
Properties and gene expression profiling of acquired radioresistance in mouse breast cancer cells
Introduction
Breast cancer has been identified as a leading cause of cancer death among women, accounting for about 500,000 deaths per year (1). Surgery, radiation and adjuvant systemic therapies (chemotherapy, endocrine therapy and tissue-targeted therapies) are common treatments for breast cancer (2). Radiotherapy (RT) is usually used to shrink large tumors before surgery or kill the postoperative residual breast cancer cells. However, different breast cancer subtypes respond differently to RT. Human epidermal growth factor receptor 2 (HER2)-positive tumors accounts for about 15–20% of breast cancers and has been reported to show much more radioresistance than HER2-negative breast cancer in clinical radiotherapy (3-5). Accumulating evidence suggests that overexpression of HER2 confers to the radioresistance mediated by activating NF-κB and PI3K/Akt pathway, promoting epithelial-to-mesenchymal transition or acquiring a stem cell phenotype. However, radioresistance of HER2-positive breast cancer still cannot be fully explained by current known mechanisms.
Substantial progress has been made in cancer radiotherapy, but radioresistance remains a main obstacle to the effective treatment (6). In clinical radiotherapy, radiotherapy occasionally is not effectively to inhibit tumor regression and accelerated reproliferation is also usually observed at the late course of radiation (7). Even though tumors almost completely disappeared on the medical images after radiotherapy, the residual tumor cells which are undetectable using conventional diagnostic procedures sometimes remained viable. In fact, repopulating tumors initiating from these residual cells often acquire radioresistance, which is an important cause of radiotherapy failure (8). Meanwhile, acquired radioresistance leads to massive differences in biological properties between the resistant cells and their parent cells. Previous studies have shown that acquired radioresistance cells undergo characteristic changes including accelerated repopulation, enhanced migration and invasion and increased anti-apoptosis activity (9-11). In addition, accumulated evidence suggests that alterations in metabolism after irradiation are associated with the acquisition of radioresistance and play an important role in tumor metastasis and tolerance (12,13). It is necessary to profile the characteristic changes in the cells with acquired radioresistance.
As far as we know, acquired radioresistance usually results from a combination of multiple signaling pathways in response to radiation. However, it is difficult to identify the key effectors in these comprehensive signaling pathways. Moreover, changes after irradiation in biological functions including cellular proliferation, migration, invasion and apoptosis also usually boil down to changes in the gene expression profile (9,11,14). Based on the technique RNA-seq (RNA-Sequencing) which can precisely measure transcripts and their isoforms levels, the involved signal pathway molecules can be identified at the same time. The RNA-seq technique has been used to investigate radiation-responsive genes. Ma et al. identified 1656 significantly changed genes in response to radiation with RNA-seq and these genes are enriched in various biological pathways including cell cycle arrest, DNA replication, DNA repair and apoptosis (15). Young et al. also found several genes associated with DNA damage repair, which play an important role in radioresistance (16).
In this study, a radioresistant breast cancer cell line (NF639R) was established from Her-2/neu-overexpressing NF639 cells by simulating clinical routine radiotherapy. Some properties of radioresistant cells were described as follows: (I) a series of morphological changes including smaller and longer cell bodies and more slender pseudopodia were observed; (II) activities of proliferation, anti-apoptosis and migration were enhanced, but the basal level of ROS (reactive oxygen species) was decreased; (III) mitochondrial respiration and glycolytic function were increased. Furthermore, a total of 490 differentially expressed genes in radioresistant cell lines were identified and the possible mechanisms associated with the above properties were also explored. In summary, we provided an ideal model for studying the identified various properties after repeated radiotherapy and preliminarily explored the related possible mechanisms. Our study gave new insights to tumor recurrence after radiotherapy.
We present the following article in accordance with the MDAR reporting checklist (available at http://dx.doi.org/10.21037/atm-20-4667).
Methods
Cell culture and culture condition
The Her-2/neu–driven mouse breast cancer cell line NF639 was purchased from American Type Culture Collection (Manassas, VA, USA) and maintained in Dulbecco Modified Eagle Medium (Cat. No.: C11875500BT, Gibco, USA) supplemented with 10% fetal bovine serum (FBS, Cat. No.: S711-001S, Lonsera, Uruguay), 100 U/mL penicillin and 100 μg/mL streptomycin (Cat. No.: 15140-122, Gibco, USA). The cells were incubated at 37 °C in a humidified atmosphere with 5% CO2 incubator and routinely checked for mycoplasma contamination.
X-ray irradiation
The cells were exposed to X-ray (SHINVA 600D) with specified doses with a dose rate of 0.189 Gy/min. Before irradiation, the culture medium was replaced with fresh medium.
Establishment of acquired radioresistant cell line
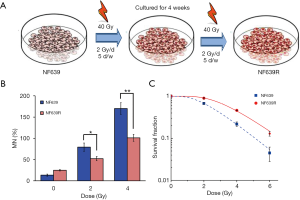
Radioresistant NF639R cells were developed from their parental NF639 cells by simulating clinical routine radiotherapy fractionated in 2 Gy per day, 5 days a week. A total dose of 80 Gy divided into two schedules (total 40 Gy/schedule) with 4 weeks interval was received over 12 weeks. Briefly, after receiving 40 Gy (the first schedule: 20 fractions over 4 weeks), the majority of cells died, and then the residual cells were incubated for 4 weeks (NF63940 Gy) and subsequently irradiated with 40 Gy (the second schedule: 20 fractions over 4 weeks) (Figure 1A). After 3 months (~30 passages) of culture following radiation (total 80 Gy), a radioresistant cell line, named NF639R, was established and then used in the follow-up experiments. The parental cells were treated in the same manner but without irradiation (IR).
Micronucleus assay
The frequency of micronucleus formation was measured with the cytokinesis block technique. Cells were trypsinized and then seeded at an initial density of 5×104 cells per 35 mm dish. Cytochalasin B (Cat. No.: C6762, Sigma, USA) was added into the culture medium at the final concentration of 1.5 μg/mL after 0, 2 or 4 Gy of irradiation. At 48 h after irradiation, the cells were fixed in 4% paraformaldehyde (Cat. No.: P6148-100G, Sigma, USA) for 30 mins and stained with 0.1% acridine orange (Cat. No.: A6014, Sigma, USA) solution. At least 1,000 cells were examined under a fluorescence microscope (Leica DMI 4000B, Germany) and only micronuclei in binucleated cells were considered for analysis.
