
Development and validation of a collagen signature-based nomogram for preoperatively predicting lymph node metastasis and prognosis in colorectal cancer
Introduction
Colorectal cancer (CRC), a major malignancy, represents the second deadliest cancer worldwide (1). Accurate assessment of lymph node (LN) metastasis is important for treatment decisions and prognostic predictions for CRC patients (2). Preoperative evaluation of LN status can help determine the need for adjuvant/neoadjuvant therapy and the adequacy of surgical resection, thereby aiding in pretreatment decision-making. Some histopathologic parameters, such as lymphovascular infiltration, are predictive of LN metastasis in CRC but are available only postoperatively (3). Several imaging methods, including computed tomography (CT), are limited in terms of detecting actual high-risk patients with LN metastasis, resulting in remarkable case understaging or overstaging (4). Therefore, a more robust biomarker is urgently required to improve the accuracy of nodal staging preoperatively.
The extracellular matrix (ECM) is a highly specialized scaffold through which cancer cells reside in tissues, and the interaction between cancer cells and the ECM regulates diverse cellular functions, such as growth, differentiation and migration (5). As the main component of the ECM, collagen is responsible for most ECM functions in the tumor microenvironment (6,7). Collagen reorganization in the tumor microenvironment has been shown to be a metastatic and prognostic biomarker in several solid tumors (8-10). In the clinic, preoperative biopsies obtained from colonoscopic examination are adequate for determining the malignancy status of CRC; thus, it would be helpful to acquire a predictive biopsy-based biomarker to improve the accuracy of nodal staging. However, the association between collagen alterations in preoperative biopsies of CRC and LN metastasis has not yet been investigated. Herein, we hypothesize that collagen alterations in the tumor microenvironment of the preoperative biopsy are associated with LN metastasis in patients with CRC.
As an imaging modality involving the combination of two-photon excitation fluorescence (TPEF) with second harmonic generation (SHG), multiphoton imaging has been increasingly applied in the field of biological medicine (11). Because of its underlying physical properties, multiphoton imaging has emerged as a powerful modality for collagen imaging in a broad range of tissues (12). The SHG signals of multiphoton imaging are highly sensitive to collagen structure and, importantly, to changes that occur in the tumor microenvironment (13). In addition, collagen features, including morphological and textural features, can be extracted from multiphoton images to comprehensively describe collagen alterations (14,15).
Integrating multiple factors into a single biomarker would yield more powerful and accurate prediction performance (16,17). The least absolute shrinkage and selection operator (LASSO) method is a popular method for variable selection with high-dimensional data (18,19). Here, we propose a collagen signature that is derived from multiple LN metastasis-associated collagen features of preoperative biopsies based on multiphoton imaging and the LASSO regression method. Therefore, in this study, we aimed to develop and validate a nomogram that incorporated the collagen signature from biopsies and clinicopathological risk factors for individual preoperative prediction of LN status in CRC. In addition, we evaluated the prognostic value of the nomogram.
We present the following article in accordance with the TRIPOD reporting checklist (available at http://dx.doi.org/10.21037/atm-20-7565).
Methods
This retrospective study was approved by the institutional review board of Nanfang Hospital, Southern Medical University, which waived the requirement to obtain written informed consent and was carried out according to the Declaration of Helsinki (as revised in 2013) for biomedical research involving human subjects.
Study design and participants
The present study was conducted in CRC patients treated between October 1, 2015, and June 30, 2018, at the Nanfang Hospital of Southern Medical University (Guangzhou, China). The inclusion criteria were as follows: (I) pathologically confirmed stage I-III CRC; (II) surgical resection with curative intent; (III) lymphadenectomy performed with at least 12 LNs harvested; (IV) complete clinicopathological and follow-up data; and (V) availability of preoperative formalin-fixed and paraffin-embedded (FFEP) biopsies. Individuals with double or multiple primary tumors or receiving neoadjuvant anticancer therapy were excluded (Figure S1). Finally, a total of 342 consecutive patients were enrolled for analysis. Subsequently, computer-generated random numbers were used to assign 70% and 30% of patients to the training and validation cohorts, respectively.
Patient clinicopathological information, including age, sex, tumor location; carcinoembryonic antigen (CEA) and cancer antigen 19-9 (CA 19-9) levels; CT-reported tumor size, T stage, and LN status; tumor differentiation and histological type from biopsies; histological type, tumor differentiation, T stage, and LN status from surgical specimens; and follow-up data (follow-up duration and survival status), was collected. The CEA and CA 19-9 levels, as well as CT-reported results, were obtained from routine preoperative examinations within one week before surgery. The primary outcome was the pathological diagnosis of LN metastasis after surgery.
