnostic and therapeutic strategies should be developed to diagnose and treat CVDs in the early stage.
At present, the roles of circRNAs in neurological diseases, immune diseases, and cancers have come to light. In addition, increasing studies have determined the expression profiles of circRNA in CVDs through high throughput sequencing or circRNA microarray, and found that some circRNAs are linked to the pathogenesis of CVDs. In the review, we systematically summarized the formation mechanisms, functional models, and research approaches of circRNAs, and several circRNAs involved in CVDs. Finally, we proposed that developing circRNAs as biomarkers or circRNA-based therapeutic strategies based on biological or physical materials may be promising to diagnose or treat CVDs in the future.
We present the following article in accordance with the Narrative Review reporting checklist (available at http://dx.doi.org/10.21037/atm-20-7929).
The biogenesis of circRNA
CircRNAs can be back-spliced from precursor mRNA (pre-mRNA) post transcription with the cooperative effects of spliceosome and RNA polymerase II (Pol II) (15); back-splicing has a much lower efficiency than canonical splicing in most human gene loci (16). However, recent studies have confirmed a subset of circRNAs have higher expression levels than their cognate linear counterparts (17,18). One possible explanation is that the expressions of these circRNAs are independent of their linear isoforms (15). In addition, a high Pol II transcription elongation rate (TER) could boost back-splicing and impede linear splicing (16), partially accounting for the higher expression levels of several circRNAs. According to intron or exon composition, circRNAs are mainly classified into three types: exonic circRNA (ecircRNA), intronic circRNA (ciRNA), and exon-intron circRNA (EIciRNA) (19) (Figure 1). Three mechanisms are currently reported to explain the model of back-splicing and biogenesis of circRNA: lariat-driven circularization, intron pairing-driven circularization, and RNA-binding protein (RBP)-driven circularization.
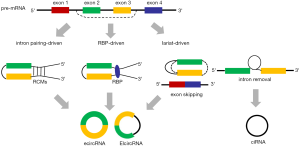
Lariat-driven circularization
Lariat-driven circularization is occurred in intron removal (20) or exon skipping (21) events. When two adjacent exons are spliced, the between intron will form a lariat which leads to the formation of an intronic circRNA. In addition, exon skipping during alternative splicing makes the skipped exon(s) form an exon-containing lariat; meanwhile, a linear transcript excluding the skipped exon(s) is also concomitantly produced (Figure 1).
Intron pairing-driven circularization
In 2013, Jeck et al. found intron flanking circularized exons contained more ALU elements than non-circularized exons, suggesting the contribution of ALU to the biogenesis of circRNA (22). In 2015, Ivanov et al. demonstrated that compared with linear RNA transcript, introns flanking circularized exons are rich in reverse complementary matches (RCMs) which are critical to circRNA production (23) (Figure 1). Adenosine deaminases acting on RNAs (ADARs) is a double-stranded RNA binding enzyme that can convert adenosine residues in double-stranded RNA into creatinine, and adenosine-to-inosine (A-to-I) editing activity of ADAR1 destroys the complementarity and thus reduces back-splicing. Through further experiments, Ivanov et al. confirmed that knockdown of ADAR1 significantly promoted the expression of circRNAs (23). Interestingly, not all introns flanking circRNAs contain RCMs, and the model of intron pairing-driven circularization can only explain part of the biogenesis of circRNAs.
RBP-driven circularization
In addition to cis-acting elements, several trans-acting factors, such as RBPs, are also linked to the formation of circRNAs (Figure 1). Conn et al. reported that Quaking (QKI) promotes the formation of circRNA by binding to specific binding sites on circRNA flanking introns during human epithelial mesenchymal transition (EMT) (24). Errichelli et al. found that fused in sarcoma (FUS) participates in the formation of circRNA by binding to exon/intron boundary in neurons (25). RNA binding motif protein 20 (RBM20) is a RBP that regulates numerous cardiac-specific gene editing processes. Khan et al. found that mutation of RMB20 participates in the pathogenesis of dilated cardiomyopathy through affecting the generation of circRNAs derived from Titin (26). Kramer et al. also demonstrated that heterogeneous nuclear ribonucleoprotein (hnRNPs) and serine-arginine (SR) proteins regulate circRNA expression in Drosophila (27).
Only one mechanism is not sufficient to explain the biogenesis of all circRNAs. It is a combination of cis-acting elements and trans-acting proteins that control or regulate the occurrence of back-splicing and biogenesis of circRNA in physiological conditions. Moreover, the kinetics of back-splicing can be altered in some pathological conditions, such as inflammation, tumor, or cardiovascular disorders. Apparently, the biogenesis of circRNA more or less affects intracellular circRNA abundance. In addition, the decay of circRNA also contributes to the regulation of circRNA abundance (15). Recent studies have suggested several endonucleases are responsible for the degradation of circRNAs. Cytoplasmic endonuclease RNase L can globally degrade circRNAs (28); ATP-dependent RNA helicase upstream frameshift 1 (UPF1) and its associated endonuclease G3BP1 can target and degrade highly structured circRNAs (29); ribonuclease complex RNase P/MRP can degrade m6A-modified circRNAs (30). Furthermore, Hansen et al. found the decay of CDR1as depends on miR-671 binding and subsequent Argonaute 2 (AGO2) cleavage (31). More detailed mechanisms are needed to explain the decay of circRNAs and their abundance regulation.
The functions of circRNA
MicroRNA sponge
Serving as microRNA (miRNA) sponge is the widely studied mechanism for circRNAs. Through adsorbing miRNAs, circRNAs release the targets of miRNAs, and thus participate in several physiological or pathological processes (Figure 2). In 2013, Memczak et al. firstly proposed the mechanism of miRNA sponge. They found circRNA cerebellar degeneration-related 1 antisense transcript (CDR1as) harbours 63 microRNA-7 (miR-7) binding sites. Further in vivo experiments indicated that overexpression of human CDR1as in zebrafish impaired midbrain development of zebrafish, which is similar to the effect of silencing miR-7, suggesting that CDR1as can serve as the sponge and antagonist of miR-7 (32). In addition, Hansen et al. identified 16 putative binding sites for miR-138 within testes-specific circRNA Sry, and proposed that the circular Sry can serve as the sponge of miR-138 (33). Of note, the expressions of most circRNAs are not sufficient to adsorb miRNAs and antagonize the effects of miRNAs. Moreover, considerable amounts of circRNAs are shorter in length and do not contain many binding sites of miRNAs. Furthermore, most of miRNAs localize in the cytoplasm, while a large part of circRNAs are located in the nucleus. Therefore, the model of miRNA sponge is available for such circRNAs with abundant expression levels or binding sites of miRNAs, for which the most typical examples are CDR1as and circular Sry. Nevertheless, we have no evidence to deny that administering a therapeutic dose of exogenous circRNAs in vivo based on the model of miRNA sponge may be promising for diseases treatment.
