
Adenosine A3 receptor activated in H2O2 oxidative stress of primary open-angle glaucoma
Introduction
Glaucoma is characterized as a function loss of retinal ganglion cells and is the second leading cause of blindness worldwide (1). High intraocular pressure (IOP) is a risk factor for glaucoma (2). Under normal conditions, IOP is maintained by the dynamic balance between the production and excretion of the aqueous humor (AH) through the human trabecular meshwork (HTM) (3,4). Dysfunction of trabecular meshwork cells (TMCs) can decrease drainage in the AH, leading to high-IOP and further inducing primary open-angle glaucoma (POAG) (5). Therefore, understanding HTM pathology may help to treat POAG.
Studies have demonstrated that TMCs are the most sensitive cell type to oxidant insults (2,6,7). The ability of TMCs to counter oxidative stress damage is critical to their survival and function (1). Under normal conditions, the trabecular meshwork (TM) is exposed to a constant low concentration of oxidative stress (8). The TMCs are relatively resistant to oxidative damage due to the expression of antioxidant enzymes that neutralize active substances (9). An imbalance between oxidants and antioxidants or excessive reactive oxygen species (ROS) accumulation may lead to oxidative stress in TMCs (10). Progression of POAG may accompany a reduction in TMC antioxidant capacity (8). Also, oxygen free radicals cause a gradual increase in oxidative damage, extracellular matrix (ECM) accumulation, and cytoskeletal changes in the structure of TMCs (11). Oxidative stress damage to ECM adhesion results in TMC adhesion, cell loss, and HTM integrity damage (12). The pathomechanism of TMCs, including oxidative stress and ECM deposition, caused resistance in AH outflow and elevated IOP (1,13-16). Therefore, ascertaining the molecular link between oxidative stress and excessive ECM production in TMCs may be a promising avenue in POAG treatment.
The analysis of diseased biosamples or tissue was based on high throughput assessment; however, sequencing in several studies has shown more credibility (17). The past decade has witnessed the development of next-generation sequencing and bioinformatics analysis methods, which have enabled genome-wide analysis to identify possible genes and biomarkers affecting disease-associated phenotypes. These have provided an accessible and straightforward way for researchers to focus on possible research targets and pathways.
In this study, we explored novel related genes and potential biomarkers affecting the pathology of TM injury. Adenosine A3 receptor (ADORA3), a protein that belongs to the family of adenosine receptors, was selected for further analysis (18-21). Further, we validated this novel selected gene in POAG patients. We hypothesized that ADORA3 might promote ECM protein expression in TMCs under oxidative stress, contributing to high IOP that finally leads to the POAG. To test this hypothesis, we validated ADORA3 expression in POAG tissues and its function in promoting ECM expression and inhibiting migratory functions in the TMCs under oxidative stress. The results provided a novel pathogenic mechanism underlying POAG and a potential therapeutic target. We present the following article following the MDAR reporting checklist (available at http://dx.doi.org/10.21037/atm-20-6154).
Methods
Ethics approval and consent to participate
All tissue samples in our study were collected according to the Declaration of Helsinki (as revised in 2013) on biomedical research involving human subjects. This study protocol was reviewed and approved by the Center for Medical Ethics Committees of the Third Xiangya Hospital of Central South University (Scientific Research Project: 2016-S147). The cells were obtained post-mortem from anonymized donor tissue and tissue after ophthalmectomy. All donors and their guardians provided written informed consent. The biosamples were collected and stored in Xiangya Biobank, Central South University.
Participants and tissue specimens
We obtained TM tissue from five healthy people and 22 POAG patients who underwent trabeculectomy between July 2018 and October 2019. The patients were diagnosed as POAG with elevated IOP according to the diagnosis and treatment guidelines of primary glaucoma in China. The control group consisted of age and sex-matched non-glaucomatous samples donated for corneoscleral rims that were unsuitable for transplant and from patients who underwent eyeball enucleation.
Data source and quality control
Our datasets were collected from the Gene Expression Omnibus (GEO) (http://www.ncbi.nlm.nih.gov/geo/) and ArrayExpress (http://www.ebi.ac.uk/arrayexpress) databases with the keywords: glaucoma, POAG, primary open-angle glaucoma, trabecular meshwork, and trabecular meshwork cell. After preliminary screening, five datasets (all from GEO) were included in our study: GSE4316, GSE27276, GSE126170, GSE27058, and GSE129183.
