
Assessment of HCMV-encoded microRNAs in plasma as potential biomarkers in pregnant women with adverse pregnancy outcomes
Introduction
Fetal structural anomaly (FSA) and spontaneous abortion (SA) are two important types of adverse pregnancy outcomes (APOs). In the clinically recognized pregnancies, the incidence of FSA is approximately 3% (1,2), and the cumulative risk of SA is approximately 11–20% (3,4). Although various types of genetic variations can cause FSA and SA, human cytomegalovirus (HCMV) is one of the most important nonhereditary causes of APO (5-8). Nearly 30–40% of primary HCMV infected pregnant women have congenitally infected fetuses due to mother-child transmission (9,10). In the United States, 4% to 5% of live births have congenital HCMV infection (11), and approximately 10–15% have birth defects (12). To date, there is no evidence that pregnant women who are immune have a lower congenital HCMV infection risk (13). Furthermore, although HCMV-DNA positive pregnant women show a 3-fold greater chance of congenital infection, there is no positive correlation between HCMV DNA positivity and APO (14). Therefore, the identification of noninvasive predictors of APO for pregnant women with HCMV infection is an important gap in our knowledge.
MicroRNAs (miRNAs) are a type of noncoding RNA nucleotide ~22 nt in length that play a critical roles in various biological processes (15). Previous studies have revealed that a variety of miRNAs derived from the host itself and other species are highly stable in plasma and serum, and may serve as noninvasive biomarkers for various diseases (16-21). In addition, small noncoding RNAs can be transmitted through the mammalian placenta and directly regulate fetal gene expression (22).
HCMV encodes at least 26 mature miRNAs during its infectious life cycle (23). HCMV miRNAs can target both host and viral genes to regulate various aspects of cellular and viral biology, such as host immune responses (24-26), cell cycle regulation (27), viral replication (28), and viral latency (29-32). HCMV-encoded miRNAs have been confirmed to exist in plasma, and they are associated with hypertension, chronic hepatitis B and oral lichen planus (33-35). However, the plasma signature of HCMV-encoded miRNAs in pregnant women with APO and its clinical relevance have not been studied.
We postulated the existence of a unique plasma profile of HCMV-encoded miRNAs in pregnant women with APO, and used quantitative reverse-transcription polymerase chain reaction (RT-qPCR) assays to test this hypothesis. Furthermore, we evaluated the diagnostic potential of the significantly altered HCMV miRNAs in the plasma as a noninvasive predictor of APO for pregnant women with HCMV infection. We present the following article in accordance with the MDAR reporting checklist (available at http://dx.doi.org/10.21037/atm-20-7354).
Methods
The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013). This study was approved by the Institutional Review Board of Qiqihar Medical University {[2017]19} and individual consent for this retrospective analysis was waived.
Patient characteristics and clinical features
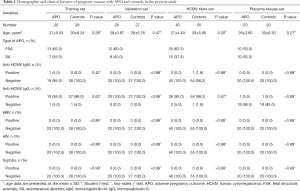
A total of 95 pregnant women were enrolled from the First, Second and Third Affiliated Hospitals of Qiqihar Medical University and Jianhua Hospital (Qiqihar, China), and obtained relevant permits. Normal or APO were determined based on clinical examinations and laboratory assessments. As shown in Table 1 and Table S1, 40 pregnant women with APO and 55 with normal pregnancy participated in this study. All blood samples were collected in EDTA tubes, and centrifuged at room temperature at 3,000 ×g for 10 min. The supernatant (plasma) and blood cells were stored separately at –80 °C until analysis.
Full table
RNA isolation and RT-qPCR
Total RNA was extracted from plasma using phenol/chloroform purification protocol. Briefly, 100 μL of plasma was diluted with 300 μL of ribonuclease-free water and 200 μL of acid phenol (Sigma, P4682), and then 20 μL of synthetic MIR2911 was added. After the mixture was vortex-mixed vigorously, 200 μL of chloroform was added. The sample was incubated at room temperature for 15 minutes and then centrifuged at 16,000 ×g for 20 minutes. The aqueous supernatant was transferred to a fresh 1.5 mL microcentrifuge tube and mixed with 2 volumes of isopropyl alcohol and 1/10-volume of 3 M sodium acetate (pH 5.3). Next, the mixture was stored at –20 °C for 2 h and then centrifuged at 16,000 ×g for 20 minutes. Subsequently, the RNA pellet was collected, washed with 75% ethanol and dried for 10 minutes at room temperature. Finally, the pellet was dissolved in 30 μL of ribonuclease-free water and stored at –80 °C until further analysis. Total RNA was extracted from placenta tissues using TRIzol reagent (Invitrogen, CA, USA).