Colony formation assay
NRF639 and NRF639R cells were seeded into 60 mm Petri dishes in a density of 300 cells per dish. Adherent cells were then irradiated with 0, 2, 4, or 6 Gy of X-ray. After 10 days, the cells were fixed with 4% paraformaldehyde and stained with 1% crystal violet. Colonies containing more than 50 cells were counted and the plating efficiency and surviving fraction (SF) were calculated. The cell survival curves were obtained by the single-hit multi-target model with Origin 8.0 software (OriginLab Corporation, Northampton, MA, USA). Each experiment was performed for three times.
Cell proliferation assay
To evaluate the proliferation rate of cells, the cell viability was determined with Cell Counter Kit-8 (CCK-8) (Cat. No.: CK04-500T, Dojindo, Japan) assay according to the manufacturer’s instructions. Briefly, the cells were seeded at a density of 4,000 cells per well in a 96-well plate. The CCK-8 solution (10 μL) was added into each well and incubated at 37 °C for 2 h. The optical density (OD) of each well at 450 nm was measured on a Varioskan Flash microplate reader (Thermo Fisher Scientific, Germany).
Cell migration assay
The cell migration assay was performed in a transwell insert with polycarbonate membrane bottom (8.0 μm pore size) (Cat. No.: 3422, Corning, NY, USA). Briefly, cells were suspended in serum-free medium and seeded into the inner chamber (5×104 cells per chamber). The complete growth medium was added into the outer chamber. After 12 hours of incubation, non-migrating cells on the upside of the membrane were carefully removed with a cotton swab and the cells migrated through the membrane were fixed and stained with 1% crystal violet. The images of stained cells were captured with a microscope (100×). In each well, the cells in at least 5 randomly selected fields were counted. Each experiment was performed in triplicate.
Intracellular ROS detection
The production of intracellular ROS was measured with a fluorescent probe, the CELLROX™ DEEP RED (Cat. No.: C10422, Thermo Fisher Scientific, Waltham, MA, USA), following the manufacturer’s instructions. Briefly, the cells were trypsinized and washed with FBS-free DMEM medium, followed by incubation with CELLROX™ DEEP RED (2.5 μM) for 30 min at 37 °C in dark. The cells were then collected and washed with PBS. The basal level of ROS was determined by using a flow cytometer (Accuri C6, BD Biosciences, Bedford, MA, USA).
Apoptosis detection
After ionizing radiation treatment, cells were harvested at 72 h and apoptosis was determined with the PI-Annexin V Apoptosis Detection Kit (Cat. No.: 559763, BD, USA) according to the manufacturer’s protocol. The cells were then immediately analyzed with a flow cytometer (Accuri C6, BD Biosciences, Bedford, MA, USA). All the data analyses were performed by using FlowJo analysis software.
Seahorse XF assays
Equal numbers of cells (2.0×104 cells per well) were seeded into quadruplicate wells of XF24 cell culture plate (Seahorse Bioscience, North Billerica, MA, USA) to reach 90% confluence. On the following day, OCR (oxygen consumption rate) and ECAR (extracellular acidification rate) were measured by using an XF24 Extracellular Flux Analyzer (Seahorse Bioscience). To determine the mitochondrial stress, cells were washed and replaced with XF medium-I (pH 7.4, 5 mM glucose, 2 mM sodium pyruvate and 2 mM L-glutamine in Seahorse XF Base Dulbecco’s Modified Eagle’s Medium; Cat. No.: 102353-100, Agilent, Santa Clara, CA, USA) followed by incubation for 1 h at 37 °C in a CO2 free incubator. Oligomycin (0.5 μM), carbonyl cyanide p-(trifluoromethoxy) phenylhydrazone (FCCP, 0.5 μM) and antimycin A/rotenone (10 μM) from the XF Cell Mito Stress Test Kit (Cat. No.: 103015-100, Agilent, Santa Clara, CA, USA) were then sequentially injected. To determine the glycolysis stress, cells were washed and the medium was replaced with XF medium-II (pH 7.4, 2 mM L-glutamine in Seahorse XF Base Dulbecco’s modified Eagle’s medium) followed by incubation for 1 h at 37 °C in a CO2 free incubator. Three compounds (10 mM glucose, 1.0 μM oligomycin and 100 mM 2-deoxyglucose) from the XF Glycolysis Stress Test Kit (Cat. No.: 103020-100, Agilent, Santa Clara, CA, USA) were then sequentially injected. The results were normalized to the total protein measured by Bradford assay and the data were analyzed with the Seahorse Wave software. Basal oxidative phosphorylation (OXPHOS) (before adding oligomycin) and ATP-linked OXPHOS (the difference between basal OCR and oligomycin-induced OCR) were calculated from the profile of OCR as the indices of mitochondrial respiratory function. Basal glycolysis (after adding glucose), maximal glycolysis (after adding oligomycin) and glycolytic capacity (the difference between oligomycin-induced ECAR and 2-deoxyglucose-induced ECAR) were calculated from the profile of ECAR as indices of glycolytic pathway activation.
RNA extraction and sequencing
Total RNA was extracted using the Qiagen RNeasy Midi Kit (Cat. No.: 75144, Qiagen, Valencia, CA, USA) following the manufacturer’s instructions. The quantity of RNA samples was determined with NanoDrop 2000 (Thermo Fisher Scientific, Waltham, MA, USA) and Agilent 2100 bioanalyzer (Agilent Technologies, Santa Clara, CA, USA). TruSeq RNA Library Prep Kit v2 (Cat. No: RS-122-2001, Illumina, San Diego, CA, USA) were used to prepare libraries following the manufacturer’s instructions. Purified libraries were quantified by Qubit 2.0 Fluorometer (Life Technologies, Carlsbad, CA, USA) and Agilent 2100 bioanalyzer. Clusters were generated by using the cBot with the library and sequenced on Illumina HiSeq 2500 platform (San Diego, CA, USA).
Bioinformatic analyses of RNA-seq
FastQC (version: 0.11.2) was conducted for quality control (QC) of RNA-Seq reads (http://www.bioinformatics.babraham.ac.uk/projects/fastqc). Trimming was performed with Trimmomatic (version: 0.36) for known Illumina TruSeq adapter sequences, poor reads and ribosome RNA reads etc. The trimmed reads were then mapped to the Mus musculus reference genome (MM10) by the HISAT2 (version: 2.1.0) (17,18). StringTie (version: 1.3.3b) was performed for each gene count from trimmed reads (19). Gene counts were normalized by trimmed mean of M-values (TMM) and Transcripts Per Million (TPM) were calculated (20). DEGseq was performed for determining differential-expressed genes, following a threshold with q value <0.05 and absolute values of |log2FC (fold change)| >1 (21). GO (Gene Ontology) and KEGG (Kyoto Encyclopedia of Genes and Genomes) pathways were enriched with R package (v3.5.1) to better understand the functions of the DEGs (differentially expressed genes) (22). A protein-protein interaction (PPI) network was constructed using STRING database. In this study, clusterProfiler was applied to analysis of GO terms and KEGG pathways and the top 30 GOs and pathways were presented.