The GSE27058 and GSE129183 datasets were excluded from the later work. The GSE27058 dataset is a customized microarray chip using a custom Affymetrix Glyco v2 GeneChips and produced only 4,682 sites resulting in 752 human genes that could not be traced back to the samples in the dataset, which made it unsuitable for this analysis. The GSE129183 dataset uses methylation analysis; the data included 1 unpaired POAG sample and 5 normal samples, which were not suitable for our study. Furthermore, the intersection of the 5 datasets estimated nonstatistical significance results, making it impossible to obtain intersecting results. After evaluation and filtering, 3 gene expression profiles GSE27276 (13 controls and 15 POAG cases), GSE126170 [3 control human TMCs (HTMCs), and 3 POAG HTMCs], and GSE4316 (3 controls and 2 POAG cases) were downloaded from the GEO database and analyzed.
Identification of differentially expressed genes (DEGs)
The DEGs between different samples were analyzed using GEO2R. Acquired data were preprocessed and analyzed using the limma package for R (linear regression models), and the packages were available from Bioconductor (https://www.bioconductor.org/). The hierarchical clustering was generated and performed by R package ggplots (vision 2.2.1, http://cran.r-progect.org/web/Packages/ggplot 2). Moreover, the common DEG was performed by Venn diagram. The expression level with log2(fold-change) [log2(FC)] ≥1.5 and false discovery rate (FDR) <0.05 were considered statistically significant.
Pathway analysis
To identify the upregulated and downregulated genes within the functional pathway, we uploaded the genes to the Database for Annotation, Visualization, and Integrated Discovery (DAVID) (https://david.ncifcrf.gov/) to analyze with the Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway and Gene Ontology (GO) function. This grouped the genes into biological processes (BP), molecular function (MF), and cellular component (CC). The pathways network was visualized using KOBAS 3.0 (http://kobas.cbi.pku.edu.cn/). Next, we performed an overrepresentation analysis with a common cut-off criterion for the level of expression changes. This took the P value and absolute log2(FC) into consideration with gene expression. We set the threshold P at <0.05. The expression results were higher than 0.58, and P<0.05 was considered a significant expression change.
KEGG pathway
The pathways were performed from the free access pathway databases. In order to obtain more comprehensive pathway coverage, we took three datasets together. The pathway databases were analyzed by KEGG enrichment. The clustering of the pathways was performed by Cytoscape (www.cytoscape.org), an open-access tool, to visualize the genes and relatives between multiple categories.
GO analysis
We performed GO analysis to assess the pathology of glaucoma. The same functional terms were clustered together into functional categories. The results were obtained and compared with other genes in functional categories and visualized using KOBAS 3.0. The clusters were analyzed using DAVID and Metascape (http://metascape.org/). A P value <0.05 was considered statistically significant.
Construction of the protein-protein interaction (PPI) network
The differential expression of genes and proteins plays a critical role in disease processes. Moreover, the function of processes performed by proteins is coded by genes. The relationship between DEGs associated with glaucoma was displayed using the PPI network with STRING (https://string-db.org/) and viewed in Metascape.
Hematoxylin and eosin (HE) staining
Thin slices of TM tissue were obtained and fixed in 4% formaldehyde solution (pH: 7.0) for more than 24 h. Tissues were processed routinely for paraffin embedding, dehydrated using an alcohol gradient, cut into 4 μm thick sections, and placed on glass slides. Samples were stained with HE (Solarbio, Shanghai, China, No. G1120) and pathologists determined the histological type.
Histology immunohistochemistry
The TM tissues were immunostained with ADORA3 antibody (Abcam, Cambridge, MA, USA; Cat.ab203298). For immunohistochemistry (IHC), antigen retrieval was performed using a standard citrate buffer protocol. The expression was graded as follows: negative (score 0), weak (score 1), moderate (score 2), and strong (score 3). Percentage scores were assigned as 1, 1–25%; 2, 26–50%; 3, 51–75%; and 4, 76–100%. The score grades were calculated by three people and evaluated by pathologists.