Acquisition of multiphoton images and selection of regions of interest
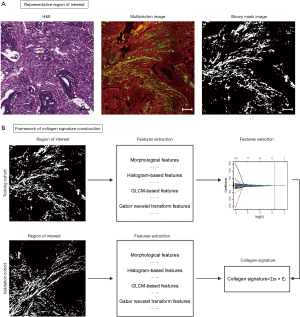
Multiphoton imaging was performed on a section of each FFPE biopsy with a 20× objective using multiphoton microscopy. The multiphoton microscopy used in this study has been described previously (20). Briefly, the system contained a high-throughput scanning inverted Axiovert 200 microscope (LSM 510 META; Zeiss, Germany) and was equipped with a mode-locked femtosecond titanium (Ti): sapphire laser (110 fs, 76 MHz), tunable from 700 to 980 nm (Mira 900-F; Coherent, America). An acousto-optic modulator was used to control the attenuation of the laser intensity. A plan-apochromat 20× objective (Zeiss) was utilized for focusing the excitation beam and for collecting the backward signals. The META detector collected the backward multiphoton signals from the tissue sample. The two-channel mode could achieve TPEF and SHG signals, which were separated by the dichroic mirror in the detection path. One channel corresponds to a wavelength range of 430 to 708 nm to show the cell morphologies from the TPEF signals, whereas another channel covered the wavelength range from 390 to 410 nm to present the microstructures of collagen from the SHG signals. The excitation wavelength (λex) was 810 nm.
The 5-µm hematoxylin-eosin (H&E)-stained slides of all enrolled patients were prepared on the other serial section. After acquisition of the multiphoton images of the biopsies, the corresponding H&E-stained slides were used for histologic assessment by a pathologist who was blinded to the LN status of each patient, and three regions of interest within the tumor tissues (field of view, 500×500 µm) were randomly selected for extraction of collagen features from multiphoton images.
Extraction of collagen features
The extraction of collagen features was performed using MATLAB 2015b (MathWorks) (14,15). For morphological features, the SHG image was first segmented into collagen pixels and background pixels using the Gaussian mixture model method (21). The binary collagen mask image was then processed using a fiber network extraction algorithm to trace each collagen fiber in the image and identify the cross-link points, which are defined as connecting points between two or more fibers (22). Moreover, we quantified an orientation index to reflect the collagen alignment based on Fourier transform spectra (23). Seven morphological features were extracted, namely, the collagen area, number, length, width, straightness, cross-link density and orientation, and the mean and variation in each morphological feature were calculated. For textural features, a histogram-based approach was first used. The mean, variation, skewness, kurtosis, energy and entropy were calculated from the histogram of the SHG pixel intensity distribution. Then, eighty gray-level cooccurrence matrix (GLCM)-based texture features were extracted (24). The contrast, correlation, energy and homogeneity were calculated from the GLCM with five different displacements of pixels at 1, 2, 3, 4 and 5 and four different directions at 0, 45, 90 and 135 degrees. Furthermore, forty-eight Gabor wavelet transform features were also extracted for analysis (25). To calculate the Gabor wavelet transform features, the SHG images were convolved with Gabor filters at five different scales and six different orientations, and the mean and variation in the magnitude of the convolution over the image at each setting were calculated. Finally, a total of 148 features were extracted (Table S1).
Construction of collagen signature
The LASSO regression method applying a 10-fold cross-validation was used for selecting a panel of the most predictive features in the training cohort, which was an accepted approach for the regression of high-dimensional data (18,19). The method used an L1 penalty to shrink some regression coefficients to exactly zero. A penalty parameter λ was selected via 1-SE (standard error) criteria, namely, the optimal λ was the largest value for which the area under the receiver operator characteristic (ROC) curve (AUROC) was within one SE of the largest value of the AUROC in the training cohort. Thereafter, a multiple-feature-based collagen signature was constructed based on a linear combination of the nonzero regression coefficients derived from the LASSO regression and the corresponding features. The collagen signature in the validation cohort was obtained directly, according to the formula used in the training cohort.
Association of collagen signature with LN status
The potential association between the collagen signature and LN status was first assessed in the training cohort and then validated in the validation cohort. Then, stratified analyses were carried out based on various subgroups. The discrimination of the collagen signature was evaluated using the AUROC.
Development and evaluation of the nomogram
All preoperative clinicopathological variables and the collagen signature were assessed by univariable analysis for their associations with LN metastasis in the training cohort, and variables showing P<0.05 were selected for the multivariate analysis. A multivariate prediction model was constructed, and a nomogram was developed. The multicollinearity of the multivariate prediction model was assessed using the tolerance and variance inflation factor (26). Discrimination and calibration were used to evaluate the performance of the nomogram (27). For quantification of the discrimination of the nomogram, the AUROC was measured. The calibration of the nomogram was evaluated by the calibration curve to assess the goodness of fit, accompanied by the Hosmer-Lemeshow test (28).
Validation of the nomogram
The bootstrap method was used for internal validation, in which random samples drawn with replacement from the original data set were the same size as the training cohort (29). One thousand bootstrap repetitions were performed, and the mean concordance index was calculated. The nomogram was then applied in the validation cohort for external validation, with the AUROC calculated and calibration curve plotted.
Clinical usefulness of the nomogram
A decision curve analysis was performed to illustrate the clinical usefulness of the nomogram by calculating the net benefits at different threshold probabilities in both the training and validation cohorts (30). The context for decision curve analysis was a situation in which individuals’ risks for an undesirable outcome were assessed, and individuals with sufficiently high risk were recommended for some intervention or treatment (31). The decision curve analysis provided a net benefit, which was calculated using the following formula:
In the formula, Pt was the threshold probability where the expected benefit of treatment was equal to the expected benefit of avoiding treatment (31). Here, Pt was the threshold probability of LN metastasis.