Real-time quantitative PCR
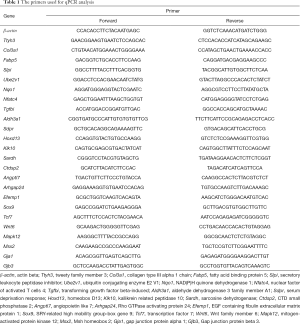
The concentration and purity of total RNA were analyzed using NanoDrop 2000 (Thermo Fisher Scientific, Waltham, MA, USA). Reverse transcription for cDNA synthesis was performed with RevertAid First Strand cDNA Synthesis Kit (Cat. No.: K1621, Thermo Scientific, USA) from 2 μg of total RNA according to the manufacturer’s instructions. The qPCR (real-time quantitative PCR) was performed with the QuantiTect SYBR Green PCR kit (Cat. No.: 204143, Qiagen, Germany) by using specified primers as listed in Table 1. The housekeeping gene, β-actin, served as an internal control and the relative mRNA levels were calculated by the 2−ΔΔCq method.
Full table
Statistical analysis
T-tests were employed to screen out differentially expressed genes between NF639R and NF639 cells, with a threshold of P<0.05 and fold change >2. All experiments were performed independently 3 times and 3 parallel samples were taken for each experiment. All data were presented as the mean ± SD (standard deviation) from triplicate independent experiments. Statistical significance was determined with t-test and P<0.05 was considered as significant difference.
Results
The acquired radioresistance of NF639R cells
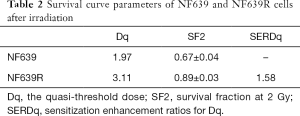
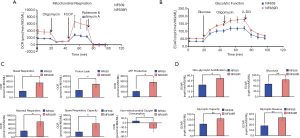
To verify the radioresistance of NF639R cells, i.e., the survived NF639 cells after total irradiation of 80 Gy, clone survival assay and micronuclei test were performed. Compared with the NF639 cells, the yield of micronucleus in NF639R cells was also significantly lower after the same dose radiation (Figure 1B), indicating the unrepaired or misrepaired DNA damage decreased. The survival fractions of NF639 and NF639R cells were fitted by the single-hit multi-target model and the dose-effect fitting curve was shown in Figure 1C. Multiple parameters including D0 (mean lethal dose), Dq (the quasi-threshold dose), SF2 (survival fraction at 2 Gy) and SERDq (sensitization enhancement ratios of Dq) values were calculated and shown in Table 2. The results showed the survival fraction of NF639R cells was significantly higher than that of NF639 cells after exposure to the same dose irradiation, suggesting a significant increase in radioresistance. In addition, D0, Dq and SF2 values of NF639R cells were all higher than those of NF639 cells, further indicating that the average lethal dose was distinctly increased in NF639R cells. The sensitizer enhancement ratio for Dq (SERDq) showed a significant increase to 1.58 (Table 2) in NF639R. Taken together, the NF639R cells exhibited significantly increased radioresistance compared with the NF639 cells.
Full table
Promotion of tumor cell malignancy accompanied by acquisition of radioresistance
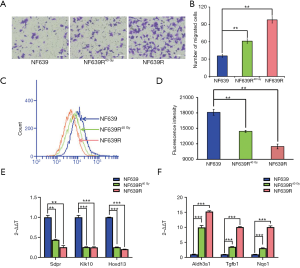
Different from the large and flat morphology of NF639 cells, smaller and spindlier cell bodies and long slender pseudopods were observed in most NF639R cells (Figure 2A). It is known that the spindle shaped morphology, a mesenchymal-like phenotype, is beneficial to migration of tumor cells (23-25). Indeed, the transwell migration assay revealed that the number of migrating NF639R cells was ~2.5-fold more than migrating NF639 cells, indicating a strong migration ability of NF639R cells (Figure 2B,C). The proliferative assay also showed a significant increase (~1.15-fold) of NF639R cells compared with NF639 cells (Figure 2D), indicating that the proliferation of tumor cells was accelerated after transferring to radioresistant cells. Accumulating evidence suggests that the acquired ability to inhibit apoptosis plays a critical role in the development of radiation resistance (11,26). The results in Figure 2E showed that NF639R cells exhibited significantly less apoptosis than NF639 cells, confirming the increased anti-apoptotic ability of NF639R cells. Finally, the basal ROS levels in NF639 and NF639R cells were detected and the results showed that the average fluorescence intensity in NF639R cells was 1.54-fold lower than that of NF639 cells (Figure 2F,G), indicating lower basal ROS level in NF639R cells.
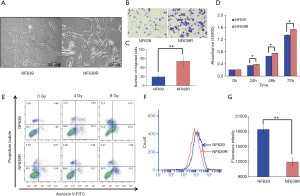
Taken together, the alteration of cell morphology, enhancement of migration ability, proliferation and anti-apoptosis ability and the decrease in basal ROS were observed in NF639R cells with the acquired radioresistance. These results indicated that NF639R cells should be more malignant than their parental NF639 cells.
Energy metabolism reprogramming in NF639R cells
Aerobic glycolysis and mitochondrial oxidative phosphorylation are the two major energy metabolism pathways to provide enough energy for cell survival. Changes in energy metabolism can profoundly influence the fate of tumor cells (27,28). Hence, the glycolysis ability and mitochondrial function in NF639 and NF639R cells were further studied. The kinetic profiles of OCR and ECAR in NF639 and NF639R cells were obtained (Figure 3A,B) and various fundamental parameters were calculated. A significant increase in basal OCR, proton leak, ATP production, spare respiratory capacity and maximal respiration levels, but a significant decrease of the non-mitochondrial oxygen consumption was observed in NF639R cells compared to NF639 cells (Figure 3C). These results suggested that the mitochondrial respiration capacity was elevated in NF639R cells compared to NF639 cells. Moreover, higher spare respiratory function might also be associated with increased energy availability depending on OXPHOS in NF639R cells. Anaerobic glycolysis, defined as the conversion of glucose to lactate, is a major ATP-producing pathway independent of mitochondrial respiration. The glycolytic activity was evaluated by measuring the extracellular acidification rate (ECAR). NF639R cells displayed higher base glycolysis, glycolytic capacity, glycolytic reserve and non-glycolytic acidification than NF639 cells (Figure 3D), indicating an increased glycolytic activity in radioresistant cell line NF639R. The increased anaerobic glycolysis may also produce more precursors for the synthesis of macromolecules, such as nucleotides, proteins and lipids, which are essential to cell survival and proliferation.