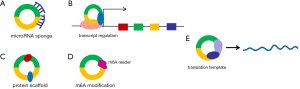
Transcriptional regulators
Intron-containing circRNAs, including EIciRNAs and ciRNAs, are mainly located in the nucleus to serve as the transcriptional regulators (Figure 2). Zhang et al. found that ci-ankrd52 derived from the second intron of ankyrin repeat domain 52 (ANKRD52) localizes in the nucleus and promotes transcription of ANKRD52 (20). In addition, Li et al. demonstrated that two EIciRNAs, circEIF3J and circPAIP2, bind with small nuclear ribonucleoprotein U1 (snRNP U1) to form EIciRNA-U1 snRNP complexes which interact with RNA polymerase II at the promoter regions of parental genes to promote their transcriptions (34).
Protein scaffolds
Sufficient evidence has indicated that circRNAs can serve as the protein scaffolds (Figure 2). For example, circ-Foxo3 was found highly expressed in mouse non-cancer cells, suggesting a role of cancer suppression. Mechanistically, circ-Foxo3 binds with cyclin-dependent kinase 2 (CDK2) and cyclin-dependent kinase inhibitor 1 (p21) to form a circFoxo3-p21-CDK2 ternary complex which can inhibit CDK2 function and impede cell-cycle progression (35). Another typical example is circ-Amotl1 which is highly expressed in neonatal heart tissue. Through binding to serine/threonine kinase AKT1 and phosphoinositide-dependent kinase 1 (PDK1), circ-Amotl1 promotes phosphorylation of AKT1 through PDK1 and subsequent AKT1 nuclear localization the process that can protect heart from destruction (36).
Translation
Although circRNAs are considered as a class of non-coding RNAs, more recent studies have shown the potentials of circRNAs to translate into proteins or peptides (Figure 2). Due to the lack of typical 5' cap for translation initiation, circRNA translation is likely dependent on internal ribosome entry site (IRES) which allows translation initiation in the absence of typical translation initiation factors (37). Several studies have revealed the protein-coding abilities of circZNF609, circFBXW7, circSHPRH and circβ-catenin, which were driven by IRES (38-41). Furthermore, studies also elucidated the pro- or anti-tumor roles of the proteins derived from these circRNAs. In addition, N(6)-methyladenosine (m6A) modification is another alternative mechanism of circRNA translation. M6A, induced by the methyltransferase-like 3/14 (METTL3/14), Wilm’s tumor associated protein (WTAP) (42,43) and suppressed by the demethylase fat mass and obesity-associated protein (FTO) and alkylated DNA repair protein alkB homolog 5 (ALKBH5) (44,45), could promote circRNA translation through recruiting eukaryotic translation initiation factor eIF4G2 and m6A reader YTH domain family protein 3 (YTHDF3) (46).
N6-methyladenosine (m6A) modification
M6A is considered as the most general modification on RNAs in eukaryotes (47,48). In 2017, Yang et al. firstly proposed that circRNAs existed m6A modification which drove the translation of circRNAs (46). Afterwards, an increasing number of studies revealed the roles of m6A modification of circRNAs in physiological or pathological states (Figure 2). Su et al. identified the m6A map of circRNAs in transcriptome level in Hypoxia mediated pulmonary hypertension (HPH), and they found that m6A abundance of total circRNAs was reduced under hypoxia exposure (49). Chen et al. demonstrated that m6A modification promoted the cytoplasmic export of circNSUN2, stabilized downstream high mobility group AT-hook 2 (HMGA2) mRNA, thereby enhancing colorectal cancer progression (50). In addition to be modified by m6A, circRNAs can also bind to m6A-modified enzymes to modulate m6A modification process. Huang et al. found that circSTAG1 was at low expression level in major depressive disorder (MDD), and it could prevent ALKBH5 entering the nucleus by binding to ALKBH5, thereby increasing the m6A modification of total RNA (51).
The research approaches of circRNA
Identification tools of circRNA
Identification of circRNA based on RNAseq libraries is the preliminary for circRNA research. Several different tools or pipelines have so far been developed to identify circRNAs, such as Find_circ (32), CIRCexplorer (52), CIRI (53), Mapsplice (54), and circRNA_finder algorithms (55). The rigor and reliability differ among these algorithms. Comprehensive comparisons of different circRNA algorithms have been performed by Zeng et al. and Hansen et al. (56,57), and are not discussed in detail here. Collectively, the combination of different algorithms was suggested when identifying a novel circRNA. Further, circRNA sequencing or microarray analysis at the transcriptome level is an essential means to screen and study differentially expressed circRNAs in a given disease.
Validation of circularization
Reverse transcription (RT) followed by polymerase chain reaction (PCR) using divergent primers which spans the back-spliced junction is the most widely method to validate circRNAs. After RT-PCR, sanger sequencing is then performed to detect the back-spliced junction. In addition, northern blotting is another strategy for which the specific probe spanning the back-spliced junction should be used; however, the low expression abundance of circRNAs limits the application of this technique. Due to the absence of 5’ and 3’ end, circRNAs are resistant to the degradation of Ribonuclease R (RNase R), while linear parental genes are digested readily (58). Therefore, RNase R treatment can also be utilized for the validation of circRNAs. Considering accuracy, most scholars combined these technologies in their studies.