We performed a TaqMan probe-based RT-qPCR assay (Table S2) to investigate the differential expression of HCMV-encoded miRNAs between normal and APO groups as described previously (33,34). The RT-qPCR assays were performed in triplicate using an ABI 7300 instrument (Applied Biosystems, Foster City, CA, USA). Expression levels of miRNAs were calculated using the CT values. We assessed the detection limits of the RT-qPCR assay by performing calibration curves developed with synthetic miR-16 oligonucleotides (Figure S1). All reactions were performed in triplicate.
HCMV titers
To investigate the copy numbers of HCMV in peripheral blood leukocytes (PBLs) and placenta tissues, RT-qPCR was performed on samples from 40 pregnant women with APO and 55 with normal pregnancy. DNA was extracted from PBLs and placenta tissues using Blood Genomic DNA Midi Kit (CWBIO, CW0541) and TIANamp Genomic DNA Midi Kit (TIANGEN, DP304-03) according to the manufacturer’s protocols. We amplified HCMV DNA by TaqMan real-time PCR with the following HCMV specific primers: HCMV DNA forward: 5'-CACGGTCCCGGTTTAGCA-3', HCMV DNA reverse: 5'-CGTAACGTGGACCTGACGTTT-3'. A 10-fold diluted recombinant plasmid containing the HCMV target sequence was used as a template for standard curve preparation (Figure S2). The procedure consisted of 45 cycles of denaturation at 95 °C for 15 seconds, annealing at 60 °C for 30 seconds and extension at 72 °C for 30 seconds. Plasmid DNA containing the HCMV target sequence was used in separate reactions serving as a positive control. Results were expressed as copies per 1 mL blood and 10 mg placenta tissues.
Enzyme-linked immunosorbent assay (ELISA)
Anti-HCMV immunoglobulin G (IgG) and immunoglobulin M (IgM) antibodies were detected with a commercially available ELISA kit (Dia.Pro Diagnostic Bioprobes s.r.l, CMVG.CE/S and CMVM.CE/S) according to the manufacturer’s instructions. For the IgG-ELISA, a value of <0.5 IU/mL was considered a negative result, and a value of >0.5 IU/mL was considered a positive result, indicating prior exposure to HCMV. For the IgM-ELISA, the test results were calculated using the optical density (OD) value at 450 nm, and the cut-off value for positivity was OD >1.2.
Statistical analysis
Clinical characteristics between the two study cohorts were statistically analyzed using a Student’s t-test and χ2 test (Table 1). Data from the RT-qPCR assays were also statistically analyzed using a Student’s t-test. The results are presented as the mean ± SEM for miRNAs and the mean ± SD for other variables. Univariate logistic regression analyses were performed to analyze associations between plasma HCMV-encoded miRNAs and APO. In addition, linear regression models were performed to analyze associations between plasma HCMV-encoded miRNAs and variables. A P value of <0.05 was considered statistically significant. All statistical analyses were performed using GraphPad Prism 7.0 software (GraphPad Software, USA).
Results
Expression profiles of HCMV-encoded miRNAs in plasma from pregnant women
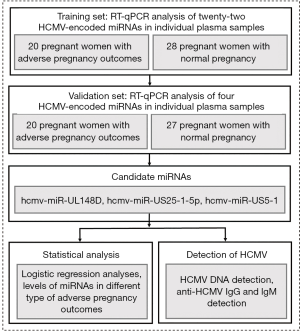
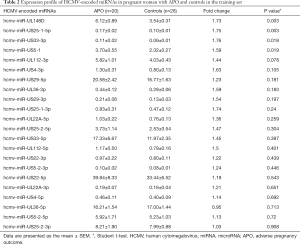
The experimental design of this study is shown in Figure 1. We enrolled 95 pregnant women to participate in our study, and the samples were randomly divided into a training set and a validation set. In the training set, we detected 22 HCMV-encoded miRNAs in plasma from 20 pregnant women with APO and 28 normal controls by TaqMan probe-based RT-qPCR. Our data showed that the plasma of all pregnant women contained a specific level of the examined HCMV-encoded miRNAs. As shown in Table 2, 4 of 22 HCMV-encoded miRNAs, including hcmv-miR-US25-1-5p, hcmv-miR-UL148D, hcmv-miR-US33-3p, and hcmv-miR-US5-1, were expressed at higher levels in plasma from pregnant women with APO than in plasma from normal controls (P<0.05). These results indicate that the altered levels of plasma HCMV-encoded miRNAs may be specific to pregnant women with APO.