Identification of differently expressed genes between NF639 and NF639R cells
In order to systematically identify the genes associated with the altered properties of NF639R cells, high-throughput RNA sequencing was employed. There was the high similarity of three independent biological repeats and the significant difference between NF639R and NF639 cells (Figure 4A). After a series of bioinformatic analyses, 490 DEGs including 187 upregulated genes and 303 downregulated genes were identified according to the strict threshold of q<0.05 and |log2FC| >1 (https://cdn.amegroups.cn/static/public/atm-20-4667-1.pdf, Figure 4B). Hierarchical clustering of DEGs was visualized in Figure 4C. After the PPIs of DEGs were predicted with the STRING database, the PPI network was visualized (Figure S1).
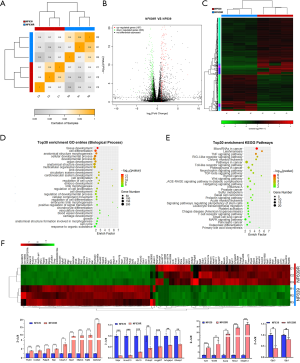
To further understand the DEGs associated with properties of NF639R cells, GO enrichment analysis was performed with 490 DEGs by clusterProfiler. A total of 1,772 significant GO terms (P<0.05) (https://cdn.amegroups.cn/static/public/atm-20-4667-2.pdf) were enriched, including 1,385 biological process terms, 176 cellular component terms and 211 molecular function terms. In TOP30 GO biological processes (Figure 4D), GO categories with highly significant p values including regulation of cell differentiation, regulation of cell proliferation, cellular component movement and regulation of cell cycle were consistent with the accelerated proliferation and altered cell morphology in NF639R cells. In keeping with the hypermigration phenotype of NF639R, migration-related terms including cell migration, cell motility, cell adhesion and mesenchymal cell development were also observed in TOP150 GO biological processes. Moreover, the apoptosis-related terms (cell death, positive and negative regulation of programmed cell death), the tumor stem cell-related terms (positive regulation of stem cell differentiation and positive regulation of stem cell proliferation) and energy metabolism-related terms (regulation of metabolic process, response to glucose and positive regulation of glucose transport) also were observed in 1,772 significant GO terms (https://cdn.amegroups.cn/static/public/atm-20-4667-2.pdf). In addition, there were some significant GO terms, such as cell surface receptor signaling pathway, MAPK cascade, Wnt signaling pathway, ERK1 and ERK2 cascade, ERBB signaling pathway, regulation of protein metabolic process, inflammatory response and Notch signaling pathway (https://cdn.amegroups.cn/static/public/atm-20-4667-2.pdf), which deserve more attention and further studies.
Pathway enrichment analysis of DEGs could provide further insights into the function of genes and their interactions. We performed KEGG pathway enrichment analysis for DEGs and found 221 pathway terms including 49 with significance (P<0.05) (https://cdn.amegroups.cn/static/public/atm-20-4667-3.pdf). In the top 30 pathways (Figure 4E) with the greatest enrichment, there are some pathways involved in proliferation, migration, immunity and tumor stem cell, including retinoic acid-inducible gene I (RIG-I)-like receptor signaling pathway, MicroRNAs in cancer, TNF signaling pathway, arginine biosynthesis, hedgehog signaling pathway, glycosaminoglycan degradation, transforming growth factor-β (TGF-β) signaling pathway, beta-alanine metabolism, Toll-like receptor signaling pathway, alanine, aspartate and glutamate metabolism, Wnt signaling pathway and nucleotide oligomerization domain (NOD)-like receptor signaling pathway. It is worth noting that several amino acid metabolism pathways have also been changed in the radioresistant NF639R cells. The sixteen genes were randomly selected in the top 100 differently expressed genes (Figure 4F) and then verified with qPCR. Consistent with RNA-seq data, higher expression of Ttyh3, Col3a1, Fabp5, Slpi, Ube2v1, Nqo1, Nfatc4, Tgfbi and Aldh3a1 (Figure 4G), but lower expression of Sdpr, Hoxd13, Klk10, Ctdsp2, Angptl7, Arhgap24 and Efemp1 were detected in NF639R cells (Figure 4H) compared to NF639 cells. These results indicated that the RNA-seq results were reliable. Considering that EMT-like (slender cells and the increased migration) and CSC-like (low basal ROS) phenotypes were observed in NF639R cells, we detected the differently expressed genes associated with EMT (Aldh3a1, Sox9, Mapk12, Wnt6 and Tcf7) and CSC (Msx2, Gja1 and Gjb3) by using qPCR (29-36). In previous studies, it has been confirmed that Aldh3a1, Sox9, Mapk12, Wnt6 and Tcf7 could promote CSC phenotypes and Msx2 could promote EMT phenotypes. Gja1 and Gjb3 have also been reported to inhibit EMT phenotypes. Our results showed that the NF639R cells exhibited the increased expression of Aldh3a1, Sox9, Mapk12, Wnt6, Tcf7 and Msx2 (Figure 4I), but the decreased expression of Gja1 and Gjb3 (Figure 4J) compared with the NF639 cells, suggesting that EMT-like and CSC-like characteristics may be acquired in the NF639R cells.
The enhance of tumor malignancy after two radiation schedules
In our study, two radiation schedules (total 40 Gy/schedule) were performed with an interval of 4 weeks. We evaluated basal level of ROS and the migration ability in the cells after one radiation schedule (NF639R40 Gy) and the cells after two radiation schedule (NF639R). Compared with NF639R40 Gy cells, NF639R cells displayed enhanced migration ability (Figure 5A,B) and lower basal level of ROS (Figure 5C,D), suggesting that two schedules of radiation (40 Gy/schedule) render NF639R cells more aggressive, Furthermore, we detected the expression of some genes including a positive prognostic indicator (Hoxd13), two inducers of radioresistance (Nqo1 and Aldh3a1) (37,38), one inducer of cell migration (Tgfbi) and two suppressors of cell migration (Sdpr and Klk10) using qPCR in NF639R40 Gy cells and NF639R cells. The results showed that the expression of Aldh3a1, Tgfbi and Nqo1 were increased, but expression of Sdpr, Klk10 and Hoxd13 were decreased in both NF639R40 Gy and NF639R cells. Compared with NF639R40 Gy cells, higher expression of Aldh3a1, Tgfbi and Nqo1 but lower expression of Sdpr and Hoxd13 were observed in NF639R cells (Figure 5E,F), further confirming the increase of malignancy after two radiation schedules.
