
Decision-support for treatment with 177Lu-PSMA: machine learning predicts response with high accuracy based on PSMA-PET/CT and clinical parameters
Introduction
Machine learning (ML) has gained essential importance in therapy planning and patient selection for certain treatments recently (1,2). The role of radiomics features for patients screening for certain therapies has been under investigation as well (3,4). Prostate cancer (PC) is one of the most common malignancies in men worldwide. If spread beyond the prostate it can lead to a significant mortality (5). Although treatment of advanced PC has improved significantly in recent years, more than 250.000 fatalities are caused by PC per year.
Radioligand therapy targeting the prostate specific membrane antigen (PSMA) gained great importance in the last years and a clear benefit for patients who do not respond to any other available treatment was shown (6). In these patients, pretherapeutic imaging is performed using PSMA analogues labeled mainly with positron emitters Gallium-68 or Fluorine-18 as theranostics approach (7). However, about 10% to 32% of the patients show progressive disease during treatment with 177Lu-PSMA (8). Therefore strategies to differentiate patients who may benefit from therapy from patients who may not benefit are of great importance. Pretherapeutic PSMA positron emission tomography/computed tomography (PET/CT) scans as well as different clinical parameters like initial Gleason score or serum levels of prostate-specific antigen (PSA) have been investigated for this purpose without clear findings (9).
In the past years, radiomics features such as textural parameters have been gaining importance in the analysis of PET/CT data. The significance of textural features analysis in diagnosis and therapy response prediction using PSMA PET/CT scan has been shown as well (3,4,10,11). Our previous findings showed that machine learning (ML) can facilitate detection of pathological uptake in 68Ga-PSMA PET/CT scans with nuclear medicine (NM) expert accuracy (12). Also, for the prediction of treatment response to 177Lu-PSMA therapy in PC patients first results have been published by Khurshid et al. showing that there is a significant correlation between the mean homogeneity and entropy of PET scans as patient-based textural features on the one hand, and the PSA level difference as a therapy response indicator on the other hand (13). While many studies aimed at analyzing the correlation between each clinical or textural parameter and tumor malignancy or therapy response, respectively (11,13), many ML methods are available that outperform independent feature analyses by combining several parameters to perform similar tasks (1,2,12).
In the presented study, we propose a method for treatment response prediction in patients undergoing 177Lu-PSMA therapy. In the first step, the baseline scans are manually annotated to detect the pathological uptakes of the whole cohort resulting in 2070 hotspots. Then, the radiomics features of all the annotated hotspots are calculated individually. Afterwards, linear regression is performed to identify best correlating features and clinical parameters with changes in PSA-level as surrogate marker for treatment response and survival (14). Finally, ML methods are applied on different combinations of the features and clinical parameters to predict response to 177Lu-PSMA treatment. We aim at quantifying the classification accuracy of different ML classifiers for the prediction task.
We present the following article in accordance with the MDAR checklist (available at http://dx.doi.org/10.21037/atm-20-6446).
Methods
Patients and Volume of interest (VoI) definition and annotation
A total of 83 male patients with advanced PC scheduled for treatment with 177Lu-PSMA were included in this retrospective analysis. The patients’ age range varied from 48 to 87 years and their Gleason score ranged from 6 to 10. The serum PSA level range of the cohort was between 4.7 and 5,910 ng/mL. All patients underwent pre-therapeutic 68Ga-PSMA PET/CT scans 5 to 21 days before the beginning of the treatment. The scans were carried out between November 2014 and August 2019. About 40 to 80 minutes after intravenous injection of 98 to 159 MBq in-house produced 68GA-HBED-CC PSMA, a Biograph 2 PET/CT system (Siemens Medical Solutions, Erlangen, Germany) was used to take the low-dose CT (16 mAs, 130 kV) from the base of skull to mid thigh. Then, the PET scan acquired over the same area with 3 or 4 minutes per bed position depending on the body weight of the patient. The PET data were reconstructed in 128 by 128 matrices with 5 mm slices thickness. The CT data were reconstructed in 512 to 512 matrices with 5 mm slice thickness. As implemented by the manufacturer, an attenuation-weighted ordered subsets expectation maximization algorithm was utilized for attenuation and scatter corrections (8 iterations, 16 subsets), a 5 mm Gaussian post-reconstruction-filter was applied afterwards. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013). All patients gave written and informed consent to the imaging procedure and for anonymized evaluation and publication of their data. Due to the retrospective character of the data analysis an ethical statement was waived by the institutional ethical review board according to the professional regulations of the medical board of Nordrheinwestfalen, Germany.