Full table
Further validation of HCMV-encoded miRNAs in the plasma from pregnant women by RT-qPCR
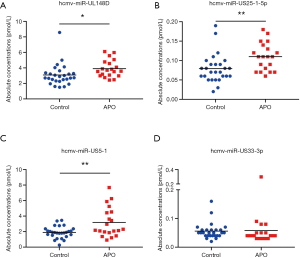
Subsequently, we confirmed the 4 altered HCMV miRNAs in an additional independent sample set (validation set) consisting of 20 pregnant women with APO and 27 normal subjects (Figure 2). As shown in Figure 2A,B,C and Table S3, the pregnant women with APO had higher plasma levels of hcmv-miR-US25-1-5p, hcmv-miR-US5-1 and hcmv-miR-UL148D than the normal controls in the validation set. However, the plasma levels of hcmv-miR-US33-3p showed no significant difference between pregnant women with APO and normal controls (Figure 2D and Table S3). The observed differences in the 3 identified miRNAs between 40 pregnant women with APO and 55 control individuals are illustrated in Figure 3A,B,C and Table S4.
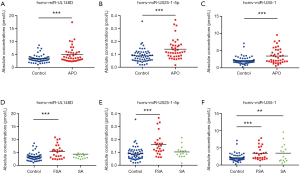
Plasma expression levels of 3 identified HCMV-encoded miRNAs in different types of APO
FSA and SA are common categories of APO. These APOs are devastating for couples, and most FSAs and SAs are unexplained. We analyzed the altered expression of the above 3 HCMV miRNAs in the pregnant women with different types of APO [FSA (n=25) and SA (n=15)] enrolled in this study. The results showed that hcmv-miR-UL148D and hcmv-miR-US25-1-5p were only elevated in pregnant women with FSA but that hcmv-miR-US5-1 was significantly upregulated in pregnant women with both FSA and SA (Figure 3D,E,F and Table S5). Additionally, the plasma level of hcmv-miR-US25-1-5p was much higher in pregnant women with FSA than in pregnant women with SA (Figure 3E and Table S5).
Diagnostic value of 3 altered HCMV-encoded miRNAs as a predictor of APO for pregnant women with HCMV infection
To assess the diagnostic accuracy of the 3 plasma HCMV miRNAs for APO, we performed receiver-operating characteristic (ROC) curve analysis. The area under the ROC curve (AUC) of the 3 altered miRNAs (hcmv-miR-UL148D, hcmv-miR-US25-1-5p, and hcmv-miR-US5-1) was 0.689, 0.735 and 0.688, respectively (Table S6). Among them, hcmv-miR-US25-1-5p presented the largest AUC (0.735; 95% CI, 0.635–0.836), with a sensitivity of 68% and specificity of 71%. Although we also investigated the diagnostic value of the combinations of the 3 selected miRNAs, the AUC of all the combinations was lower than that of hcmv-miR-US25-1-5p alone (Table S6). Our data indicate that plasma hcmv-miR-US25-1-5p is a potential predictor of APO in pregnant women with HCMV infection.
Increased plasma levels of HCMV-encoded miRNAs in pregnant women correlate with APO
To further evaluate the ability of the three increased HCMV miRNAs in plasma from pregnant women to predict risk, we used APO as the dependent variable and the risk score as the covariate to conduct a univariate logistic regression analysis. Hcmv-miR-US25-1-5p and hcmv-miR-US5-1 correlated independently with APO (Table S7). The odds ratios (ORs) of these 2 HCMV miRNAs for pregnant women with APO were as follows: hcmv-miR-US25-1-5p (OR =26500.657, 95% CI, 2.848–246,567,602.202, P=0.029) and hcmv-miR-US5-1 (OR =1.406, 95% CI, 1.024–1.931, P=0.035). The results suggest that elevated plasma levels of hcmv-miR-US25-1-5p and hcmv-miR-US5-1 are potential risk factors for APO.