The maximum Youden index was selected as the optimal cutoff value in the training cohort, and then, all 342 patients were divided into high-risk and low-risk subgroups. The sensitivity, specificity, accuracy, negative predictive value (NPV), and positive predictive value (PPV) of the nomogram were calculated in the training, validation, and total cohorts, respectively.
Incremental value of the collagen signature to clinicopathological risk factors
To evaluate the incremental value of the collagen signature to the clinicopathological risk factors, a clinicopathological model that included only the preoperative clinicopathological risk factors was constructed for comparison. The incremental value of the collagen signature in the clinicopathological model was assessed with respect to the AUROC and net benefits.
Association of the collagen signature-based nomogram with prognosis
To determine the prognostic value of the collagen signature-based nomogram, the associations between the nomogram-predicted LN metastasis and disease-free survival (DFS), recurrence-free survival (RFS) and overall survival (OS) were investigated for all patients. DFS was defined as the interval from surgery to first recurrence at any site or all-cause death, whichever came first. RFS was defined as the interval from surgery to first recurrence. OS was defined as the interval between surgery and death or the last date of follow-up. The follow-up was censored on December 31, 2018.
Statistical analysis
Categorical variables were compared by using the chi-square test or Fisher’s exact test. Continuous variables, where appropriate, were compared by using Student’s t test or the Mann-Whitney U test. Binary logistic regression analysis was used to calculate the odds ratio (OR) and the corresponding 95% confidence interval (CI). Survival curves were generated by using the Kaplan-Meier method and compared by the log-rank test. Univariate and multivariate analyses with Cox proportional hazards regression determined the hazard ratio (HR) of preoperative predictors for DFS, RFS and OS. All statistical analyses were performed using R software (version 3.6.2) and SPSS (version 19.0), and a two-sided P<0.05 was considered statistically significant. The LASSO regression analysis was performed using the “glmnet” package. The ROC curves were plotted, and the AUROC was calculated using the “pROC” package. The nomogram and calibration curve were generated using the “rms” package. The Hosmer-Lemeshow test was conducted using the “generalhoslem” package. The decision curve analysis was performed using the function of “dca.R”, and the survival analysis was conducted using the “survminer” package.
Results
Clinicopathological characteristics
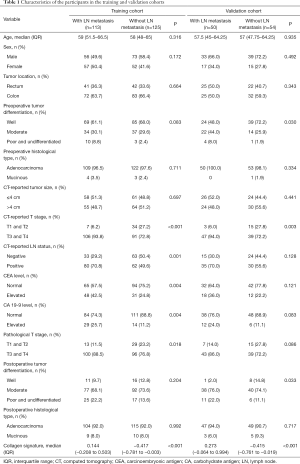
The clinicopathological characteristics of the participants in the training (N=238) and validation (N=104) cohorts are summarized in Table 1. Of the 342 enrolled patients, 58.8% (201/342) were male, and the median (interquartile range, IQR) age was 58 (48–65) years. The overall incidence of LN metastasis was 47.66% (163/342), with 47.48% (113/238) in the training cohort and 48.07% (50/104) in the validation cohort (P=0.919). The diagnostic accuracy of the subjective CT-reported LN status was 59.06% (202/342), with a sensitivity of 70.55% (115/163), a specificity of 48.60% (87/179), an NPV of 64.44% (87/135), and a PPV of 55.56% (115/207).
Full table
Collagen signature construction and its association with LN metastasis
The framework for the construction of the collagen signature is presented in Figure 1. A seven-feature-based collagen signature was constructed from the 148 features using the LASSO regression method (Figure S2). The collagen signature was calculated using the following formula: collagen signature = −1.70 + 0.25 × mean of collagen number + 0.08 × mean of cross-link density + 1.34 × mean of collagen orientation – 0.30 × variation of collagen orientation – 0.005 × kurtosis + 2.72 × GLCM_constrast_0°_4 pixel + 1.21 × Gabor_mean_orientiation 1_scale 1.
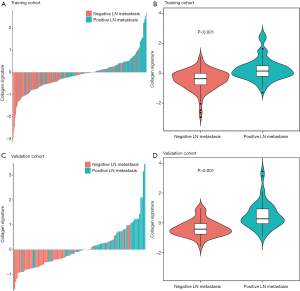
A significant difference in the collagen signature [median (IQR)] was found between patients with LN metastasis [0.144 (−0.208 to 0.503)] and patients without LN metastasis [−0.417 (−0.781 to −0.003)] in the training cohort (median difference: 0.570; 95% CI: 0.417–0.737; P<0.001) (Figure 2A,B). This finding was confirmed in patients with LN metastasis [0.273 (−0.064 to 0.994)] and patients without LN metastasis [−0.415 (−0.761 to −0.019)] in the validation cohort (median difference: 0.766; 95% CI, 0.548–1.041; P<0.001) (Figure 2C,D). The collagen signature yielded an AUROC of 0.759 (95% CI: 0.699–0.819) in the training cohort and 0.824 (95% CI: 0.743–0.904) in the validation cohort for LN metastasis, which indicated favorable predictive efficacy (Figure S3). Stratified analyses indicated that the collagen signature in patients with LN metastasis was also significantly higher than that in patients without LN metastasis under different preoperative variables in the training cohort, accompanied by satisfactory discrimination, which was also validated in the validation cohort (Table S2). In particular, in patients with negative CT-reported LN status (cN0), the collagen signature still showed a robust ability to discriminate the patients who were more likely to suffer from LN metastasis [AUROC: 0.788 (95% CI: 0.692–0.885) in the training cohort; 0.789 (95% CI: 0.639–0.939) in the validation cohort].