For each scan, all the pathological hotspots have been identified and delineated by a trained nuclear medicine physician (NM) (board certified with 7 years’ experience in PET/CT analysis) using InterView Fusion software (Mediso Medical Imaging, Hungary, Version 3.08.005). The hotspots include the primary tumor if present as well as metastatic uptakes in any organs. Per hotspot, a total of 73 (37 PET-based + 36 CT-based) features were calculated (Table 1). The features include first and higher order statistics features (mean, max, kurtosis, etc.), shape based features (max diameter and volume), textural features (entropy, contrast, homogeneity, etc.), and volumetric zone and run length statistics (grey-level non-uniformity, short run emphasis, etc.).

Full table
In addition to the radiomics features, fourteen numerical clinical parameters have been taken into account for each individual patient. These clinical parameters include age, weight, height as well as therapeutic parameters such as Gleason score, ALP1 and base-line serum PSA level. For the detailed list of the clinical parameters, see Table 2.

Full table
According to previous findings (14) and as surrogate markers for treatment response, prostate specific antigen (PSA) serum values have been collected at the time point of the PET/CT examination and seven to eight weeks after the treatment. Changes in PSA levels (∆PSA) between these time-points have been used for further analyses. Based on the calculated ∆PSA values, out of the 83 patients, 59 and 24 patients have been classified as responders and non-responders respectively.
Statistical analysis
Linear regression
After accumulating the data from all the scans, radiomics features and clinical parameters of individual patients were combined to form feature vectors for further analyses. To achieve these, the values of the radiomics features of the individual pathological hotspots of each patient were averaged to calculate the mean values of the features. The clinical parameters of the individual patients were then merged with their corresponding radiomics features.
To correlate individual features and clinical parameters with ∆PSA, linear regression has been used for all the 73 features and 14 numerical clinical parameters. The ∆PSA is calculated by subtracting the PSA level at the post therapy scan from the corresponding PSA level at the pre-therapy scan. Therefore, a negative value of ∆PSA means the patient had responded to the 177Lu-PSMA therapy and vice versa.
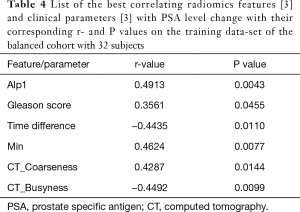
As the numbers of responders and non-responders (59 and 24 patients respectively) to the 177Lu-PSMA therapy in the original cohort did not match, for the linear regression task, a balanced subset of the cohort with 24 patients in each category of responders or non-responder was formed. The 24 responders have been randomly selected out of the whole 59 responders (the demographic and physiological distributions were maintained during the sub-sampling). As will be described in the classification and cross-validation (CV) sub-sections, each of the balanced and unbalanced cohorts were sub-divided into training and validation data-sets to assess the prediction performance for the classification task. Hence, the linear regression analyses have been conducted on training data-sets of balanced and unbalanced cohorts separately. As a result, best sets of radiomics features and clinical parameters which had strong correlations (P value <0.05) with ∆PSA were identified for both balanced and unbalanced groups. These best correlating features and parameters were used for the analyses of treatment response prediction in the further steps. This strategy of identifying the best correlating parameters by only considering training cohorts helps to avoid over-fitting (15).