The levels of hcmv-miR-UL148D, hcmv-miR-US25-1-5p, and hcmv-miR-US5-1 in the placenta tissues of 20 pregnant women with APO and 20 with normal pregnancy were further assessed. Our results showed that hcmv-miR-US25-1-5p in the placenta tissues were dramatically increased in APO compared with normal controls (Figure 4A), but hcmv-miR-UL148D and hcmv-miR-US5-1 could not be detected in the placenta tissues. Pearson correlation scatter plot analysis showed significant correlation between the levels of hcmv-miR-US25-1-5p in plasma and placenta tissues (P=0.0009) (Figure 4B). These data indicated that hcmv-miR-US25-1-5p could be transfer through placenta to the fetal side.
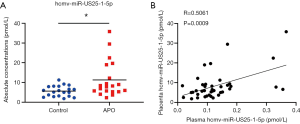
Detection of HCMV in pregnant women
Because expression of the 3 HCMV miRNAs was significantly increased in the plasma from pregnant women with APO, we investigated the discrepancy in HCMV load between pregnant women with APO and normal controls. A previous study reported that HCMV DNA in PBLs is more sensitive for diagnosing HCMV infection than that in plasma (36). Therefore, we used RT-qPCR to detect HCMV DNA in PBLs from both the training and validation sets. Nevertheless, we found no significant differences in HCMV DNA levels between pregnant women with APO and normal controls, with HCMV virus titers of (33.46–2,307) ±678.9 copies/mL in APO and (9.08–6,370) ±1,088.0 copies/mL in normal controls (P=0.60, Figure S3A). However, HCMV DNA in placenta tissues was negative in participants (20 pregnant women with APO and 20 with normal pregnancy). We further examined concentrations of anti-HCMV IgM and anti-HCMV IgG in plasma from all 40 pregnant women with APO and 55 control individuals by ELISA. Our results indicated no significant difference in the positivity rates of anti-HCMV IgM and anti-HCMV IgG or in the plasma concentrations of anti-HCMV IgG between the two study cohorts (0% vs. 1.8%, P>0.99; 95.0% vs. 98.2%, P=0.57, respectively) (Table 1 and Figure S3B). In addition, linear regression was used to examine associations between clinical characteristics and plasma hcmv-miR-US25-1-5p. Nevertheless, plasma hcmv-miR-US25-1-5p was not correlated with age of pregnant women, gestational age, HCMV DNA titers in PBLs and anti-HCMV IgG concentrations (Table S8).
Discussion
Although various types of genetic variations can cause FSA and SA, congenital HCMV infection is one of the most important nongenetic causes of APO. However, there are no reliable qualitative or quantitative methods for the early prediction of APO due to HCMV infection in pregnant women. This study examined for the first time the relationship between HCMV-encoded miRNAs in maternal plasma and APO. Furthermore, our results indicated that plasma hcmv-miR-US25-1-5p may serve as a noninvasive biomarker of APO for pregnant women with HCMV infection.
Our study identified 3 of the 22 HCMV-encoded miRNAs, hcmv-miR-US25-1-5p, hcmv-miR-US5-1, and hcmv-miR-UL148D, to be markedly increased in pregnant women with APO. More importantly, we observed a significant difference in the concentrations of detected miRNAs among pregnant women with different types of APO. Compared with normal controls, the 3 identified HCMV miRNAs in plasma were all significantly increased in pregnant women with FSA, whereas only hcmv-miR-US5-1 was significantly upregulated in pregnant women with SA. Most notably, pregnant women with FSA had higher hcmv-miR-US25-1-5p levels than did pregnant women with SA. These findings suggest that increased expression of these three HCMV miRNAs may have a greater impact on FSA. Among the 3 miRNAs, hcmv-miR-US25-1-5p and hcmv-miR-US5-1 were significantly related to APO, suggesting that these miRNAs are potential risk factors for APO. Hcmv-miR-US25-1-5p presented the largest AUC (0.735; 95% CI, 0.635–0.836), indicating that plasma hcmv-miR-US25-1-5p may serve as a predictor of APO for pregnant women with HCMV infection. Additionally, hcmv-miR-US25-1-5p in the placenta tissues were dramatically increased in APO and correlated with plasma hcmv-miR-US25-1-5p. These results suggest that hcmv-miR-US25-1-5p can be transferred to the fetal side through the placenta, which may affect the development of the fetus.