Silencing and overexpression of circRNA
Eliminating non-specific effects on cognate linear RNA is the major concern when silencing circRNA using small interfering RNA (siRNA) or short-hairpin RNA (shRNA). Considering the sequence of siRNA or shRNA targeting back-spliced sites is partially complementary to parental mRNA, Li et al. proposed that the control sequence should be partially replaced (~10 nt) by the back-spliced sites-targeting siRNA sequence (59). The strategy of CRISPR-Cas9 is another possibility. Of note, caution is needed when applying the strategy to silence circRNAs, because deletion of the back-spliced exons in the genome simultaneously affects the expression of the cognate linear RNA. Therefore, aiming at deleting RCMs within introns flanking circularized exon(s) may be a preferred solution. Using this strategy, Zhang et al. and Zheng et al. achieved the silence of circGCN1LI in human PA1 cells (16) and circHIPK3 in HEK-293T cells (60), respectively.
Flanking RCMs-containing vectors are commonly used to overexpress circRNAs. The overexpressing vectors are classified into two types based on the source of the flanking RCMs. One is universal framework vector within which flanking complementary sequence is universal, not gene-specific. pcDNA3.1(+) CircRNA Mini Vector is a typical example for it (61). The other is gene-specific flanking sequence vector which simulates the natural process of circRNA biogenesis in vivo. Of note, because overexpression vectors cannot achieve complete circularization, linear RNA will also be concomitantly formed, which seems to be an additional challenge for circRNA overexpression.
CircRNAs in CVDs
In 2016, Jakobi et al. identified 575 circRNAs in adult murine heart through RNase R enrichment strategy (62). They further found many of these circRNAs were produced from cardiovascular disease-associated gene loci, revealing disease-relevant functions of cardiac circRNAs. Immediately afterwards, Werfel et al. determined more than 9,000 circRNAs in human, mouse, and rat hearts, respectively. Approximately 30% are conserved between mouse and rat; 10% are conserved in the three species (63). Moreover, the authors found circRNAs in neonatal rat cardiomyocytes (NRCMs) were tended to localize in the cytoplasm (63). In addition, by performing ribosomal-depleted RNA-sequencing, Tan et al. found 15,318 and 3,017 circRNAs in human and mouse, respectively, and the abundance of these circRNAs was generally positively correlated with that of the cognate linear RNAs (64). Other than in physiological conditions, increasing roles of circRNAs in CVDs are gradually highlighted. We have now summarized the circRNAs and their roles in different types of CVDs (Table 1).

Full table
CircRNAs in myocardial infarction
Geng et al. found that the expression of circRNA Cdr1as was upregulated in both mouse MI injury model and hypoxia-treated cardiomyocytes. Overexpression of Cdr1as promoted cardiomyocyte apoptosis, which was reversed by overexpression of miR-7a. Further, they determined that poly ADP ribose polymerase (PARP) and transcription factor specialized protein-1 (SP1) are the functional targets of miR-7a. Therefore, the authors proposed Cdr1as functioned as the sponge of miR-7a and then promoted PARP/SP1 expression, thereby aggravating MI injury (65). Zeng et al. reported that circ-Amotl1 overexpression increased cardiomyocyte survival and reduced apoptosis. In addition, circ-Amotl1 overexpression was found to be cardioprotective with reduced left ventricular dilatation, collagen deposition, and apoptotic cells in doxorubicin-induced cardiomyopathy mouse model. Mechanistically, circ-Amotl1 promoted phosphorylation of AKT1 through PDK1 and subsequent AKT1 nuclear localization the process that can protect heart from destruction (36).
CircRNAs in cardiac hypertrophy and heart failure
Wang et al. found miR-223 overexpression induced cardiac hypertrophy and HF in mice and hypertrophy in cardiomyocytes; miR-223 knockdown presented a protective effect. Moreover, they identified apoptosis repressor with caspase recruitment domain (ARC), an apoptosis repressor which was reported to be associated with cardiomyocyte hypertrophy (74-76), as the downstream target of miR-223. Furthermore, they demonstrated heart related circRNA HRCR which showed a markedly decreased expression in failing mouse hearts inhibited cardiomyocytes hypertrophy in vitro and cardiac hypertrophy and HF in vivo through serving as the sponge of miR-223 to increase the expression of ARC (66).
CircRNAs in atherosclerosis
A genome-wide association study (GWAS) identified chromosome 9p21.3 as a risk loci of AS (77). Long non-coding RNA (lncRNA) ANRIL transcribed from chromosome 9p21.3 was reported to be associated with the progression of AS (78). Interestingly, a circRNA generated from lncRNA ANRIL, termed circANRIL, can competitively bind to pescadillo homologue 1 (PES1) to impair precursor ribosomal RNA (pre-rRNA) maturation, leading to nucleolar stress and p53 activation, thereby promoting apoptosis and inhibiting proliferation in vascular smooth muscle cells (VSMCs) and macrophages which are pivotal components in the pathogenesis of AS, and thus confer a protective effect on AS (67). In addition, Chen et al. determined circRNA expression profile in high glucose-induced VSMCs, and identified an upregulated circRNA, circWDR77. Subsequent results showed silence of circWDR77 inhibited the excessive proliferation and migration of high glucose-induced VSMCs. Bioinformatic and experimental evidence supported miR-124 and fibroblast growth factor 2 (FGF-2) were the downstream targets of circWDR77 regarding the function model of microRNA sponge, and revealed the role of circWDR77/miR-124/FGF-2 axis in the proliferation and migration of VSMCs and the progression of AS (68).
CircRNAs in cardiac senescence
Du et al. found the expression of circ-Foxo3 was significantly higher in hearts of aged patients and mice, and displayed a positive correlation with senescence-associated β-galactosidase (SA-β-gal) which indicates the senescence of cardiomyocytes, in hydrogen peroxide-treated cardiomyocytes. In doxorubicin-induced cardiomyopathy, circ-Foxo3 silencing reversed cardiomyopathy, showing the recovery of left ventricular function and the reduction of the left ventricular end-diastolic diameter, etc. Mechanistically, circ-Foxo3 functioned as the protein sponge of senescence-associated proteins inhibitor of differentiation-1(ID-1), transcription factor E2 promoter binding factor 1 (E2F-1), anti-stress protein focal adhesion kinase (FAK), and hypoxia inducible factor-1 (HIF-1) to prevent these proteins entering the nucleus, thereby inhibiting anti-senescence and anti-stress effects, and leading to heart senescence (69).