Classification
As support vector machines (SVMs) and decision tree based methods are widely used for clinical treatment outcome prediction [e.g., prediction outcome of chemotherapy (16), prediction of optimal cancer drug therapies (17), and risk stratification in primary prostate cancer (18)], we have applied several classifiers from these groups for the therapy response prediction task. The five ML classifiers [linear, radial basis function (RBF), and polynomial kernel SVM (19), ExtraTrees (20), and RandomForest (21)] were used to investigate the relative importance of different groups of radiomics features and clinical parameters. The accuracy measures [area under the curve (AUC), sensitivity (SE), and specificity (SP)] are averaged to calculate the total precision for each of the tasks. Thus, for each pair of classifier and feature group, we calculate AUC, SE, and SP separately.
Cross-validation (CV)
It is essential to have separate data for hyperparameter tuning and for quantifying final accuracy to achieve generalizable results and to avoid over-fitting. To this end, two different CV steps are taken. In the first step, the whole data-set with 83 patients, including 59 responders and 24 non-responders, is taken into account. In the second step, a balanced subset of the cohort with 48 subjects (the same subset as used for the prior linear regression task) is used for CV. This strategy of having an extra CV step based on a balanced cohort helps to identify if the classifiers’ scores on the unbalanced cohort were realistic.
Unbalanced cohort
First, the whole cohort of 83 patients was randomly sub-divided into two subsets: (I) the training cohort with 56 subjects, and (II) the validation or hold-out set with 27 subjects. The demographics and clinical states of the cohorts were similar. The ratios of responders to non-responders in the training and validation sets were also comparable. To standardize and normalize the data, MinMaxScaler method (22) was used. Stratified-KFold CV with 3 folds applied to the training cohort for hyperparameter tuning.
In each CV step, a grid search has been performed to find the best set of parameters for each of the ML algorithms to predict the true labels for each category. For the grid search, several parameters with wide ranges of values (C=[1, 10, 100, 1000, 2-5, 2-3, ..., 215], gamma=[1e-3, 1e-4, 2-15, 2-13, 2-11..., 23], etc.) were used to fine-tune the ML classifiers.
After tuning the best set of hyperparameters for each ML method based on the accuracies achieved on the training cohort, the prediction performances of the ML classifiers were quantified, comparing with the ground truth labels from the hold-out cohort. Again, the relative importance of different radiomics features groups and clinical parameters were analyzed individually.
Balanced cohort
As the numbers of the responder and non-responder groups did not match, additional CV steps have been taken based on a balanced subset of the cohort with 48 patients (including 24 responders and 24 non-responders). This balanced cohort was separated into training and validation sets as well. This time, the training cohort consisted of 32 subjects and the validation or hold-out set consisted of 16 patients. Again, the responder to non-responder ratio was equal in both of the training and validation subsets. Similar to the first CV step (for the unbalanced cohort), stratified KFold with 3 folds have been applied on the training set to fine-tune the hyperparameters, including standardization of the feature values as well as grid search on each CV iteration. Afterwards, as the final validation step and for each classifier applied to each group of features or clinical parameters, prediction accuracies were calculated on the validation subset. Finally, the accuracy measures of each classifier on each feature group applied to the validation (hold-out) cohort will be reported as the achieved performance.
Permutation test
To assure that the results are significant, a permutation test is performed. The permutation test rejected the null hypothesis which stated that permuted distribution of ground truth labels could have resulted in similar prediction scores. Hence, a separate three-fold CV on the cohort with 32 patients from the second CV step is conducted. There were 80,000 total iterations with exactly similar groups of radiomics features and clinical parameters as well as ML classifiers as for the prior CV steps. In each CV step, the ground truth binary labels were permuted. All the AUCs equal to or higher than the threshold of 0.61 (the worst AUC achieved by our classifiers on the hold-out set) are counted. Then, to calculate the P value of the permutation test, the resulting number is divided by the total number of iterations [80,000]:
where p is the P value of the permutation test, n() is the number of the test scores over the given threshold (thr), AUCs are the calculated areas under the ROC curves for each classifier on each feature group at each iteration, and N is the total number of iterations (Eq. [1]).