Development, performance evaluation, and validation of the nomogram
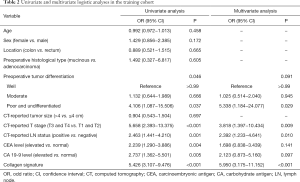
Univariate analyses were performed for each preoperative variable in the training cohort, among which the preoperative tumor differentiation (P=0.046), CT-reported T stage (P<0.001), CT-reported LN status (P=0.001), CEA level (P=0.004), CA 19-9 level (P=0.005) and collagen signature (P<0.001) had significant associations with LN metastasis (Table 2). Compared with the other five predictors, the collagen signature showed the most powerful ability to predict LN status preoperatively (Figure S3). A prediction model was subsequently constructed based on the multivariate analysis of these preoperative predictors. The variance inflation factor of each predictor was less than 10, with the corresponding tolerance exceeding 0.1, indicating no multicollinearity among all predictors (Table S3) (26). A nomogram was developed on the basis of these six predictors (Figure 3A).
Full table
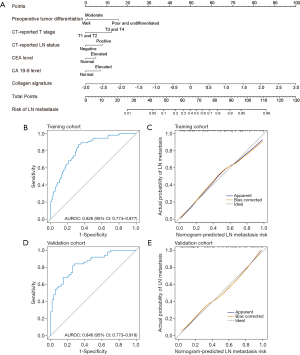
The nomogram demonstrated good accuracy in estimating the risk of LN metastasis (AUROC: 0.826, 95% CI: 0.774–0.877) (Figure 3B). The calibration curve graphically indicated good agreement on the presence of LN metastasis between the risk estimation based on the nomogram and histopathologic findings on surgical specimens (Figure 3C). The Hosmer-Lemeshow test demonstrated a P value of 0.135, indicating no departure from a good fit.
The abovementioned bootstrap method was used for internal validation, and the results remained largely unaltered between iterations, with a mean concordance index of 0.822. Good discrimination with an AUROC of 0.846 (95% CI: 0.773–0.918) was validated in the validation cohort, and a good calibration curve was also obtained for risk evaluation (Figure 3D,E). A Hosmer-Lemeshow test also yielded a nonsignificant P value of 0.766.
Clinical usefulness of the nomogram
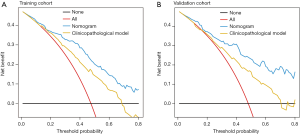
The decision curve analysis of the nomogram in the training and validation cohorts is depicted in Figure 4. The x- and y-axes represent threshold probability and net benefit, respectively. The black and red lines represent the assumptions that no case and all cases had LN metastasis, respectively. The decision curve analysis demonstrated that using the collagen signature-based nomogram to detect the LN status could result in a higher net benefit than the treat-all or treat-none scheme.
In addition, the maximum Youden index of 0.384 in the training cohort was selected as the optimal cutoff value, and then, all patients were divided into high-risk and low-risk subgroups. The sensitivity, specificity, accuracy, NPV and PPV of the nomogram in detecting the presence or absence of LN metastasis in the training cohort were 87.6%, 68.0%, 77.3%, 86.2% and 71.4%, respectively. In the validation cohort, the nomogram had a sensitivity of 84.0%, a specificity of 70.4%, an accuracy of 76.9%, a NPV of 82.7% and a PPV of 72.7%. Among all 342 patients, the sensitivity was 86.5%, the specificity was 68.2%, the accuracy was 76.9%, the NPV was 84.9%, and the PPV was 71.2% (Table 3).
Full table
Incremental value of the collagen signature to clinicopathological risk factors
To assess the incremental value of the collagen signature to clinicopathological risk factors, we excluded the collagen signature and constructed a clinicopathological model based on preoperative tumor differentiation, CT-reported T stage, CT-reported LN status, CEA level and CA 19-9 level (Table S4). The clinicopathological model showed AUROCs of 0.726 (95% CI: 0.662–0.798) and 0.727 (95% CI: 0.623–0.824) in the training and validation cohorts, respectively (Figure S4).