CircRNAs in hypertension
By analysing plasma circRNA expression profile, Wu et al. demonstrated hsa-circ-0005870 was significantly downregulated in hypertensive patients. Subsequently, the authors screened top five putative miRNAs which might bind to hsa-circ-0005870, including miR-6807-3p, miR-5096, miR-5095, miR-1273g-3p, and miR-619-5p. Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway and gene oncology (GO) analyses showed that the target mRNAs of these miRNAs were correlated to HT, suggesting the potential roles of hsa_circ_0005870 in HT (70). In addition, Bao et al. found the expression of hsa_circ_0037911 was significantly higher in hypertensive patients compared to the healthy controls in a case-control study on 200 essential hypertension (EH) patients, and they subsequently revealed that hsa_circ_0037911 might serve as the sponge of miR-637, thus contributing to the pathogenesis of EH (71).
CircRNAs in cardiac fibrosis
Tang et al. found that in diabetic mouse myocardium and Ang-II-induced mouse cardiac fibroblasts, the expression of circRNA_000203 was significantly upregulated, and further research showed that circRNA_000203 could act as miR-26b-5p sponge to to promote the expression of fibrosis-related protein collagen type I alpha 2 (Col1a2) and connective tissue growth factor (CTGF) which are the known targets of miR-26b-5p, thus accelerating the proliferation of cardiac fibroblasts and procession of cardiac fibrosis (72). Similar to circRNA_000203, circRNA_010567 was also reported to be remarkably upregulated in diabetic mouse myocardium and mouse cardiac fibroblasts treated with Ang-II. Mechanistically, circRNA_010567 promoted the expression of transforming growth factor β (TGF-β), fibrosis-related protein collagen type I (Col I), collagen type III (Col III), and α-smooth muscle actin (α-SMA) by sponging miR-141 (73).
CircRNAs as diagnostic biomarkers for CVDs
Up to date, CVDs were generally detected by various biomarkers, such as creatine kinase-myocardial band isoenzymes (CK-MB) (79) and cardiac troponins T/I (cTnT/I) (80). However, the diagnostic effectiveness of current biomarkers might be affected by several noncardiac causes, such as heart-related diseases, lifestyles, and age (81). CircRNAs are characterized with relatively high stability, conservation, and tissue- or developmental-specificity (22,32). In addition, circRNAs are reproducible, easily detectable in blood samples, and have high sensitivity (82). Therefore, these outstanding advantages confer circRNAs more potentials to serve as novel biomarkers for CVDs.
Existed studies have suggested circRNAs can serve as biomarkers of CVDs. Salgado-Somoza et al. demonstrated that the expression of circRNA myocardial infarction-associated circular RNA (MICRA) was related to outcome of MI, and might be a potential biomarker of left ventricular dysfunction in the acute MI patients (83). Zhao et al. showed that hsa_circ_0124644 is upregulated in the peripheral blood of CHD patients and can serve as a diagnostic biomarker for CHD (84). Sonnenschein et al. showed that circulating circRNAs DNAJC6, MBOAT2, and TMEM56 can distinguish healthy controls from hypertrophic cardiomyopathy (HCM) patients; the expression of circTMEM56 and circDNAJC6 represent disease severity in patients with obstructive form (HOCM), implying their potentials as novel biomarkers for the diagnosis and prognosis of HCM (85). Moreover, a circRNA microarray expression profile showed that the expression level of hsa_circ_0014243 was higher in the blood of EH patients than paired controls, and was correlated with age, high density lipoprotein, and glucose levels. Therefore, hsa_circ_0014243 might be a potential biomarker to diagnose HT (86).
CircRNA-based therapeutic strategies for CVDs
Current therapeutic strategies against CVD range from lifestyle adjustments and pharmacological treatments to surgery and transplant, with the distinct type and severity of CVDs. Despite advancements in treatment strategies, mortality of CVDs still remains high (87). Gene therapy, delivering siRNA, miRNA, or plasmids by virtue of adenovirus, retrovirus, and so forth to the body, is now an emerging therapeutic strategy. Considering the involvements of circRNAs in CVDs and their outstanding characteristics, circRNA-based gene therapy may be a promising approach to treat CVDs. For those circRNAs which play protective roles in CVDs, overexpression vector delivery in vivo might achieve the goal of alleviating corresponding CVDs; conversely, for those which may promote the occurrence or development of CVDs, siRNA, shRNA, or other loss of function-related strategies could take effect. We have now summarized several circRNA-based therapeutic strategies based on biological or physical materials for CVDs (Table 2).

Full table
Transient transfection of siRNA or overexpression vector in vivo
A study by Du et al. showed intraperitoneal injection of circ-Foxo3 plasmid induced senescence and aggravated doxorubicin-induced cardiomyopathy. As expected, intraperitoneal injection of circ-Foxo3-targeted siRNA inhibited senescence and relieved cardiomyopathy (69). In addition, Geng et al. achieved the overexpression of Cdr1as in vivo via intracardial injection of pcDNA-Cdr1as vector in mouse MI injury model (65). These studies suggest that direct siRNA or overexpression vector delivery of circRNA in vivo might be an effective therapeutic strategy for CVDs (Figure 3). Despite availability and time saving, the effects of transient transfection could just last for a short time on one hand, and on the other hand, the introduction of exogenous RNA or DNA molecules can trigger an innate immune response. Hopefully, several chemical modifications, such as cholesterol, phosphorothioate, and phosphate, can improve the anti-ribonuclease ability and cell adsorption of siRNAs.