Discussion
In the present study, radioresistant cells were obtained via irradiating the parent cell lines with repeated daily dose consistent with clinical routine radiotherapy (39). Although high dose-fraction irradiation (10 Gy) was also used to induce the radioresistance in previous studies, some researchers suggested high-dose irradiation favors the decrease in radioresistance instead of induction of radioresistance because high-dose irradiation may overwhelm the radioresistance mechanisms (40). The majority of literature still confirmed that 2 Gy or lower dose fractions are appropriate for selecting radioresistant cells (41,42). In a previous study, the acquired radioresistant cells established by fractionated radiation also could reverted to wild-type after sub-cultured for several passages (10–20 passages) in the absence of radiation (10). However, it was interesting to note that the radioresistant phenotype in NF639R cells was still maintained after 30 passages. In fact, some researchers observed that 40 Gy irradiation is enough to induce radioresistance in some breast cancer cells such as MCF7 and MDA-MB-231 (43,44). However, radioresistant cells established by 40 Gy or less total dose radiation may be instable due to transient genetic changes. In contrast, two discontinuous schedules (40 Gy in each schedule) in our study may induce the permanent genetic changes, similar to the observation that short-term drug treatment leads to reversible drug resistance, while long-term drug treatment leads to permanent drug resistance (45). Furthermore, NF639R cells also exhibited lower basal level of ROS and enhanced migration ability compared with NF639R40 Gy, confirming two schedules of radiation (40 Gy/schedule) render NF639R cells more aggressive. This would have potential clinical implications since the restart of radiotherapy with short-term recurrence after radiotherapy may lead to the increased radioresistance and promotion of tumor progress. Certainly, further study is needed to draw a conclusion. In addition, mouse NF639R cells will also be an ideal research tools in vivo to study the interaction between radioresistant and immunologic microenvironment.
After irradiation of 80 Gy, the NF639R cells not only acquired radioresistance but also some other properties distinct from the parental cell line NF639. Results from other groups confirmed that migration, invasion, proliferation and anti-apoptotic ability is changed accompanied by the acquired radioresistance (10,12,15). Different from the previous studies showing reduced proliferation ability of radioresistant cells (46,47), we found significant increase in proliferation of resistant cells compared to their parental cells. Recently, Tahmasebi-Birgani et al. also observed that fractionated irradiation induces the radioresistance, along with the inhibition or enhancement of the proliferative ability, depending on the cellular context (48). Moreover, our results are also consistent with the reported accelerated repopulation after clinical radiotherapy (49). In our study, radioresistant cells also exhibited increased migration and anti-apoptotic ability, consistent with previous reports (50). Interestingly, the morphology changes observed in NF639R cells, including reduction and elongation of cell bodies, are similar to the cells undergoing epithelial-mesenchymal transition (EMT). Previous studies have demonstrated that radiation could induce EMT, which enhance cell migration in various cancer cell lines (48,51). In addition, the basal level of ROS in NF639R cells was lower than that of NF639 cells. Given the lower level of ROS in cancer stem cells (CSCs) than their non-tumorigenic progeny, we proposed that radiation induced generation of CSCs in the transformation from NF639 to NF639R and the increased CSCs could enhance the anti-oxidative ability due to increased expression of ROS antioxidant enzymes, resulting in radioresistance (52). Moreover, different expression of several genes associated with EMT and CSC including Aldh3a1, Sox9, Mapk12, Wnt6, Tcf7, Msx2, Gja1 and Gjb3 between NF639 and NF639R cells also supported our speculation of acquired EMT-like and of CSC-like characteristics in NF639R cells.
In response to increased activities of migration, proliferation and radioresistance, more ATP, a main source of energy, should be needed. In fact, we found more ATP production in NF639R cells compared to NF639 cells. Furthermore, our data also showed that the two main pathways for ATP generation, mitochondrial oxidative phosphorylation and aerobic glycolysis, were both enhanced in NF639R cells. As the most efficient process of ATP generation, mitochondrial oxidative phosphorylation could meet the increased energy requirements for survival of radioresistant cells from irradiation (53). A previous report also showed that chemoresistant breast cancer cells also exhibit the elevated mitochondrial OXPHOS capacity (54), perhaps suggesting a common mechanism of resistance. Additionally, NF639R cells exhibited the increased spare respiratory capacity, which was confirmed to have a strong physiologic correlation with death-resistance (55,56). Interestingly, there is accumulated evidence to suggest that glycolytic metabolism is involved in the acquisition of radioresistance (12,56). Since ECAR is an indicator of lactate production through glycolysis, increased ECAR also indicates elevated production of lactate in NF639R cells (57). Groussard et al. reported that lactate, a main product of glycolytic metabolism, could effectively scavenge ROS and is positively correlated with radioresistance (58,59). Thus, increased lactate may be a reason for the generation of acquired radioresistance after the fractionated radiation. Moreover, enhancement of NF639R cell motility might also be due to the elevated production of lactate, which was found to induce the migration of cancer cells (60). Taken together, energy metabolism reprogramming during the evolution of acquired radioresistance not only supplies the ATP to meet the energy requirements of proliferation, migration and resistance, but also produces many metabolites to promote tumor progression.
With the bioinformation analysis of RNA-seq results, we found that irradiation affected some important biological processes, such as cell proliferation, regulation of cell cycle, cell migration, cell motility, response to oxidative stress, morphogenesis of a branching structure, epithelial to mesenchymal transition, phosphate-containing compound metabolic process, superoxide metabolic process, stem cell maintenance and apoptosis etc. These dysregulated biological processes are associated with radioresistance and other phenotypic changes. Moreover, the signaling cascades such as the TGF-β, Wnt, Hedgehog, vascular endothelial growth factor (VEGF), RIG-I-like receptor, Toll-like receptor and NOD-like receptor pathways are significantly altered in NF639R cells. Previous studies have confirmed that these dysregulated pathways are closely related to phenotypic changes in radioresistant cells. TGF-β and Wnt-β-catenin pathways play the important role in activating EMT (61,62). Moreover, activating TGF-β signaling could increase malignancy of Neu-induced mammary tumors and Wnt/β-catenin signaling was reported to mediate the radioresistance (63-65). Accumulating evidence demonstrates that Hedgehog (HH) and Wnt pathways are involved in regulating self-renewal of CSCs which are resistant to chemotherapy and/or radiotherapy (66). Activation of VEGF pathway contributes to the proliferation and radioresistance of breast cancer (67,68). RIG-I-like receptor, Toll-like receptor and NOD-like receptor are belonging to pattern recognition receptors (PRRs) which could be activated by damage-associated molecular patterns (DAMPs), such as HMGB1, ATP, DNA and RNA, released from damaged or dying cells (69). The activation of PRR-induced signaling by DAMPs is known to promote tumor proliferation, migration and neo-vascularization (70). In addition, PRRs pathways also paly the key role in regulating the immunosuppressive tumor microenvironment (71,72). In our study, PRRs pathways such as RIG-I-like receptor, Toll-like receptor and NOD-like receptor in NF639R cells are dysregulated, indicating the acquired radioresistant cells may also acquire immune tolerance, which helps in evading the antitumor immunity. Another interesting observation in our study is that MicroRNAs in cancer are the most enrichment KEGG pathway, indicating dysregulation of miRNAs in NF639R cells. Thus, miRNA may play an essential role in the radioresistance and progression of breast cancer, agreeing with previous studies showing that miRNAs are involved in various biological processes of cancer, such as proliferation, migration, apoptosis, energy metabolism, radiotherapy resistance and immune response (73,74). Certainly, the correlation between these pathways and phenotypic changes needs to be further verified. Overall, the acquired radioresistant NF639R cells exhibited different mRNA expression profiles from those of their parental cells. This provided support for our in-depth interpretation of radioresistance.