Compared with the clinicopathological model, the collagen signature-based nomogram, which included the collagen signature in the clinicopathological model, exhibited a significant improvement in discriminative ability for LN metastasis (Table S5, Figure S5), with an increased AUROC of 0.832 (95% CI: 0.789–0.874), which was over 0.726 (95% CI: 0.671–0.778), for all 342 patients (P<0.001). The decision curve analysis also indicated that the net benefit would be higher with the collagen signature-based nomogram than with the clinicopathological model for estimating the risk of LN metastasis (Figure 4).
Association between the collagen signature-based nomogram and prognosis
The median follow-up time of all enrolled patients was 21 (IQR: 13.75–37) months, with an estimated 3-year DFS of 68.9% (95% CI: 63.1–75.4%), 3-year DFS of 70.6% (95% CI: 64.7–77.0%) and 3-year OS of 79.3% (95% CI: 73.6–85.3%) (Figure S6).
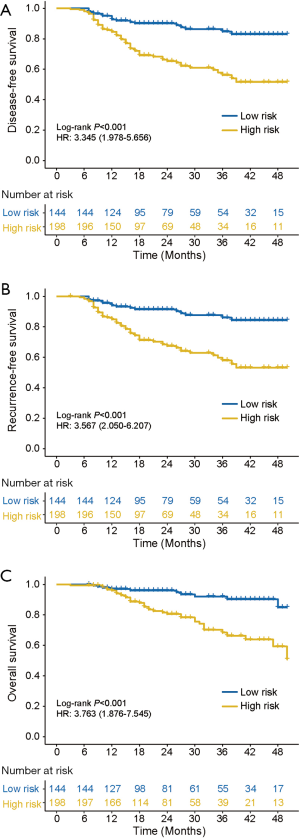
Among patients in the nomogram-predicted high-risk subgroup, DFS was significantly worse than that among patients in the nomogram-predicted low-risk subgroup [3-year DFS: high risk, 56.2% (95% CI: 47.6–66.2%); low risk, 84.7% (78.0–92.1%); log-rank P<0.001] (Figure 5A). Additionally, a worse RFS was observed in patients with high-risk subgroup compared to patients with low-risk subgroup [3-year RFS: high risk, 57.9% (95% CI: 49.3–68.1%); low risk: 86.1% (95% CI: 79.4–93.3%); log-rank P<0.001] (Figure 5B). Similarly, the OS of patients in the high-risk subgroup was worse than that of the patients in the low-risk subgroup [3-year OS: high risk, 68.4% (95% CI: 59.6–78.5%); low risk, 90.3% (84.2–97.0%); log-rank P<0.001] (Figure 5C). Univariate Cox regression analysis revealed that the nomogram-predicted high-risk subgroup was significantly associated with an unfavorable DFS (HR: 3.345, 95% CI: 1.978–5.656, P<0.001), RFS (HR: 3.567, 95% CI: 2.050–6.207; P<0.001) and OS (HR: 3.763, 95% CI: 1.876–7.545; P<0.001). Nomogram-predicted LN metastasis remained an independent preoperative predictor of DFS and OS even upon adjustment for the preoperative clinicopathological risk factors (Table S6). In addition, in 77 patients who suffered from recurrence, 32 patients were diagnosed as local recurrence and 45 patients were diagnosed as distant metastasis, respectively. Among the 32 patients with local recurrence, the percentage of low- and high-risk subgroup were 18.8% (6/32) and 81.2% (26/32), respectively. In the 45 patients with distant metastasis, 22.2% (10/45) patients were low-risk subgroup, and 77.8% (35/45) patients were high-risk subgroup (Figure S7). No significant difference was found between pattern of recurrence and nomogram-predicted subgroup (P=0.711).
Discussion
Accurate estimation of the risk of LN metastasis in CRC before surgery is vital for decision-making and prognostic predictions. In this study, we built a multi-feature-based collagen signature from preoperative biopsies, and this signature showed a significant association with LN metastasis. Additionally, we developed and validated a collagen signature-based nomogram that showed a robust ability to predict LN status. Furthermore, our decision curve analysis revealed that the net benefits would be higher with the nomogram than with the treat-all or treat-none scheme. Compared with that of the clinicopathological model, the predictive performance of the collagen signature-based nomogram was improved. In addition, the nomogram prediction of LN metastasis was relevant to patient prognosis and was an independent preoperative prognostic predictor for DFS and OS.
Collagen in the tumor microenvironment is considered a promising biomarker with great potential for clinical translation in the era of personalized medicine (7,32). However, despite some advances in the application of collagen in the clinic, most studies have been based on qualitative or semiquantitative results (9,33,34). To our knowledge, this is the first analysis of the predictive biomarker for LN metastasis based on the fully quantitative collagen signature obtained from the multiphoton images of preoperative biopsies in patients with CRC.
There were two main factors that determined the construction of the collagen signature. The first was an appropriate imaging method for specific visualization of collagen. Herein, multiphoton imaging was used because of its underlying physical origin (11,13). The second was a fully quantitative approach to comprehensively evaluate collagen alterations in the tumor microenvironment. For this purpose, we have established a framework for the quantification of collagen (8,14,15). Thus, it is feasible to construct the collagen signature of biopsies in CRC.