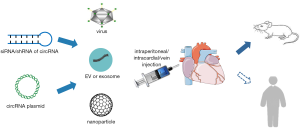
Viral vector-based gene therapy in vivo
Viral vector-based circRNA therapy has been applied to treat CVDs (Figure 3). Adenoviral vectors have many advantages, such as high titer and transduction efficiency, low pathogenicity, wide range of infection tissue, and no host cell genome integration. In a recent study, adenoviral circRNA HRCR was administered by direct injection to the jugular vein, which significantly attenuated hypertrophic responses in isoproterenol-induced cardiac hypertrophy and heart failure mouse model (66). Recombinant adeno-associated virus vector (rAAV), with the advantages of high transduction efficiency, stable, safe, and tissue- or organ-specific expression, was regarded as one of the most promising gene transfer vectors and has been widely used in gene therapy and vaccine research worldwide. Different AAV serotypes have different tissue affinities, and among them, AAV9 vector has myocardial tropism, which can be used to achieve cardiac-specific expression (88). Through intravenous injection of AAV9 vectors packaging circRNA vector, Meganck et al. achieved robust circRNA overexpression in the hearts of mice (89). Lim et al. showed AAV9-mediated RNAi knockdown of highly abundant circular RNA cricSlc8a1 attenuated cardiac hypertrophy and HF; AAV9-mediated forced overexpression of circSlc8a1 induced HF in vivo (90). Han et al. found AAV9 vector-based overexpression of CircITCH improved doxorubicin-induced cardiotoxicity manifestations in mice (91). In addition, Garikipati et al. generated AAV9 viral particles expressing circFndc3b under control of a CMV promoter to delivery circFndc3b in vivo, and found that overexpression of circrFndc3b post-MI in the hearts of mice reduced cardiomyocyte apoptosis, promoted neovascularization, and further improved heart function (92). Moreover, Ni et al. showed intravenous injection of AAV-9 containing sh-circHIPK3 vector 2 weeks prior to minipumps implantation indeed silenced the expression of circHIPK3, and alleviated Ang II-induced cardiac hypertrophy and functional impairment in the mice (93). Even though, caution should be remained due to the activation of innate immune responses and genomic integration. In conclusion, choosing the optimal virus in the context of certain CVD is conductive to minimize these shortcomings.
Extracellular vesicle- or exosome-based gene delivery in vivo
Yang et al. constructed extracellular vesicles (EVs) that target the central nervous system and encapsulate circSCMH1 and demonstrated that circSCMH1-EVs significantly promoted functional recovery of motor after stroke in mice and nonhuman primate ischemic stroke models (94). Exosomes can be rapidly taken up by target cells via specific surface markers. Despite the lack of relevant research, exosomes or EVs modified by cardiovascular cell-specific surface markers might be a promising circRNA carrier to treat CVDs (Figure 3). However, the strategy is still in its infancy with several unknown points, such as the optimal dose, route of administration, etc. More animal or preclinical experiments are needed to maximize the therapeutic effects of this strategy.
Nanoparticle-based gene delivery in vivo
Nanoparticle can improve the delivery of drug to intracellular targets that are hard to reach by common manner, which makes nanoparticle-packaged deliver to be a promising strategy to perform circRNA-based gene therapy (Figure 3). Zeng et al. constructed a nanoparticle circAmotl1 plasmid by packaging circAmotl1 plasmid in polyethylene glycol and then mixing with gold nanoparticles, and observed that intraperitoneal injection of nanoparticle circAmotl1 plasmid improved doxorubicin-induced cardiomyopathy in mice (36). However, the lack of popularity of nanotechnology is a great challenge to generalize this emerging in vivo gene delivery strategy. Therefore, improving the popularity of nanotechnology and the cooperation of interdisciplinary laboratories is indispensable in the future.
Conclusions and perspectives
Recent studies have revealed the involvement of circRNAs in the pathogenesis of CVDs. In the review, we systematically summarized the formation mechanisms, functional models, and research approaches of circRNAs, and several circRNAs involved in CVDs. Finally, we proposed that developing circRNAs as biomarkers or circRNA-based therapeutic strategies based on biological or physical materials may be promising to diagnose or treat CVDs in the future.
Notably, even though laboratory work and computational technology have revealed several circRNAs involved in CVDs, most of studies only suggested a relevance between circRNAs and specific CVD, and the detailed functions and mechanisms are still in their infancy, which puts up an obstacle for developing circRNA-based therapeutic strategies. Therefore, future circRNA studies on CVDs would be directed at the following points. First, the functions of circRNAs from large-scale bioinformatics and high-throughput sequencing analyses should be identified in vitro or in vivo. Second, more potential mechanisms besides miRNA sponge should be explored to explain the contribution of circRNAs in the development and pathogenesis of CVDs. Finally, developing circRNAs as biomarkers or circRNA-based therapeutic strategies by virtue of biological or physical materials would lower CVDs burden in the long run.
Acknowledgments
Funding: This work was supported by grants from the project: Assessment and intervention of balanced gait in elderly after physical disability (191592HS), Department of Ageing Health, National Health Commission.
Footnote
Reporting Checklist: The authors have completed the Narrative Review reporting checklist. Available at http://dx.doi.org/10.21037/atm-20-7929
Peer Review File: Available at http://dx.doi.org/10.21037/atm-20-7929
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/atm-20-7929). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Hsu MT, Coca-Prados M. Electron microscopic evidence for the circular form of RNA in the cytoplasm of eukaryotic cells. Nature 1979;280:339-40. [Crossref] [PubMed]
- Sanger HL, Klotz G, Riesner D, et al. Viroids are single-stranded covalently closed circular RNA molecules existing as highly base-paired rod-like structures. Proc Natl Acad Sci U S A 1976;73:3852-6. [Crossref] [PubMed]
- Cocquerelle C, Mascrez B, Hetuin D, et al. Mis-splicing yields circular RNA molecules. FASEB J 1993;7:155-60. [Crossref] [PubMed]
- Pasman Z, Been MD, Garcia-Blanco MA. Exon circularization in mammalian nuclear extracts. RNA 1996;2:603-10. [PubMed]
- Cocquerelle C, Daubersies P, Majerus MA, et al. Splicing with inverted order of exons occurs proximal to large introns. EMBO J 1992;11:1095-8. [Crossref] [PubMed]
- Nigro JM, Cho KR, Fearon ER, et al. Scrambled exons. Cell 1991;64:607-13. [Crossref] [PubMed]
- Zheng LL, Li JH, Wu J, et al. deepBase v2.0: identification, expression, evolution and function of small RNAs, LncRNAs and circular RNAs from deep-sequencing data. Nucleic Acids Res 2016;44:D196-202. [Crossref] [PubMed]
- Barrett SP, Wang PL, Salzman J. Circular RNA biogenesis can proceed through an exon-containing lariat precursor. Elife 2015;4e07540.