Results
Linear regression-unbalanced cohort
Among all the 73 radiomics features and 14 numerical clinical parameters, the linear regression tests on the training set of the unbalanced cohort illustrated that 5 radiomics features from both PET (Min and Correlation) and CT (CT_Min, CT_Coarseness, and CT_Busyness) modalities (named best correlating features or Best-Radiomics from now on) have the best correlation scores with PSA level difference (P values <0.05) as the surrogate marker for therapy response. Figure 1A shows the regression diagrams of the 5 best correlating features with ∆PSA. Table 3 shows these 5 features and their corresponding r- and P values of the regression tests on the unbalanced group.
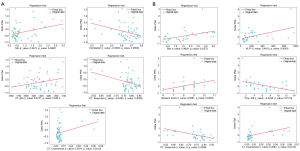
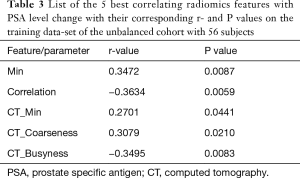
Full table
Linear regression-balanced cohort
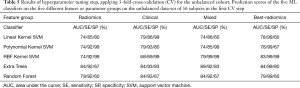
As for the unbalanced cohort, the linear regression analyses on the training set of the balanced cohort resulted in a group of 3 radiomics features (PET_Min, CT_Busyness, and CT_Coarseness) and 3 clinical parameters (Alp1, Time difference, and Gleason score). The results are shown in Figure 1B and Table 4. For further analyses, two different groups of best correlating parameters are created. First group (Best-Radiomics) includes only the best correlating radiomics features and the second group (Best-Mixed) includes features or parameters which had strong correlation with ∆PSA from both of the radiomics and clinical groups.
Full table
Classification-unbalanced cohort
As shown in Table 5, the SVM classifier with RBF kernel had the best performance (83% AUC, 99% SE, and 99% SP) on the best correlating radiomics features with ∆PSA (named Best-Radiomics group) in the first CV step on the unbalanced training cohort with 56 subjects. The relatively low values of specificity for some classifiers applied to all the radiomics features or the mixture of all the 73 radiomics features and 14 numerical clinical parameters (named the Mixed group) reflect the unbalanced characteristic of the cohort.
Full table
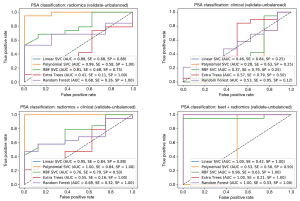
Based on the grid search results on the CV step, hyperparameters of each classifier have been tuned (Table 6). These tuned values for the parameters have been used in the validation step to calculate the prediction score of the classifiers as applied to the hold-out set. In the validation step, the cohort of 56 subjects was used as the training data-set and the cohort of 27 subjects was used as the test set. The results of this validation step is shown in Table 7 and Figure 2. Here, the clinical parameters group showed relatively weak scores, compared to the scores achieved by the other groups. The results reveal that the polynomial kernel SVM with parameters degree =3 and C=1 had the best performance as applied to the mixture of all radiomics and clinical values (99% AUC, 84% SE, and 99% SP). Also, the SVM classifier with linear and RBF kernels achieved reasonable scores (95% AUC, 84% SE, and 88% SP and 96% AUC, 63% SE, and 99% SP respectively) as applied to the Mixed and Best-Radiomics groups respectively.