Currently, even with the combination of CT with other imaging examinations, the overall accuracy of CT for the preoperative evaluation of LN metastasis is not more than 60% (4). In our study, the diagnostic accuracy of CT for LN status was only 59.06%, with a sensitivity of 70.55% and a specificity of 48.60%. The unfavorable performance for nodal staging constantly resulted in under- or overtreatment. According to the collagen signature-based nomogram, the overall accuracy for nodal staging was increased to 76.9%, with improved sensitivity and specificity values of 86.5% and 68.2%, respectively, which would favor clinical decision-making regarding the treatment of CRC.
Compared to patients with a nomogram-predicted low risk of LN metastasis, those with a high risk of LN metastasis had greatly decreased predicted DFS and OS rates, even upon receiving curative-intent surgery. The high-risk patients had a 3.345-fold increased risk of a poor DFS and a 3.763-fold increased risk of a poor OS compared to low-risk patients. Multivariable Cox regression analyses also indicated that the nomogram-predicted LN status was an independent prognostic predictor of survival. In these high-risk cases, neoadjuvant chemotherapy might be considered for the potential improvement of survival (35).
To improve the predictive performance of LN metastasis in CRC, researchers have discovered several biomarkers. Qu et al. (36) constructed a miRNA panel based on four differentially expressed miRNAs, namely, miR-122-5p, miR-146b-5p, miR-186-5p and miR-193a-5p, in serum samples of CRC patients and found that the miRNA panel could increase the LN prediction capability compared with CT. Furthermore, a nomogram encompassing the miRNA panel and CT-reported LN status was developed, and this nomogram performed well in predicting the LN status. Huang et al. (37) presented a radiomics nomogram that incorporated the radiomics signature and other preoperative risk factors involving the CT-reported LN status and CEA, with a desirable AUROC of 0.736. Wei et al. (38) established a gene-related nomogram, which included 59 hub genes, for the prediction of LN metastasis in CRC and demonstrated its good diagnostic value. Although these biomarkers have not yet been translated to clinical application, we still envision that various biomarkers from radiomics, histology, serology and genomics can be considered together to improve the accuracy of estimating the risk of LN metastasis in CRC preoperatively in the future.
Multiphoton imaging is a useful approach for visualizing the tissue structure and cell morphology of samples based on their intrinsic signals without the need for additional fluorescent dyes (11,39). Additionally, multiphoton imaging provides important information on stromal collagen when its features are robustly measured and quantified (40). Because of the comparable results of multiphoton imaging to those of H&E staining, it is possible for investigators to define the regions of interest using only multiphoton imaging, after training (39). Considering the negligible effect of tissue fixation and paraffin embedding, additional removal of the paraffin in biopsies is not needed, as it would not affect multiphoton imaging (41). Multiphoton imaging of each biopsy takes approximately ten minutes, and the collagen signature can be calculated from the formula; therefore, this approach does not significantly increase the work burden imposed on clinicians.
Nomograms are widely used as predictive or prognostic tools in oncology. They have the ability to generate an individual probability of a clinical event by integrating diverse predictive or prognostic variables, thereby meeting the need for biologically and clinically integrated models and getting us closer to our goal of precision medicine (42). In this study, the following six predictors always contributed to the estimation of LN metastasis: preoperative tumor differentiation, CT-reported T stage, CT-reported LN status, CEA level and CA 19-9 level. These data were obtained from routine examinations in the clinic, and the collagen signature could be acquired after multiphoton imaging, which made calculating the individual risk of LN metastasis convenient.
There are three aspects where the nomogram might change the approach to therapy of CRC. First, the feasibility and potential survival benefits from neoadjuvant chemotherapy for locally advanced colon cancer have been proved, thus, patients with nomogram-predicted high risk of LN metastasis might also be candidates for neoadjuvant chemotherapy (43,44). Second, for rectal cancer, patients with nomogram-predicted high risk of LN metastasis would be suggested to receive preoperative chemoradiotherapy. Third, for submucosally invasive colorectal cancer (T1), additional surgical resection might be required for patients with nomogram-predicate high risk of LN metastasis after endoscopic resection.
Despite the satisfactory ability of the collagen signature-based nomogram to predict LN status, some limitations in our study should not be ignored. First, due to the retrospective nature of this study, potential selection bias was inevitable. Thus, a prospective CRC cohort is needed to verify the performance of the nomogram. Second, all enrolled participants came from a single institution. Hence, cohorts from other medical centers, especially from those in Western countries, are needed to further validate these findings.
In conclusion, this study revealed that the fully quantitative collagen signature in the tumor microenvironment of preoperative biopsies is an independent predictor for LN metastasis in CRC. Additionally, the collagen signature-based nomogram we developed and validated is helpful for individual estimations of the risk of LN metastasis among patients with CRC. Moreover, the nomogram is useful for prognostic predictions before surgery, which might facilitate decision-making and improve survival among CRC patients.