Conclusions
In summary, we successfully established a cell model of acquired radioresistance with a new method and revealed some phenotypic changes including smaller and longer cell body, enhanced activity of proliferation and migration, increased anti-apoptosis activity, reduced basal ROS and upregulated glycolysis ability and mitochondrial function. Moreover, 490 DEGs including 187 upregulated genes and 303 downregulated genes were identified and multiple signal pathways were enriched by RNA-seq data analysis, providing evidence on the molecular level for these phenotypic changes. Some potential signal pathways also provided a basis for further research. Our study provided insights into radioresistance and clinical suggestion that treatment for acquired-radioresistant cancers should not only focus on the radioresistance but also comprehensively consider changes in other properties which promote cancer progression.
Acknowledgments
Funding: This research was supported by the Chinese National Natural Science Foundation (Nos. 81703168, U1632145 and 81227902), Innovative Program of Development Foundation of Hefei Center for Physical Science and Technology (No. 2019HSC-CIP010), the Natural Science Fund of Anhui Province (No. 1608085QH181), and project funded by the Priority Academic Program Development of Jiangsu Higher Education Institutions (PAPD) and Jiangsu Provincial Key Laboratory of Radiation Medicine and Protection. This research was also supported by the research grant IRF/0024 from the State Key Laboratory in Marine Pollution, City University of Hong Kong.
Footnote
Reporting Checklist: The authors have completed the MDAR reporting checklist. Available at http://dx.doi.org/10.21037/atm-20-4667
Data Sharing Statement: Available at http://dx.doi.org/10.21037/atm-20-4667
Peer Review File: Available at http://dx.doi.org/10.21037/atm-20-4667
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/atm-20-4667). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. This was an in vitro study using cell lines. There was no need for ethical approval or patient consent.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Bray F, Ferlay J, Soerjomataram I, et al. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin 2018;68:394-424. [Crossref] [PubMed]
- Maughan KL, Lutterbie MA, Ham PS. Treatment of breast cancer. Am Fam Physician 2010;81:1339-46. [PubMed]
- Liang K, Jin W, Knuefermann C, et al. Targeting the phosphatidylinositol 3-kinase/Akt pathway for enhancing breast cancer cells to radiotherapy. Mol Cancer Ther 2003;2:353-60. [PubMed]
- Duru N, Candas D, Jiang G, et al. Breast cancer adaptive resistance: HER2 and cancer stem cell repopulation in a heterogeneous tumor society. J Cancer Res Clin Oncol 2014;140:1-14. [Crossref] [PubMed]
- Candas-Green D, Xie B, Huang J, et al. Dual blockade of CD47 and HER2 eliminates radioresistant breast cancer cells. Nat Commun 2020;11:4591. [Crossref] [PubMed]
- Kim SH, Lee WH, Kim SW, et al. EphA3 maintains radioresistance in head and neck cancers through epithelial mesenchymal transition. Cell Signal 2018;47:122-30. [Crossref] [PubMed]
- Xie P, Li X, Tan X, et al. Sequential serum let-7 is a novel biomarker to predict accelerated reproliferation during fractional radiotherapy in lung cancer. Clin Lung Cancer 2016;17:e95-101. [Crossref] [PubMed]
- Dong Q, Sharma S, Liu H, et al. HDAC inhibitors reverse acquired radio resistance of KYSE-150R esophageal carcinoma cells by modulating Bmi-1 expression. Toxicol Lett 2014;224:121-9. [Crossref] [PubMed]
- Gray M, Turnbull AK, Ward C, et al. Development and characterisation of acquired radioresistant breast cancer cell lines. Radiat Oncol 2019;14:64. [Crossref] [PubMed]
- Russell J, Wheldon TE, Stanton P. A Radioresistant Variant Derived from a Human Neuroblastoma Cell Line Is Less Prone to Radiation-induced Apoptosis. Cancer Res 1995;55:4915. [PubMed]
- Su WH, Chuang PC, Huang EY, et al. Radiation-Induced Increase in Cell Migration and Metastatic Potential of Cervical Cancer Cells Operates Via the K-Ras Pathway. Am J Pathol 2012;180:862-71. [Crossref] [PubMed]
- Tang L, Wei F, Wu Y, et al. Role of metabolism in cancer cell radioresistance and radiosensitization methods. J Exp Clin Cancer Res 2018;37:87. [Crossref] [PubMed]
- Lehuédé C, Dupuy F, Rabinovitch R, et al. Metabolic plasticity as a determinant of tumor growth and metastasis. Cancer Res 2016;76:5201-8. [Crossref] [PubMed]
- Jiang H, Xu Y, Li W, et al. Low-dose radiation does not induce proliferation in tumor cells in vitro and in vivo. Radiat Res 2008;170:477-87. [Crossref] [PubMed]
- Ma H, Rao L, Wang HL, et al. Transcriptome analysis of glioma cells for the dynamic response to gamma-irradiation and dual regulation of apoptosis genes: a new insight into radiotherapy for glioblastomas. Cell Death Dis 2013;4:e895 [Crossref] [PubMed]
- Young A, Berry R, Holloway AF, et al. RNA-seq profiling of a radiation resistant and radiation sensitive prostate cancer cell line highlights opposing regulation of DNA repair and targets for radiosensitization. Bmc Cancer 2014;14:808. [Crossref] [PubMed]
- Bolger AM, Lohse M, Usadel B. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 2014;30:2114-20. [Crossref] [PubMed]
- Kim D, Langmead B, Salzberg SL. HISAT: a fast spliced aligner with low memory requirements. Nat Methods 2015;12:357-60. [Crossref] [PubMed]