Full table

Full table
Classification-balanced cohort
Similar to the analyses of the unbalanced cohort, another CV step followed by a validation step has been conducted on the balanced training and test cohorts including 32 and 16 subjects respectively. The results of the CV step are shown in Table 8. Here, as compared to the CV step for the unbalanced cohort, more consistent results are achieved. The highest scores (up to 99% AUC, 99% SE, and 99% SP) are achieved by almost all of the pairs of classifier-parameter groups. These extremely high scores are achieved by the grid search for the purpose of hyperparameter tuning and are not considered as final accuracies.
Full table
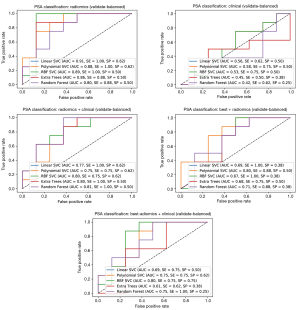
The results of the hyperparameter tuning for the balanced cohort are presented in Table 9 and the results of applying the classifiers with the tuned parameters to the validation cohort are shown in Table 10 and Figure 3. Here, except for the clinical parameters group which showed insufficient prediction accuracies, the linear, polynomial, and RBF kernel SVM classifiers showed the most consistent performances (91% AUC, 99% SE, and 62% SP for linear SVM on radiomics group, 88% AUC, 99% SE, and 62% SP for polynomial SVM on radiomics group, and 80% AUC, 75% SE, and 75% SP for RBF SVM on Best-Mixed group).
Full table

Full table
The final step was the permutation test which has resulted in a P value of 0.0043 that assures the significance of the results.
Discussion
We showed that parameters of PSMA PET (Min and correlation) have statistically significant correlations with the PSA level difference as a surrogate marker for therapy response prediction, which is in accordance with the findings by Khurshid et al. (13). Furthermore, it was shown that some of the features from the low-dose CT (CT_Min, CT-Busyness, and CT-Coarseness) as well as three clinical parameters (Alp1, Time Difference, and Gleason score as defined in Table 2) have strong correlations with PSA level difference. In addition, by applying ML classifiers with tuned hyperparameters, we showed that features from baseline 68Ga-PSMA scan can help to predict responders to 177Lu-PSMA therapy with reasonable certainty.
Due to the retrospective characteristic of the study and because of the fortunate fact that most of the PC patients examined at our 68Ga-PSMA PET/CT center are responders to the 177Lu-PSMA therapy, our original cohort was unbalanced with regard to response to the therapy. Thus, the whole cohort consisted of 59 responders and 24 non-responders. Although the unbalanced cohort achieved reasonable results in terms of prediction accuracies, similar analyzes were conducted on the balanced cohort to check if the accuracy scores could be maintained. However, as the size of the hold-out set for the balanced cohort (16 subjects) was relatively small, relatively low specificities were achieved in the corresponding validation step. This important observation urges for studies on bigger cohorts in the future.
Although the clinical parameters have shown insufficient prediction performances in the validation steps on balanced or unbalanced data-sets, however; the overall best accuracy scores (up to 80% AUC, 75% SE, and 75% SP) are achieved on the combination of best correlating radiomics features and clinical parameters with ∆PSA (Best-Mixed) by SVM classifier with RBF kernel (Table 10).
As the results suggest, ML methods have shown their potential for further, automated algorithms for treatment response prediction in prostate cancer patients based on 68Ga-PSMA PET/CT data and therefore for decision-support tools. To implement this goal, our next steps include automated segmentation of hotspots, which was beyond the scope of this first study. Although in the presented study, the additional value of including clinical parameters could not be shown, in our opinion this is still an important topic and should be part of further studies.
Drawbacks of the study are for sure, that as gold-standard visual image analysis was used instead of histopathology as a real gold-standard. However, biopsies of more than one or two hotspots are hardly possible in patients, so this is actually the best option for ground truth data acquisition. Although we had just 83 patients included in this first study, we analyzed 2070 pathological hotspots in total, so that we could show statistical significance in our results. However, larger studies need to be performed in the future to enhance the predictive performances of the algorithms. Also beyond the scope of this study was the analysis of how the results can be applied on 68Ga-PSMA PET scans with different protocols [such as PET/MRI (23)] or obtained with other PET scanners (24). This is an important topic in this field and needs to be investigated in further studies.