In the present study, we assessed HCMV DNA in PBLs from our studied pregnant women and found no significant difference between pregnant women with APO and normal controls. HCMV DNA in maternal blood and/or serological investigation has become a key indicator for the diagnosis of primary infection (36). However, the correlation between HCMV DNA and congenital infection remains controversial (14,36-38). Additionally, HCMV DNA does not correlate with APO (14), consistent with our finding. Moreover, HCMV DNA in placenta tissues of pregnant women was negative in this study, while hcmv-miR-US25-1-5p was upregulated in placenta tissues of pregnant women with APO, indicating hcmv-miR-US25-1-5p is easier to pass through the placenta than HCMV. Our results showed that more than 90% of the pregnant women in both study cohorts were anti-HCMV IgG positive, with no significant difference in positivity rates for anti-HCMV IgG or IgM. There is also no strong evidence that antiviral antibody responses provide protection against maternal infection, intrauterine transmission, or APO (39). Currently, predictors of APO in pregnant women with HCMV infection are not well defined, and solving this problem has important clinical value. Our results show that HCMV-encoded miRNAs are able to distinguish pregnant women with APO from normal controls.
In general, prognosis can only be determined until at late pregnancy if the fetus has no ultrasound features or severe ultrasound abnormalities (40,41). Sometimes invasive operations are required to obtain fetal blood for some prognostic parameter measurements (42-44). Therefore, the use of plasma HCMV miRNAs as a noninvasive predictor of APO for pregnant women with HCMV infection has great clinical significance.
Our study identified three altered HCMV-encoded miRNAs in pregnant women. Based on the fact that exogenous/endogenous small noncoding RNAs in the maternal system can be transferred through the mammalian placenta and influence fetal development and health by directly regulating fetal gene expression (22), determining whether the three HCMV-encoded miRNAs can influence fetal development warrants future study.
The validated downstream target genes for hcmv-miR-UL148D include IER5, ACVR1B, RANTES, and IEX-1 (23,29,45,46), and those for hcmv-miR-US25-1-5p include CD147, cyclin E2, BRCC3, YWHAE, UBB, NPM1, and HSP90AA (27,47-49); those for hcmv-miR-US5-1 include GMNN, IKKα, IKKβ, and US7 (50-53). These validated target genes can be divided into four categories: viral latency (IER5, ACVR1B, and RANTES), viral replication (YWHAE, UBB, NPM1, HSP90AA, GMNN, and US7), immune evasion (ACVR1B, RANTES, IKKα, and IKKβ), and cell processes (IEX-1, CD147, cyclin E2, BRCC3). Downregulation of these genes by HCMV-encoded miRNAs may interfere with host immune responses and the cell cycle, leading to abnormalities in embryonic development. The above results indicate that the HCMV-encoded miRNAs identified in our study may contribute to the pathogenesis of HCMV infection-related diseases as well as APO, but further research is needed.
Overall, our study shows that HCMV-encoded miRNAs in plasma are safe, stable, and specific biomarker candidates for APO monitoring and prognosis. Moreover, the required testing costs are relatively low. As the factors leading to APO in pregnant women are complex, it is necessary to further exclude the known confounding factors that affect the results of the maternal cohort study. This study only compared the two major types of APO (FSA and SA) with normal pregnant women, and more detailed classification and larger sample sizes for prospective studies are needed. Although we clarified the relationship between plasma HCMV-encoded miRNAs and APO in pregnant women, the function of these identified HCMV miRNAs on intrauterine transmission and the pathogenic mechanism have not yet been elucidated. Therefore, further careful design and implementation of prospective studies are required.
In summary, we first report a distinctive plasma signature of HCMV-encoded miRNAs in pregnant women with APO and that plasma levels of hcmv-miR-US25-1-5p and hcmv-miR-US5-1 in pregnant women are associated with APO. Furthermore, hcmv-miR-US25-1 may constitute a noninvasive biomarker for predicting APO in pregnant women during HCMV infection. These findings indicate that HCMV-encoded miRNAs may provide pathophysiological clues for the development of APO and warrant further study.