- Broadbent KM, Broadbent JC, Ribacke U, et al. Strand-specific RNA sequencing in Plasmodium falciparum malaria identifies developmentally regulated long non-coding RNA and circular RNA. BMC Genomics 2015;16:454. [Crossref] [PubMed]
- Lu T, Cui L, Zhou Y, et al. Transcriptome-wide investigation of circular RNAs in rice. RNA 2015;21:2076-87. [Crossref] [PubMed]
- Wang PL, Bao Y, Yee MC, et al. Circular RNA Is Expressed across the Eukaryotic Tree of Life. PLoS One 2014;9:e90859 [Crossref] [PubMed]
- Lyu D, Huang S. The emerging role and clinical implication of human exonic circular RNA. RNA Biol 2017;14:1000-6. [Crossref] [PubMed]
- Salzman J, Chen RE, Olsen MN, et al. Cell-type specific features of circular RNA expression. PLoS Genet 2013;9:e1003777 [Crossref] [PubMed]
- de Mestral C, Stringhini S. Socioeconomic Status and Cardiovascular Disease: an Update. Curr Cardiol Rep 2017;19:115. [Crossref] [PubMed]
- Chen LL. The expanding regulatory mechanisms and cellular functions of circular RNAs. Nat Rev Mol Cell Biol 2020;21:475-90. [Crossref] [PubMed]
- Zhang Y, Xue W, Li X, et al. The Biogenesis of Nascent Circular RNAs. Cell Rep 2016;15:611-24. [Crossref] [PubMed]
- Maass PG, Glazar P, Memczak S, et al. A map of human circular RNAs in clinically relevant tissues. J Mol Med (Berl) 2017;95:1179-89. [Crossref] [PubMed]
- Guo JU, Agarwal V, Guo H, et al. Expanded identification and characterization of mammalian circular RNAs. Genome Biol 2014;15:409. [Crossref] [PubMed]
- Yang L, Fu JR, Zhou YF. Circular RNAs and Their Emerging Roles in Immune Regulation. Front Immunol 2018;9:2977. [Crossref] [PubMed]
- Zhang Y, Zhang XO, Chen T, et al. Circular Intronic Long Noncoding RNAs. Mol Cell 2013;51:792-806. [Crossref] [PubMed]
- Zaphiropoulos PG. Circular RNAs from transcripts of the rat cytochrome P450 2C24 gene: Correlation with exon skipping. Proc Natl Acad Sci U S A 1996;93:6536-41. [Crossref] [PubMed]
- Jeck WR, Sorrentino JA, Wang K, et al. Circular RNAs are abundant, conserved, and associated with ALU repeats (vol 19, pg 141, 2013). RNA 2013;19:426. [Crossref]
- Ivanov A, Memczak S, Wyler E, et al. Analysis of intron sequences reveals hallmarks of circular RNA biogenesis in animals. Cell Rep 2015;10:170-7. [Crossref] [PubMed]
- Conn SJ, Pillman KA, Toubia J, et al. The RNA Binding Protein Quaking Regulates Formation of circRNAs. Cell 2015;160:1125-34. [Crossref] [PubMed]
- Errichelli L, Modigliani SD, Laneve P, et al. FUS affects circular RNA expression in murine embryonic stem cell-derived motor neurons. Nat Commun 2017;8:14741. [Crossref] [PubMed]
- Khan MAF, Reckman YJ, Aufiero S, et al. RBM20 Regulates Circular RNA Production From the Titin Gene. Circ Res 2016;119:996-1003. [Crossref] [PubMed]
- Kramer MC, Liang DM, Tatomer DC, et al. Combinatorial control of Drosophila circular RNA expression by intronic repeats, hnRNPs, and SR proteins. Genes Dev 2015;29:2168-82. [Crossref] [PubMed]
- Liu CX, Li X, Nan F, et al. Structure and Degradation of Circular RNAs Regulate PKR Activation in Innate Immunity. Cell 2019;177:865-80.e21. [Crossref] [PubMed]
- Fischer JW, Busa VF, Shao Y, et al. Structure-Mediated RNA Decay by UPF1 and G3BP1. Mol Cell 2020;78:70-84.e6. [Crossref] [PubMed]
- Park OH, Ha H, Lee Y, et al. Endoribonucleolytic Cleavage of m(6)A-Containing RNAs by RNase P/MRP Complex. Mol Cell 2019;74:494-507.e8. [Crossref] [PubMed]
- Hansen TB, Wiklund ED, Bramsen JB, et al. miRNA-dependent gene silencing involving Ago2-mediated cleavage of a circular antisense RNA. EMBO J 2011;30:4414-22. [Crossref] [PubMed]
- Memczak S, Jens M, Elefsinioti A, et al. Circular RNAs are a large class of animal RNAs with regulatory potency. Nature 2013;495:333-8. [Crossref] [PubMed]
- Hansen TB, Jensen TI, Clausen BH, et al. Natural RNA circles function as efficient microRNA sponges. Nature 2013;495:384-8. [Crossref] [PubMed]
- Li Z, Huang C, Bao C, et al. Exon-intron circular RNAs regulate transcription in the nucleus (vol 22, pg 256, 2015). Nat Struct Mol Biol 2017;24:194. [Crossref] [PubMed]
- Du WW, Yang WN, Liu E, et al. Foxo3 circular RNA retards cell cycle progression via forming ternary complexes with p21 and CDK2. Nucleic Acids Res 2016;44:2846-58. [Crossref] [PubMed]
- Zeng Y, Du WW, Wu YY, et al. A Circular RNA Binds To and Activates AKT Phosphorylation and Nuclear Localization Reducing Apoptosis and Enhancing Cardiac Repair. Theranostics 2017;7:3842-55. [Crossref] [PubMed]
- Diallo LH, Tatin F, David F, et al. How are circRNAs translated by non-canonical initiation mechanisms? Biochimie 2019;164:45-52. [Crossref] [PubMed]
- Legnini I, Di Timoteo G, Rossi F, et al. Circ-ZNF609 Is a Circular RNA that Can Be Translated and Functions in Myogenesis. Mol Cell 2017;66:22. [Crossref] [PubMed]
- Yang Y, Gao X, Zhang M, et al. Novel Role of FBXW7 Circular RNA in Repressing Glioma Tumorigenesis. J Natl Cancer Inst 2018;110:304-15. [Crossref] [PubMed]
- Zhang M, Huang N, Yang X, et al. A novel protein encoded by the circular form of the SHPRH gene suppresses glioma tumorigenesis. Oncogene 2018;37:1805-14. [Crossref] [PubMed]