Conclusions
Machine learning based on pretherapeutic 68Ga-PSMA-PET/CT radiomics features has shown high potential to predict response to treatment with 177Lu-PSMA. The application of combination of best correlating radiomic features with PSA level change showed its superiority compared to clinical parameters for the treatment response prediction task.
Acknowledgments
Funding: None.
Footnote
Provenance and Peer Review: This article was commissioned by the Guest Editor (Dr. Steven P. Rowe) for the series “Artificial Intelligence in Molecular Imaging” published in Annals of Translational Medicine. The article has undergone external peer review.
Reporting Checklist: The authors have completed the MDAR checklist. Available at http://dx.doi.org/10.21037/atm-20-6446
Data Sharing Statement: Available at http://dx.doi.org/10.21037/atm-20-6446
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/atm-20-6446). The series “Artificial Intelligence in Molecular Imaging” was commissioned by the editorial office without any funding or sponsorship. Dr. Essler reports personal fees from Bayer Healthcare (Leverkusen, Germany), personal fees from Eisai GmbH (Frankfurt, Germany), personal fees from Ipsen GmbH (Germany), personal fees from Novartis AG (Swiss), outside the submitted work. Dr. Bundschuh reports personal fees from Bayer Healthcare, personal fees from Eisai GmbH, non-financial support from Mediso Medical Imaging Ltd., outside the submitted work. The authors have no other conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013). All patients gave written and informed consent to the imaging procedure and for anonymized evaluation and publication of their data. Due to the retrospective character of the data analysis an ethical statement was waived by the institutional ethical review board according to the professional regulations of the medical board of Nordrheinwestfalen, Germany.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Parmar C, Grossmann P, Rietveld D, et al. Radiomic machine-learning classifiers for prognostic biomarkers of head and neck cancer. Front Oncol 2015;5:272. [Crossref] [PubMed]
- Wang H, Zhou Z, Li Y, et al. Comparison of machine learning methods for classifying mediastinal lymph node metastasis of non-small cell lung cancer from 18f-fdg pet/ct images. EJNMMI Res 2017;7:11. [Crossref] [PubMed]
- Hatt M, Tixier F, Pierce L, et al. Characterization of pet/ct images using texture analysis: the past, the present... any future? Eur J Nucl Med Mol Imaging 2017;44:151-65. [Crossref] [PubMed]
- Bates A, Miles K. Prostate-specific membrane antigen pet/mri validation of mr textural analysis for detection of transition zone prostate cancer. Eur Radiol 2017;27:5290-8. [Crossref] [PubMed]
- Lozano R, Naghavi M, Foreman K, et al. Global and regional mortality from 235 causes of death for 20 age groups in 1990 and 2010: A systematic analysis for the Global Burden of Disease Study 2010. Lancet 2012;380:2095-128. [Crossref] [PubMed]
- Emmett L, Willowson K, Violet J, et al. Lutetium-177 PSMA radionuclide therapy for men with prostate cancer: a review of the current literature and discussion of practical aspects of therapy. J Med Radiat Sci 2017;64:52-60. [Crossref] [PubMed]
- Yordanova A, Eppard E, Kurpig S, et al. Theranostics in nuclear medicine practice. Onco Targets Ther 2017;10:4821-8. [Crossref] [PubMed]
- Pfestroff A, Luster M, Jilg C, et al. Current status and future perspectives of PSMA-targeted therapy in Europe: opportunity knocks. Eur J Nucl Med Mol Imaging 2015;42:1971-5. [Crossref] [PubMed]
- Ferdinandus J, Eppard E, Gaertner FC, et al. Predictors of Response to Radioligand Therapy of Metastatic Castrate-Resistant Prostate Cancer with Lu-177-PSMA-617. J Nucl Med 2017;58:312-9. [Crossref] [PubMed]
- Chicklore S, Goh V, Siddique M, et al. Quantifying tumour heterogeneity in 18f-fdg pet/ct imaging by texture analysis. Eur J Nucl Med Mol Imaging 2013;40:133-40. [Crossref] [PubMed]
- Bundschuh RA, Dinges J, Neumann L, et al. Textural parameters of tumor heterogeneity in 18f-fdg pet/ct for therapy response assessment and prognosis in patients with locally advanced rectal cancer. J Nucl Med 2014;55:891-7. [Crossref] [PubMed]
- Moazemi S, Khurshid Z, Erle A, et al. Machine Learning Facilitates Hotspot Classification in PSMA-PET/CT with Nuclear Medicine Specialist Accuracy. Diagnostics 2020;10:622. [Crossref] [PubMed]
- Khurshid Z, Ahmadzadehfar H, Gaertner F, et al. Role of textural heterogeneity parameters in patient selection for 177lu-psma therapy via response prediction. Oncotarget 2018;9:09.