Acknowledgments
Funding: This study was supported by grants from the National Natural Science Foundation of China (Grant No. 81773117, 81771881, and 81772964), the Special Scientific Research Fund of Public Welfare Profession of National Health and Family Planning Commission (Grant No. 201502026), the Guangdong Gastrointestinal Disease Research Center (Grant No. 2017B020209003), the Guangdong Provincial Key Laboratory of Precision Medicine for Gastrointestinal Cancer (Grant No. 2020B121201004), the Natural Science Foundation of Guangdong Province (Grant No. 2020A1515010035), the Clinical Research Project of Nanfang Hospital (Grant No. 2018CR034, 2020CR001 and 2020CR011), and the China Postdoctoral Science Foundation (Grant No. 2020M682789).
Footnote
Reporting Checklist: The authors have completed the TRIPOD reporting checklist. Available at http://dx.doi.org/10.21037/atm-20-7565
Data Sharing Statement: Available at http://dx.doi.org/10.21037/atm-20-7565
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/atm-20-7565). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. This retrospective study was approved by the institutional review board of Nanfang Hospital, Southern Medical University, which waived the requirement to obtain written informed consent and was carried out according to the Declaration of Helsinki (as revised in 2013) for biomedical research involving human subjects.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Bray F, Ferlay J, Soerjomataram I, et al. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin 2018;68:394-424. [Crossref] [PubMed]
- Hashiguchi Y, Muro K, Saito Y, et al. Japanese Society for Cancer of the Colon and Rectum (JSCCR) guidelines 2019 for the treatment of colorectal cancer. Int J Clin Oncol 2020;25:1-42. [Crossref] [PubMed]
- Glasgow SC, Bleier JIS, Burgart LJ, et al. Meta-analysis of histopathological features of primary colorectal cancers that predict lymph node metastases. J Gastrointest Surg 2012;16:1019-28. [Crossref] [PubMed]
- Brouwer NP, Stijns R, Lemmens V, et al. Clinical lymph node staging in colorectal cancer; a flip of the coin? Eur J Surg Oncol 2018;44:1241-6. [Crossref] [PubMed]
- Theocharis AD, Skandalis SS, Gialeli C, et al. Extracellular matrix structure. Adv Drug Deliv Rev 2016;97:4-27. [Crossref] [PubMed]
- Yamauchi M, Barker TH, Gibbons DL, et al. The fibrotic tumor stroma. J Clin Invest 2018;128:16-25. [Crossref] [PubMed]
- Xu S, Xu H, Wang W, et al. The role of collagen in cancer: from bench to bedside. J Transl Med 2019;17:309. [Crossref] [PubMed]
- Chen D, Chen G, Jiang W, et al. Association of the collagen signature in the tumor microenvironment with lymph node metastasis in early gastric cancer. Jama Surg 2019;154:e185249 [Crossref] [PubMed]
- Toss MS, Miligy IM, Gorringe KL, et al. Geometric characteristics of collagen have independent prognostic significance in breast ductal carcinoma in situ: an image analysis study. Mod Pathol 2019;32:1473-85. [Crossref] [PubMed]
- Saito T, Uzawa K, Terajima M, et al. Aberrant collagen cross-linking in human oral squamous cell carcinoma. J Dent Res 2019;98:517-25. [Crossref] [PubMed]
- Zipfel WR, Williams RM, Christie R, et al. Live tissue intrinsic emission microscopy using multiphoton-excited native fluorescence and second harmonic generation. Proc Natl Acad Sci U S A 2003;100:7075-80. [Crossref] [PubMed]
- Chen X, Nadiarynkh O, Plotnikov S, et al. Second harmonic generation microscopy for quantitative analysis of collagen fibrillar structure. Nat Protoc 2012;7:654-69. [Crossref] [PubMed]
- Campagnola P. Second harmonic generation imaging microscopy: applications to diseases diagnostics. Anal Chem 2011;83:3224-31. [Crossref] [PubMed]
- Xu S, Wang Y, Tai D, et al. qFibrosis: a fully-quantitative innovative method incorporating histological features to facilitate accurate fibrosis scoring in animal model and chronic hepatitis B patients. J Hepatol 2014;61:260-9. [Crossref] [PubMed]
- Xu S, Kang CH, Gou X, et al. Quantification of liver fibrosis via second harmonic imaging of the Glisson's capsule from liver surface. J Biophotonics 2016;9:351-63. [Crossref] [PubMed]
- Zhang JX, Song W, Chen ZH, et al. Prognostic and predictive value of a microRNA signature in stage II colon cancer: a microRNA expression analysis. Lancet Oncol 2013;14:1295-306. [Crossref] [PubMed]
- Jiang Y, Zhang Q, Hu Y, et al. ImmunoScore signature: a prognostic and predictive tool in gastric cancer. Ann Surg 2018;267:504-13. [Crossref] [PubMed]
- Tibshirani R. Regression shrinkage and selection via the lasso: a retrospective. J R Stat Soc B 2011;73:273-82. [Crossref]
- Ranstam J, Cook JA. LASSO regression. Br J Surg 2018;105:1348. [Crossref]