- Liang WC, Wong CW, Liang PP, et al. Translation of the circular RNA circbeta-catenin promotes liver cancer cell growth through activation of the Wnt pathway. Genome Biol 2019;20:84. [Crossref] [PubMed]
- Liu J, Yue Y, Han D, et al. A METTL3-METTL14 complex mediates mammalian nuclear RNA N-6-adenosine methylation. Nat Chem Biol 2014;10:93-5. [Crossref] [PubMed]
- Ping XL, Sun BF, Wang L, et al. Mammalian WTAP is a regulatory subunit of the RNA N6-methyladenosine methyltransferase. Cell Res 2014;24:177-89. [Crossref] [PubMed]
- Jia G, Fu Y, Zhao X, et al. N6-Methyladenosine in nuclear RNA is a major substrate of the obesity-associated FTO. Nat Chem Biol 2011;7:885-7. [Crossref] [PubMed]
- Zheng G, Dahl JA, Niu YM, et al. ALKBH5 Is a Mammalian RNA Demethylase that Impacts RNA Metabolism and Mouse Fertility. Mol Cell 2013;49:18-29. [Crossref] [PubMed]
- Yang Y, Fan X, Mao M, et al. Extensive translation of circular RNAs driven by N(6)-methyladenosine. Cell Res 2017;27:626-41. [Crossref] [PubMed]
- Jia G, Fu Y, He C. Reversible RNA adenosine methylation in biological regulation. Trends Genet 2013;29:108-15. [Crossref] [PubMed]
- Zhang C, Chen Y, Sun B, et al. m(6)A modulates haematopoietic stem and progenitor cell specification. Nature 2017;549:273-6. [Crossref] [PubMed]
- Su H, Wang G, Wu L, et al. Transcriptome-wide map of m(6)A circRNAs identified in a rat model of hypoxia mediated pulmonary hypertension. BMC Genomics 2020;21:39. [Crossref] [PubMed]
- Chen RX, Chen X, Xia LP, et al. N(6)-methyladenosine modification of circNSUN2 facilitates cytoplasmic export and stabilizes HMGA2 to promote colorectal liver metastasis. Nat Commun 2019;10:4695. [Crossref] [PubMed]
- Huang R, Zhang Y, Bai Y, et al. N-6-Methyladenosine Modification of Fatty Acid Amide Hydrolase Messenger RNA in Circular RNA STAG1-Regulated Astrocyte Dysfunction and Depressive-like Behaviors. Biol Psychiatry 2020;88:392-404. [Crossref] [PubMed]
- Zhang XO, Wang HB, Zhang Y, et al. Complementary Sequence-Mediated Exon Circularization. Cell 2014;159:134-47. [Crossref] [PubMed]
- Gao Y, Wang J, Zhao F. CIRI: an efficient and unbiased algorithm for de novo circular RNA identification. Genome Biol 2015;16:4. [Crossref] [PubMed]
- Wang K, Singh D, Zeng Z, et al. MapSplice: Accurate mapping of RNA-seq reads for splice junction discovery. Nucleic Acids Res 2010;38:e178 [Crossref] [PubMed]
- Chen L, Yu Y, Zhang X, et al. PcircRNA_finder: a software for circRNA prediction in plants. Bioinformatics 2016;32:3528-9. [Crossref] [PubMed]
- Zeng X, Lin W, Guo M, et al. A comprehensive overview and evaluation of circular RNA detection tools. PLoS Comput Biol 2017;13:e1005420 [Crossref] [PubMed]
- Hansen TB, Veno MT, Damgaard CK, et al. Comparison of circular RNA prediction tools. Nucleic Acids Res 2016;44:e58 [Crossref] [PubMed]
- Suzuki H, Zuo YH, Wang JH, et al. Characterization of RNase R-digested cellular RNA source that consists of lariat and circular RNAs from pre-mRNA splicing. Nucleic Acids Res 2006;34:e63 [Crossref] [PubMed]
- Li X, Yang L, Chen LL. The Biogenesis, Functions, and Challenges of Circular RNAs. Mol Cell 2018;71:428-42. [Crossref] [PubMed]
- Zheng Q, Bao C, Guo W, et al. Circular RNA profiling reveals an abundant circHIPK3 that regulates cell growth by sponging multiple miRNAs. Nat Commun 2016;7:11215. [Crossref] [PubMed]
- Liang D, Wilusz JE. Short intronic repeat sequences facilitate circular RNA production. Genes Dev 2014;28:2233-47. [Crossref] [PubMed]
- Jakobi T, Czaja-Hasse LF, Reinhardt R, et al. Profiling and Validation of the Circular RNA Repertoire in Adult Murine Hearts. Genomics Proteomics Bioinformatics 2016;14:216-23. [Crossref] [PubMed]
- Werfel S, Nothjunge S, Schwarzmayr T, et al. Characterization of circular RNAs in human, mouse and rat hearts. J Mol Cell Cardiol 2016;98:103-7. [Crossref] [PubMed]
- Tan WLW, Lim BTS, Anene-Nzelu CGO, et al. A landscape of circular RNA expression in the human heart. Cardiovasc Res 2017;113:298-309. [Crossref] [PubMed]
- Geng HH, Li R, Su YM, et al. The Circular RNA Cdr1as Promotes Myocardial Infarction by Mediating the Regulation of miR-7a on Its Target Genes Expression. PLoS One 2016;11:e0151753 [Crossref] [PubMed]
- Wang K, Long B, Liu F, et al. A circular RNA protects the heart from pathological hypertrophy and heart failure by targeting miR-223. Eur Heart J 2016;37:2602-11. [Crossref] [PubMed]
- Holdt LM, Stahringer A, Sass K, et al. Circular non-coding RNA ANRIL modulates ribosomal RNA maturation and atherosclerosis in humans. Nat Commun 2016;7:12429. [Crossref] [PubMed]
- Chen J, Cui L, Yuan J, et al. Circular RNA WDR77 target FGF-2 to regulate vascular smooth muscle cells proliferation and migration by sponging miR-124. Biochem Biophys Res Commun 2017;494:126-32. [Crossref] [PubMed]
- Du WW, Yang W, Chen Y, et al. Foxo3 circular RNA promotes cardiac senescence by modulating multiple factors associated with stress and senescence responses. Eur Heart J 2017;38:1402-12. [PubMed]
- Wu N, Jin L, Cai J. Profiling and bioinformatics analyses reveal differential circular RNA expression in hypertensive patients. Clin Exp Hypertens 2017;39:454-9. [Crossref] [PubMed]