Acknowledgments
Funding: This project was funded by the Heilongjiang Province Department of Education (UNPYSCT-2018035) and by the Research Foundation of Qiqihar Medical University (QY20147-01).
Footnote
Reporting Checklist: The authors have completed the MDAR reporting checklist. Available at http://dx.doi.org/10.21037/atm-20-7354
Data Sharing Statement: Available at http://dx.doi.org/10.21037/atm-20-7354
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/atm-20-7354). All authors reports grants and non-financial support from Heilongjiang Province Department of Education, grants and non-financial support from Qiqihar Medical University, during the conduct of the study.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013). This study was approved by the Institutional Review Board of Qiqihar Medical University {[2017]19} and individual consent for this retrospective analysis was waived.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Persson M, Cnattingius S, Villamor E, et al. Risk of major congenital malformations in relation to maternal overweight and obesity severity: cohort study of 1.2 million singletons. BMJ 2017;357:j2563. [Crossref] [PubMed]
- Lord J, McMullan DJ, Eberhardt RY, et al. Prenatal exome sequencing analysis in fetal structural anomalies detected by ultrasonography (PAGE): a cohort study. Lancet 2019;393:747-57. [Crossref] [PubMed]
- Nathan NO, Mørch LS, Wu CS, et al. Rheumatoid arthritis and risk of spontaneous abortion: a Danish nationwide cohort study. Rheumatology (Oxford) 2020;59:1984-91. [Crossref] [PubMed]
- Ammon Avalos L, Galindo C, Li DK. A systematic review to calculate background miscarriage rates using life table analysis. Birth Defects Res A Clin Mol Teratol 2012;94:417-23. [Crossref] [PubMed]
- Pereira L. Have we overlooked congenital cytomegalovirus infection as a cause of stillbirth? J Infect Dis 2011;203:1510-2. [Crossref] [PubMed]
- Morton CC, Nance WE. Newborn hearing screening--a silent revolution. N Engl J Med 2006;354:2151-64. [Crossref] [PubMed]
- Fowler KB, Boppana SB. Congenital cytomegalovirus (CMV) infection and hearing deficit. J Clin Virol 2006;35:226-31. [Crossref] [PubMed]
- Gabrielli L, Bonasoni MP, Santini D, et al. Congenital cytomegalovirus infection: patterns of fetal brain damage. Clin Microbiol Infect 2012;18:E419-27. [Crossref] [PubMed]
- Manicklal S, Emery VC, Lazzarotto T, et al. The "silent" global burden of congenital cytomegalovirus. Clin Microbiol Rev 2013;26:86-102. [Crossref] [PubMed]
- Kenneson A, Cannon MJ. Review and meta-analysis of the epidemiology of congenital cytomegalovirus (CMV) infection. Rev Med Virol 2007;17:253-76. [Crossref] [PubMed]
- Boppana SB, Ross SA, Shimamura M, et al. Saliva polymerase-chain-reaction assay for cytomegalovirus screening in newborns. N Engl J Med 2011;364:2111-8. [Crossref] [PubMed]
- Boppana SB, Ross SA, Fowler KB. Congenital cytomegalovirus infection: clinical outcome. Clin Infect Dis 2013;57:S178-81. [Crossref] [PubMed]
- Britt WJ. Maternal immunity and the natural history of congenital human cytomegalovirus infection. Viruses 2018;10:405. [Crossref] [PubMed]
- Simonazzi G, Cervi F, Zavatta A, et al. Congenital cytomegalovirus infection: prognostic value of maternal DNAemia at amniocentesis. Clin Infect Dis 2017;64:207-10. [Crossref] [PubMed]
- Chen K, Rajewsky N. The evolution of gene regulation by transcription factors and microRNAs. Nat Rev Genet 2007;8:93-103. [Crossref] [PubMed]
- Chen X, Hu Z, Wang W, et al. Identification of ten serum microRNAs from a genome-wide serum microRNA expression profile as novel noninvasive biomarkers for nonsmall cell lung cancer diagnosis. Int J Cancer 2012;130:1620-8. [Crossref] [PubMed]