- Pertea M, Pertea GM, Antonescu CM, et al. StringTie enables improved reconstruction of a transcriptome from RNA-seq reads. Nat Biotechnol 2015;33:290-5. [Crossref] [PubMed]
- Robinson MD, Oshlack A. A scaling normalization method for differential expression analysis of RNA-seq data. Genome Biol 2010;11:R25. [Crossref] [PubMed]
- Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol 2014;15:550. [Crossref] [PubMed]
- Ashburner M, Ball CA, Blake JA, et al. Gene ontology: tool for the unification of biology. The Gene Ontology Consortium. Nat Genet 2000;25:25-9. [Crossref] [PubMed]
- Odero-Marah V, Hawsawi O, Henderson V, et al. Epithelial-Mesenchymal Transition (EMT) and Prostate Cancer. Adv Exp Med Biol 2018;1095:101-10. [Crossref] [PubMed]
- Creighton CJ, Rosen CJM. Epithelial-Mesenchymal Transition (EMT) in Tumor-Initiating Cells and Its Clinical Implications in Breast Cancer. J Mammary Gland Biol Neoplasia 2010;15:253-60. [Crossref] [PubMed]
- Pang MF, Georgoudaki AM, Lambut L, et al. TGF-beta1-induced EMT promotes targeted migration of breast cancer cells through the lymphatic system by the activation of CCR7/CCL21-mediated chemotaxis. Oncogene 2016;35:748-60. [Crossref] [PubMed]
- Ruth AC, Roninson IB. Effects of the multidrug transporter P-glycoprotein on cellular responses to ionizing radiation. Cancer Res 2000;60:2576-8. [PubMed]
- Amoedo ND, El-Bacha T, Rodrigues MF, et al. Cell cycle and energy metabolism in tumor cells: strategies for drug therapy. Recent Pat Anticancer Drug Discov 2011;6:15-25. [Crossref] [PubMed]
- Moreno-Sánchez R, Rodriguez-Enriquez S, Marin-Hernandez A, et al. Energy metabolism in tumor cells. Febs J 2007;274:1393-418. [Crossref] [PubMed]
- Terzuoli E, Bellan C, Aversa S, et al. ALDH3A1 Overexpression in Melanoma and Lung Tumors Drives Cancer Stem Cell Expansion, Impairing Immune Surveillance through Enhanced PD-L1 Output. Cancers (Basel) 2019;11:1963. [Crossref] [PubMed]
- Kadaja M, Keyes BE, Lin M, et al. SOX9: a stem cell transcriptional regulator of secreted niche signaling factors. Genes Dev 2014;28:328-41. [Crossref] [PubMed]
- Qi X, Yin N, Ma S, et al. P38gamma MAPK Is a Therapeutic Target for Triple-Negative Breast Cancer by Stimulation of Cancer Stem-Like Cell Expansion. Stem Cells 2015;33:2738-7. [Crossref] [PubMed]
- Gonçalves CS, Castro JV, Pojo M, et al. WNT6 is a novel oncogenic prognostic biomarker in human glioblastoma. Theranostics 2018;8:4805-23. [Crossref] [PubMed]
- Wang Y, He L, Du Y, et al. The long noncoding RNA lncTCF7 promotes self-renewal of human liver cancer stem cells through activation of Wnt signaling Cell Stem Cell 2015;16:413-25. [Crossref] [PubMed]
- di Bari MG, Ginsburg E, Plant J, et al. Msx2 induces epithelial-mesenchymal transition in mouse mammary epithelial cells through upregulation of Cripto-1. J Cell Physiol 2009;219:659-66. [Crossref] [PubMed]
- Li Y, Wang J, Asahina K. Mesothelial cells give rise to hepatic stellate cells and myofibroblasts via mesothelial-mesenchymal transition in liver injury. Proc Natl Acad Sci U S A 2013;110:2324-9. [Crossref] [PubMed]
- Oberstein A, Shenk T. Cellular responses to human cytomegalovirus infection: Induction of a mesenchymal-to-epithelial transition (MET) phenotype Proc Natl Acad Sci U S A 2017;114:E8244-53. [Crossref] [PubMed]
- Huang W, Shi G, Yong Z, et al. Downregulation of RKIP promotes radioresistance of nasopharyngeal carcinoma by activating NRF2/NQO1 axis via downregulating miR-450b-5p. Cell Death Dis 2020;11:504. [Crossref] [PubMed]
- Voulgaridou GP, Kiziridou M, Mantso T, et al. Aldehyde dehydrogenase 3A1 promotes multi-modality resistance and alters gene expression profile in human breast adenocarcinoma MCF-7 cells. Int J Biochem Cell Biol 2016;77:120-8. [Crossref] [PubMed]
- Bartelink H, Horiot JC, Poortmans PM, et al. Impact of a higher radiation dose on local control and survival in breast-conserving therapy of early breast cancer: 10-year results of the randomized boost versus no boost EORTC 22881-10882 trial. J Clin Oncol 2007;25:3259-65. [Crossref] [PubMed]
- Wergin MC, Ballmer-Hofer K, Roos M, et al. Preliminary study of plasma vascular endothelial growth factor (VEGF) during low- and high-dose radiation therapy of dogs with spontaneous tumors. Vet Radiol Ultrasound 2004;45:247-54. [Crossref] [PubMed]
- Niedbala M, Alsbeih G, Ng CE, et al. Equivalence of pulsed-dose-rate to low-dose-rate irradiation in tumor and normal cell lines. Radiat Res 2001;155:297-303. [Crossref] [PubMed]
- Kuwahara Y, Mori M, Kitahara S, et al. Targeting of tumor endothelial cells combining 2 Gy/day of X-ray with Everolimus is the effective modality for overcoming clinically relevant radioresistant tumors. Cancer Med 2014;3:310-21. [Crossref] [PubMed]
- Perez-Añorve IX, Gonzalez-De la Rosa CH, Soto-Reyes E, et al. New insights into radioresistance in breast cancer identify a dual function of miR-122 as a tumor suppressor and oncomiR Mol Oncol 2019;13:1249-67. [Crossref] [PubMed]
- Lin LC, Lee HT, Chien PJ, et al. ChienNAD(P)H:quinone oxidoreductase 1 determines radiosensitivity of triple negative breast cancer cells and is controlled by long non-coding RNA NEAT1. Int J Med Sci 2020;17:2214-24. [Crossref] [PubMed]
- Russo M, Crisafulli G, Sogari A, et al. Adaptive mutability of colorectal cancers in response to targeted therapies. Science 2019;366:1473-80. [Crossref] [PubMed]