- Ahmadzadehfar H, Wegen S, Yordanova A, et al. Overall survival and response pattern of castration-resistant metastatic prostate cancer to multiple cycles of radioligand therapy using [177Lu]Lu-PSMA-617. Eur J Nucl Med Mol Imaging 2017;44:1448-54. [Crossref] [PubMed]
- O'Donnell L, Schultz T. Statistical and Machine Learning Methods for Neuroimaging: Examples, Challenges, and Extensions to Diffusion Imaging Data. In: Visualization and Processing of Higher Order Descriptors for Multi-Valued Data. Springer, 2015:299-319.
- Deist TM, Dankers FJWM, Valdes G, et al. Machine learning algorithms for outcome prediction in (chemo)radiotherapy: An empirical comparison of classifiers Med Phys 2018;45:3449-59. [published correction appears in Med Phys 2019 Feb;46(2):1080-1087]. [Crossref] [PubMed]
- Huang C, Mezencev R, McDonald JF, et al. Open source machine-learning algorithms for the prediction of optimal cancer drug therapies. PLoS One 2017;12:e0186906 [Crossref] [PubMed]
- Cysouw MCF, Jansen BHE, van de Brug T, et al. Machine learning-based analysis of [18F]DCFPyL PET radiomics for risk stratification in primary prostate cancer. Eur J Nucl Med Mol Imaging 2021;48:340-9. [Crossref] [PubMed]
- SVC method. Available online: https://scikit-learn.org/stable/modules/generated/sklearn.svm.SVC.html. Accessed Sept 11, 2020.
- ExtraTrees Classifier method. Available online: https://scikit-learn.org/stable/modules/generated/sklearn.ensemble.ExtraTreesClassifier.html. Accessed Sept 11, 2020.
- RandomForest Classifier method. Available online: https://scikit-learn.org/stable/modules/generated/sklearn.ensemble.RandomForestClassifier.html. Accessed Sept 11, 2020.
- MinMaxScaler normalization method. Available online: https://scikit-learn.org/stable/modules/generated/sklearn.preprocessing.MinMaxScaler.html. Accessed Sept 11, 2020.
- Grubmüller B, Senn D, Kramer G, et al. Response assessment using 68Ga-PSMA ligand PET in patients undergoing 177Lu-PSMA radioligand therapy for metastatic castration-resistant prostate cancer. Eur J Nucl Med Mol Imaging 2019;46:1063-72. [Crossref] [PubMed]
- Kuten J, Sarid D, Yossepowitch O, et al. [68Ga]Ga-PSMA-11 PET/CT for monitoring response to treatment in metastatic prostate cancer: is there any added value over standard follow-up?. EJNMMI Res 2019;9:84. [Crossref] [PubMed]