- Zhang C, Wang C, Chen X, et al. Expression profile of microRNAs in serum: a fingerprint for esophageal squamous cell carcinoma. Clin Chem 2010;56:1871-9. [Crossref] [PubMed]
- Calin GA, Croce CM. MicroRNA signatures in human cancers. Nat Rev Cancer 2006;6:857-66. [Crossref] [PubMed]
- Mitchell PS, Parkin RK, Kroh EM, et al. Circulating microRNAs as stable blood-based markers for cancer detection. Proc Natl Acad Sci U S A 2008;105:10513-8. [Crossref] [PubMed]
- Guo Y, Chen Z, Zhang L, et al. Distinctive microRNA profiles relating to patient survival in esophageal squamous cell carcinoma. Cancer Res 2008;68:26-33. [Crossref] [PubMed]
- Zhang YP, Ye L, Yu N, et al. Serum microRNAs as novel biomarkers for early prediction of disease severity in patients with acute pancreatitis. ExRNA 2020;2:10. [Crossref]
- Li J, Zhang Y, Li D, et al. Small non-coding RNAs transfer through mammalian placenta and directly regulate fetal gene expression. Protein Cell 2015;6:391-6. [Crossref] [PubMed]
- Kim Y, Lee S, Kim S, et al. Human cytomegalovirus clinical strain-specific microRNA miR-UL148D targets the human chemokine RANTES during infection. PLoS Pathog 2012;8:e1002577 [Crossref] [PubMed]
- Nachmani D, Lankry D, Wolf DG, et al. The human cytomegalovirus microRNA miR-UL112 acts synergistically with a cellular microRNA to escape immune elimination. Nat Immunol 2010;11:806-13. [Crossref] [PubMed]
- Hook LM, Grey F, Grabski R, et al. Cytomegalovirus miRNAs target secretory pathway genes to facilitate formation of the virion assembly compartment and reduce cytokine secretion. Cell Host Microbe 2014;15:363-73. [Crossref] [PubMed]
- Landais I, Pelton C, Streblow D, et al. Human cytomegalovirus miR-UL112-3p targets TLR2 and modulates the TLR2/IRAK1/NFkappaB signaling pathway. PLoS Pathog 2015;11:e1004881 [Crossref] [PubMed]
- Grey F, Tirabassi R, Meyers H, et al. A viral microRNA down-regulates multiple cell cycle genes through mRNA 5'UTRs. PLoS Pathog 2010;6:e1000967 [Crossref] [PubMed]
- Grey F, Meyers H, White EA, et al. A human cytomegalovirus-encoded microRNA regulates expression of multiple viral genes involved in replication. PLoS Pathog 2007;3:e163 [Crossref] [PubMed]
- Pan C, Zhu D, Wang Y, et al. Human cytomegalovirus miR-UL148D facilitates latent viral infection by targeting host cell immediate early response gene 5. PLoS Pathog 2016;12:e1006007 [Crossref] [PubMed]
- Murphy E, Vanicek J, Robins H, et al. Suppression of immediate-early viral gene expression by herpesvirus-coded microRNAs: implications for latency. Proc Natl Acad Sci U S A 2008;105:5453-8. [Crossref] [PubMed]
- Meshesha MK, Bentwich Z, Solomon SA, et al. In vivo expression of human cytomegalovirus (HCMV) microRNAs during latency. Gene 2016;575:101-7. [Crossref] [PubMed]
- O'Connor CM, Vanicek J, Murphy EA. Host microRNA regulation of human cytomegalovirus immediate early protein translation promotes viral latency. J Virol 2014;88:5524-32. [Crossref] [PubMed]
- Ding M, Wang X, Wang C, et al. Distinct expression profile of HCMV encoded miRNAs in plasma from oral lichen planus patients. J Transl Med 2017;15:133. [Crossref] [PubMed]
- Pan Y, Wang N, Zhou Z, et al. Circulating human cytomegalovirus-encoded HCMV-miR-US4-1 as an indicator for predicting the efficacy of IFNalpha treatment in chronic hepatitis B patients. Sci Rep 2016;6:23007. [Crossref] [PubMed]
- Li S, Zhu J, Zhang W, et al. Signature microRNA expression profile of essential hypertension and its novel link to human cytomegalovirus infection. Circulation 2011;124:175-84. [Crossref] [PubMed]