- Shi D, Shi G, Gang H. Chemosensitivity of radioresistant cells in the multicellular spheroids of A549 lung adenocarcinoma. J Exp Clin Cancer Res 2009;28:72. [Crossref] [PubMed]
- Saito Y, Abiko R, Kishida A, et al. Loss of EGF-dependent cell proliferation ability on radioresistant cell HepG2-8960-R. Cell Biochemistry Function 2015;33:73-9. [Crossref] [PubMed]
- Tahmasebi-Birgani MJ, Teimoori A, Ghadiri A, et al. Fractionated radiotherapy might induce epithelial-mesenchymal transition and radioresistance in a cellular context manner. J Cell Biochem 2018; [Epub ahead of print]. [PubMed]
- Yom SS. Accelerated Repopulation as a Cause of Radiation Treatment Failure in Non–Small Cell Lung Cancer: Review of Current Data and Future Clinical Strategies. Semin Radiat Oncol 2015;25:93-9. [Crossref] [PubMed]
- Mao G, Yao Y, Kong Z. Long term exposure to gammarays induces radioresistance and enhances the migration ability of bladder cancer cells. Mol Med Rep 2018;18:5834-40. [PubMed]
- Tsukamoto H, Shibata K, Kajiyama H, et al. Irradiation-induced epithelial-mesenchymal transition (EMT) related to invasive potential in endometrial carcinoma cells. Gynecol Oncol 2007;107:500-4. [Crossref] [PubMed]
- Diehn M, Cho RW, Lobo NA, et al. Association of reactive oxygen species levels and radioresistance in cancer stem cells. Nature 2009;458:780-3. [Crossref] [PubMed]
- Lynam-Lennon N, Maher SG, Maguire A, et al. Altered mitochondrial function and energy metabolism is associated with a radioresistant phenotype in oesophageal adenocarcinoma. PLoS One 2014;9:e100738 [Crossref] [PubMed]
- Lee KM, Giltnane JM, Balko JM, et al. MYC and MCL1 cooperatively promote chemotherapy-resistant breast cancer stem cells via regulation of mitochondrial oxidative phosphorylation. Cell Metabolism 2017;26:633-47.e7. [Crossref] [PubMed]
- Nickens KP, Wikstrom JD, Shirihai OS, et al. A bioenergetic profile of non-transformed fibroblasts uncovers a link between death-resistance and enhanced spare respiratory capacity. Mitochondrion 2013;13:662-7. [Crossref] [PubMed]
- Lee KM, Giltnane JM, Balko JM, et al. MYC and MCL1 Cooperatively Promote Chemotherapy-Resistant Breast Cancer Stem Cells via Regulation of Mitochondrial Oxidative Phosphorylation. Cell Metab 2017;26:633-47.e7. [Crossref] [PubMed]
- Yizhak K, Devedec LS, Rogkoti VM, et al. A computational study of the Warburg effect identifies metabolic targets inhibiting cancer migration. Mol Syst Biol 2014;10:744. [Crossref] [PubMed]
- Groussard C, Morel I, Chevanne M, et al. Free radical scavenging and antioxidant effects of lactate ion: an in vitro study. J Appl Physiol (1985) 2000;89:169-75. [Crossref] [PubMed]
- Sattler UG, Meyer SS, Quennet V, et al. Glycolytic metabolism and tumour response to fractionated irradiation. Radiother Oncol 2010;94:102-9. [Crossref] [PubMed]
- Goetze K, Walenta S, Ksiazkiewicz M, et al. Lactate enhances motility of tumor cells and inhibits monocyte migration and cytokine release. Int J Oncol 2011;39:453-63. [Crossref] [PubMed]
- Yook JI, Li XY, Ota I, et al. Wnt-dependent regulation of the E-cadherin repressor snail. J Biol Chem 2005;280:11740-8. [Crossref] [PubMed]
- Thiery JP, Acloque H, Huang RY, et al. Epithelial-mesenchymal transitions in development and disease. Cell 2009;139:871-90. [Crossref] [PubMed]
- Siegel PM, Shu W, Cardiff RD, et al. Transforming growth factor beta signaling impairs Neu-induced mammary tumorigenesis while promoting pulmonary metastasis. Proc Natl Acad Sci U S A 2003;100:8430-5. [Crossref] [PubMed]
- Muraoka RS, Koh Y, Roebuck LR, et al. Increased malignancy of Neu-induced mammary tumors overexpressing active transforming growth factor beta1. Mol Cell Biol 2003;23:8691-703. [Crossref] [PubMed]
- Jun S, Jung YS, Suh HN, et al. LIG4 mediates Wnt signalling-induced radioresistance. Nat Commun 2016;7:10994. [Crossref] [PubMed]
- Takebe N, Miele L, Harris PJ, et al. Targeting Notch, Hedgehog and Wnt pathways in cancer stem cells: clinical update. Nat Rev Clin Oncol 2015;12:445-64. [Crossref] [PubMed]
- Liang Y, Brekken RA, Hyder SM. Vascular endothelial growth factor induces proliferation of breast cancer cells and inhibits the anti-proliferative activity of anti-hormones. Endocr-relat Cancer 2006;13:905-19. [Crossref] [PubMed]
- Jameel JK, Rao VS, Cawkwell L, et al. Radioresistance in carcinoma of the breast. Breast 2004;13:452-60. [Crossref] [PubMed]
- Takeuchi O, Akira S. Pattern recognition receptors and inflammation. Cell 2010;140:805-20. [Crossref] [PubMed]
- Pandey S, Singh S, Anang V, et al. Pattern Recognition Receptors in Cancer Progression and Metastasis. Cancer growth and metastasis 2015;8:25-34. [Crossref] [PubMed]
- Huang B, Zhao J, Unkeless JC, et al. TLR signaling by tumor and immune cells: a double-edged sword. Oncogene 2008;27:218-24. [Crossref] [PubMed]
- Kent A, Blander JM. Nod-like receptors: key molecular switches in the conundrum of cancer Front Immunol 2014;5:185. [Crossref] [PubMed]
- Kasinski AL, Slack FJ. MicroRNAs en route to the clinic: progress in validating and targeting microRNAs for cancer therapy. Nat Rev Cancer 2011;11:849-64. [Crossref] [PubMed]
- Stahlhut C, Slack FJ. MicroRNAs and the cancer phenotype: profiling, signatures and clinical implications. Genome Med 2013;5:111. [Crossref] [PubMed]