- Zhuo S, Chen J, Luo T, et al. Multimode nonlinear optical imaging of the dermis in ex vivo human skin based on the combination of multichannel mode and Lambda mode. Opt Express 2006;14:7810-20. [Crossref] [PubMed]
- Dempster A, Laird N, Rubin D. Maximum likelihood from incomplete data via the EM algorithm. J. Royal Statistical Society, Series B 1977;39:1-38. [Crossref]
- Stein AM, Vader DA, Jawerth LM, et al. An algorithm for extracting the network geometry of three-dimensional collagen gels. J Microsc 2008;232:463-75. [Crossref] [PubMed]
- Frisch KE, Duenwald-Kuehl SE, Kobayashi H, et al. Quantification of collagen organization using fractal dimensions and Fourier transforms. Acta Histochem 2012;114:140-4. [Crossref] [PubMed]
- Haralick RM, Shanmugam K, Dinstein I. Textural features for image classification. IEEE Trans Syst Man Cybern 1973;3:610-21. [Crossref]
- Daugman GJ. Complete discrete 2-D Gabor transforms by neural networks for image analysis and compression. IEEE Trans Acoust 1988;36:1169-79. [Crossref]
- Dormann CF, Elith J, Bacher S, et al. Collinearity: a review of methods to deal with it and a simulation study evaluating their performance. Ecography 2013;36:27-46. [Crossref]
- Collins GS, Reitsma JB, Altman DG, et al. Transparent Reporting of a multivariable prediction model for Individual Prognosis or Diagnosis (TRIPOD). Ann Intern Med 2015;162:735-6. [Crossref] [PubMed]
- Kramer AA, Zimmerman JE. Assessing the calibration of mortality benchmarks in critical care: The Hosmer-Lemeshow test revisited. Crit Care Med 2007;35:2052-6. [Crossref] [PubMed]
- Iasonos A, Schrag D, Raj GV, et al. How to build and interpret a nomogram for cancer prognosis. J Clin Oncol 2008;26:1364-70. [Crossref] [PubMed]
- Vickers AJ, Elkin EB. Decision curve analysis: a novel method for evaluating prediction models. Med Decis Making 2006;26:565-74. [Crossref] [PubMed]
- Kerr KF, Brown MD, Zhu K, et al. Assessing the clinical impact of risk prediction models with decision curves: guidance for correct interpretation and appropriate use. J Clin Oncol 2016;34:2534-40. [Crossref] [PubMed]
- Martins Cavaco AC, Damaso S, Casimiro S, et al. Collagen biology making inroads into prognosis and treatment of cancer progression and metastasis. Cancer Metastasis Rev 2020;39:603-23. [Crossref] [PubMed]
- Ma HP, Chang HL, Bamodu OA, et al. Collagen 1A1 (COL1A1) is a reliable biomarker and putative therapeutic target for hepatocellular carcinogenesis and metastasis. Cancers (Basel) 2019;11:786. [Crossref] [PubMed]
- Conklin MW, Eickhoff JC, Riching KM, et al. Aligned collagen is a prognostic signature for survival in human breast carcinoma. Am J Pathol 2011;178:1221-32. [Crossref] [PubMed]
- Jin M, Frankel WL. Lymph node metastasis in colorectal cancer. Surg Oncol Clin N Am 2018;27:401-12. [Crossref] [PubMed]
- Qu A, Yang Y, Zhang X, et al. Development of a preoperative prediction nomogram for lymph node metastasis in colorectal cancer based on a novel serum miRNA signature and CT scans. Ebiomedicine 2018;37:125-33. [Crossref] [PubMed]
- Huang YQ, Liang CH, He L, et al. Development and validation of a radiomics nomogram for preoperative prediction of lymph node metastasis in colorectal cancer. J Clin Oncol 2016;34:2157-64. [Crossref] [PubMed]
- Wei S, Zang J, Jia Y, et al. A gene-related nomogram for preoperative prediction of lymph node metastasis in colorectal cancer. J Invest Surg 2020;33:715-22. [Crossref] [PubMed]
- Yan J, Zheng X, Liu Z, et al. Multiphoton imaging provides a superior optical biopsy to that of confocal laser endomicroscopy imaging for colorectal lesions. Endoscopy 2019;51:174-8. [Crossref] [PubMed]
- Westreich J, Khorasani M, Jones B, et al. Novel methodology to image stromal tissue and assess its morphological features with polarized light: towards a tumour microenvironment prognostic signature. Biomed Opt Express 2019;10:3963-73. [Crossref] [PubMed]
- Kakkad SM, Solaiyappan M, Argani P, et al. Collagen I fiber density increases in lymph node positive breast cancers: pilot study. J Biomed Opt 2012;17:116017 [Crossref] [PubMed]
- Balachandran VP, Gonen M, Smith JJ, et al. Nomograms in oncology: more than meets the eye. Lancet Oncol 2015;16:e173-80. [Crossref] [PubMed]
- Cheong CK, Nistala K, Ng CH, et al. Neoadjuvant therapy in locally advanced colon cancer: a meta-analysis and systematic review. J Gastrointest Oncol 2020;11:847-57. [Crossref] [PubMed]
- Foxtrot Collaborative Group. Feasibility of preoperative chemotherapy for locally advanced, operable colon cancer: the pilot phase of a randomised controlled trial. Lancet Oncol 2012;13:1152-60. [Crossref] [PubMed]