- Revello MG, Furione M, Zavattoni M, et al. Human cytomegalovirus (HCMV) DNAemia in the mother at amniocentesis as a risk factor for iatrogenic HCMV infection of the fetus. J Infect Dis 2008;197:593-6. [Crossref] [PubMed]
- Revello MG, Zavattoni M, Sarasini A, et al. Human cytomegalovirus in blood of immunocompetent persons during primary infection: prognostic implications for pregnancy. J Infect Dis 1998;177:1170-5. [Crossref] [PubMed]
- Zavattoni M, Furione M, Lanzarini P, et al. Monitoring of human cytomegalovirus DNAemia during primary infection in transmitter and non-transmitter mothers. J Clin Virol 2016;82:89-93. [Crossref] [PubMed]
- Britt WJ. Congenital human cytomegalovirus infection and the enigma of maternal immunity. J Virol 2017;91:e02392-16. [Crossref] [PubMed]
- Bodéus M, Hubinont C, Bernard P, et al. Prenatal diagnosis of human cytomegalovirus by culture and polymerase chain reaction: 98 pregnancies leading to congenital infection. Prenat Diagn 1999;19:314-7. [Crossref] [PubMed]
- Donner C, Liesnard C, Brancart F, et al. Accuracy of amniotic fluid testing before 21 weeks' gestation in prenatal diagnosis of congenital cytomegalovirus infection. Prenat Diagn 1994;14:1055-9. [Crossref] [PubMed]
- Noyola DE, Demmler GJ, Nelson CT, et al. Early predictors of neurodevelopmental outcome in symptomatic congenital cytomegalovirus infection. J Pediatr 2001;138:325-31. [Crossref] [PubMed]
- Ancora G, Lanari M, Lazzarotto T, et al. Cranial ultrasound scanning and prediction of outcome in newborns with congenital cytomegalovirus infection. J Pediatr 2007;150:157-61. [Crossref] [PubMed]
- Leruez-Ville M, Stirnemann J, Sellier Y, et al. Feasibility of predicting the outcome of fetal infection with cytomegalovirus at the time of prenatal diagnosis. Am J Obstet Gynecol 2016;215:342.e1-9. [Crossref] [PubMed]
- Lau B, Poole E, Krishna B, et al. The expression of human cytomegalovirus microRNA miR-UL148D during latent infection in primary myeloid cells inhibits activin A-triggered secretion of IL-6. Sci Rep 2016;6:31205. [Crossref] [PubMed]
- Wang YP, Qi Y, Huang YJ, et al. Identification of immediate early gene X-1 as a cellular target gene of hcmv-mir-UL148D. Int J Mol Med 2013;31:959-66. [Crossref] [PubMed]
- Chen J, Xia S, Yang X, et al. Human cytomegalovirus encoded miR-US25-1-5p attenuates CD147/EMMPRIN-mediated early antiviral response. Viruses 2017;9:365. [Crossref] [PubMed]
- Fan J, Zhang W, Liu Q. Human cytomegalovirus-encoded miR-US25-1 aggravates the oxidised low density lipoprotein-induced apoptosis of endothelial cells. Biomed Res Int 2014;2014:531979 [Crossref] [PubMed]
- Jiang S, Qi Y, He R, et al. Human cytomegalovirus microRNA miR-US25-1-5p inhibits viral replication by targeting multiple cellular genes during infection. Gene 2015;570:108-14. [Crossref] [PubMed]
- Hancock MH, Hook LM, Mitchell J, et al. Human cytomegalovirus microRNAs miR-US5-1 and miR-UL112-3p block proinflammatory cytokine production in response to NF-kappaB-activating factors through direct downregulation of IKKalpha and IKKbeta. mBio 2017;8:e00109-17. [Crossref] [PubMed]
- Yurochko AD. New mechanism by which human cytomegalovirus microRNAs negate the proinflammatory response to infection. mBio 2017;8:e00505-17. [Crossref] [PubMed]
- Tirabassi R, Hook L, Landais I, et al. Human cytomegalovirus US7 is regulated synergistically by two virally encoded microRNAs and by two distinct mechanisms. J Virol 2011;85:11938-44. [Crossref] [PubMed]
- Jiang S, Huang Y, Qi Y, et al. Human cytomegalovirus miR-US5-1 inhibits viral replication by targeting Geminin mRNA. Virol Sin 2017;32:431-9. [Crossref] [PubMed]