- Bao X, Zheng S, Mao S, et al. A potential risk factor of essential hypertension in case-control study: Circular RNA hsa_circ_0037911. Biochem Biophys Res Commun 2018;498:789-94. [Crossref] [PubMed]
- Tang CM, Zhang M, Huang L, et al. CircRNA_000203 enhances the expression of fibrosis-associated genes by derepressing targets of miR-26b-5p, Col1a2 and CTGF, in cardiac fibroblasts. Sci Rep 2017;7:40342. [Crossref] [PubMed]
- Zhou B, Yu JW. A novel identified circular RNA, circRNA_010567, promotes myocardial fibrosis via suppressing miR-141 by targeting TGF-beta1. Biochem Biophys Res Commun 2017;487:769-75. [Crossref] [PubMed]
- Donath S, Li P, Willenbockel C, et al. Apoptosis repressor with caspase recruitment domain is required for cardioprotection in response to biomechanical and ischemic stress. Circulation 2006;113:1203-12. [Crossref] [PubMed]
- Murtaza I, Wang HX, Feng X, et al. Down-regulation of catalase and oxidative modification of protein kinase CK2 lead to the failure of apoptosis repressor with caspase recruitment domain to inhibit cardiomyocyte hypertrophy. J Biol Chem 2008;283:5996-6004. [Crossref] [PubMed]
- Zaiman AL, Damico R, Thoms-Chesley A, et al. A critical role for the protein apoptosis repressor with caspase recruitment domain in hypoxia-induced pulmonary hypertension. Circulation 2011;124:2533-42. [Crossref] [PubMed]
- Wellcome Trust Case Control Consortium. Genome-wide association study of 14,000 cases of seven common diseases and 3,000 shared controls. Nature 2007;447:661-78. [Crossref] [PubMed]
- Burd CE, Jeck WR, Liu Y, et al. Expression of linear and novel circular forms of an INK4/ARF-associated non-coding RNA correlates with atherosclerosis risk. PLoS Genet 2010;6:e1001233 [Crossref] [PubMed]
- Jang JS, Jin HY, Seo JS, et al. Prognostic value of creatine kinase-myocardial band isoenzyme elevation following percutaneous coronary intervention: a meta-analysis. Catheter Cardiovasc Interv 2013;81:959-67. [Crossref] [PubMed]
- Nguyen DV, Alfraidi H, Esmaiel A. Letter by Nguyen et al Regarding Article, "B-Type Natriuretic Peptides and Cardiac Troponins for Diagnosis and Risk-Stratification of Syncope Circulation 2019;140:e729-30. [Crossref] [PubMed]
- Rodríguez-González M, Castellano-Martinez A, Alonso-Ojembarrena A. Usefulness of age-adjusted N-terminal prohormone of brain natriuretic peptide level as a diagnostic marker for incomplete Kawasaki disease in children. Emergencias 2019;31:111-4. [PubMed]
- Li Y, Zheng Q, Bao C, et al. Circular RNA is enriched and stable in exosomes: a promising biomarker for cancer diagnosis. Cell Res 2015;25:981-4. [Crossref] [PubMed]
- Salgado-Somoza A, Zhang L, Vausort M, et al. The circular RNA MICRA for risk stratification after myocardial infarction. Int J Cardiol Heart Vasc 2017;17:33-6. [Crossref] [PubMed]
- Zhao Z, Li X, Gao C, et al. Peripheral blood circular RNA hsa_circ_0124644 can be used as a diagnostic biomarker of coronary artery disease. Sci Rep 2017;7:39918. [Crossref] [PubMed]
- Sonnenschein K, Wilczek AL, de Gonzalo-Calvo D, et al. Serum circular RNAs act as blood-based biomarkers for hypertrophic obstructive cardiomyopathy. Sci Rep 2019;9:20350. [Crossref] [PubMed]
- Zheng S, Gu T, Bao X, et al. Circular RNA hsa_circ_0014243 may serve as a diagnostic biomarker for essential hypertension. Exp Ther Med 2019;17:1728-36. [PubMed]
- Bonsu KO, Owusu IK, Buabeng KO, et al. Review of novel therapeutic targets for improving heart failure treatment based on experimental and clinical studies. Ther Clin Risk Manag 2016;12:887-906. [Crossref] [PubMed]
- Guenther CM, Brun MJ, Bennett AD, et al. Protease-Activatable Adeno-Associated Virus Vector for Gene Delivery to Damaged Heart Tissue. Mol Ther 2019;27:611-22. [Crossref] [PubMed]
- Meganck RM, Borchardt EK, Castellanos Rivera RM, et al. Tissue-Dependent Expression and Translation of Circular RNAs with Recombinant AAV Vectors In Vivo. Mol Ther Nucleic Acids 2018;13:89-98. [Crossref] [PubMed]
- Lim TB, Aliwarga E, Luu TDA, et al. Targeting the highly abundant circular RNA circSlc8a1 in cardiomyocytes attenuates pressure overload induced hypertrophy. Cardiovasc Res 2019;115:1998-2007. [Crossref] [PubMed]
- Han D, Wang Y, Wang Y, et al. The Tumor-Suppressive Human Circular RNA CircITCH Sponges miR-330-5p to Ameliorate Doxorubicin-Induced Cardiotoxicity Through Upregulating SIRT6, Survivin, and SERCA2a. Circ Res 2020;127:e108-25. [Crossref] [PubMed]
- Garikipati VNS, Verma SK, Cheng Z, et al. Circular RNA CircFndc3b modulates cardiac repair after myocardial infarction via FUS/VEGF-A axis. Nat Commun 2019;10:4317. [Crossref] [PubMed]
- Ni H, Li W, Zhuge Y, et al. Inhibition of circHIPK3 prevents angiotensin II-induced cardiac fibrosis by sponging miR-29b-3p. Int J Cardiol 2019;292:188-96. [Crossref] [PubMed]
- Yang L, Han B, Zhang Z, et al. Extracellular Vesicle-Mediated Delivery of Circular RNA SCMH1 Promotes Functional Recovery in Rodent and Nonhuman Primate Ischemic Stroke Models. Circulation 2020;142:556-74. [Crossref] [PubMed]