Cell culture and treatment
Primary HTMCs were isolated and expanded from donors, as mentioned previously. Rings of TM were cut into pieces after dissection from corneoscleral rims and placed with 0.2% collagen coated beads (Sigma-Aldrich, St. Louis, MO, USA) incubating with Dulbecco’s Modified Eagle Medium/Nutrient Mixture F-12 (DMEM/F12) (Gibco, Grand Island, NY, USA), 15% fetal bovine serum (FBS; Gibco), 2 mM L-glutamine, and 1% penicillin-streptomycin (100 U/mL penicillin and 100 ng/mL streptomycin, P/S) (22). An additional primary HTMC line was obtained from ScienCell Research Laboratories Inc. (Catalog#6590; Carlsbad, CA, USA) and cultured in DMEM/F12, 15% FBS and 1% P/S, incubating at 37 °C in a 5% CO2 environment. We analyzed the protein level of myocilin after treatment with dexamethasone to identify the TM cells (Figure S1A). All experiments were conducted using primary HTMCs and cell lines within 6 passages. We named the cells as hTM-6, N49TM3, and N55TM4. The transwell migration assay was prepared as described previously (23).
H2O2 treatment model establishment
The HTMCs were seeded onto a 96-well plate (8×103 cells/well) and cultured in DMEM/F12 with 12% FBS and 1% PS for 24 h at 37 °C. The oxidative stress injury model was established using H2O2. The HTMCs were treated with DMEM/F12 containing H2O2 for 2 h in incrementally different concentrations from 50–800 μM (50, 100, 200, 300, 400, 600, 700, 800 μM). These cells were then compared with the non-treated cells. Next, the media was removed and replaced with 100 μL DMEM/F12 with 10 μL Cell Counting Kit-8 (CCK-8) reagent (Dojindo Laboratories, Kumamoto, Japan), and cells were incubated at 37 °C for 2 h. Absorbance was measured at 450 nm with the Tecan Infinite M200 multimode microplate reader (Tecan Group Ltd., Männedorf, Switzerland). Cell viability was calculated by percentage, with normal controls representing 100%.
ADORA3 plasmid and transfection
The open reading frames of the human ADORA3 sequence (CCDS839.1) were downloaded from the National Center for Biotechnology (NCBI) and cloned into the pcDNATM 3.1(+) by Tsingke technology company (Beijing, China). According to the manufacturer’s protocols, transfection was performed using Lipofectamine 3000 reagents (L3000015, Invitrogen, Eugene, OR, USA).
Total RNA isolation and quantitative reverse transcription-polymerase chain reaction (qRT-PCR)
Trizol Reagent (Invitrogen, USA) was used to extract total RNA. This was reverse transcribed to cDNA using a Super First-Strand Synthesis System (Takara, Shangdong, China). RT-PCR assays followed the protocol of the SYBR Prime Script TM RT-PCR Kit (Takara). The relative expression was calculated as relative expression = 2 – ΔCT × 10,000 (ΔCT = CTAODRA3 – CTGAPDH). The primer pairs were listed as follows: ADORA3: 5'-CCTGGGCATCACAATCCACT-3', 5'-ACCCTCTTGTATCTGACGGTA-3'; fibronectin (FN): 5'-GAGAATAAGCTGTACCATCGCAA-3', 5'-CGACCACATAGGAAGTCCCAG-3'; collagen-I (Col-I): 5'-ATCAACCGGAGGAATTTCCGT-3’; 5'-CACCAGGACGACCAGGTTTTC-3'; α-smooth muscle actin (α-SMA): 5'-CACCAGGACGACCAGGTTTTC-3'; 5'-AGAGACAGAGAGGAGCAGGA-3'; GAPDH: 5'-AGAGACAGAGAGGAGCAGGA-3'; 5'-GTCTTCTGGGTGGCAGTGAT-3'.
Western blotting (WB)
Cells were rinsed and lysed using lysis buffer [Cell Signaling Technology (CST), Cat. 9803] that included a protease inhibitor cocktail (CST, Danvers, MA, USA; Cat.5871) and PhosSTOP phosphatase inhibitor cocktail [MedChemExpress (MCE), Monmouth Junction, NJ, USA; Cat.4906845001]. A total of 20 mg protein was separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE). Proteins were transferred to a polyvinylidene fluoride (PVDF) membrane (Millipore, Boston, MA, USA), incubated with 5% milk for 1 h at room temperature, and then incubated with a primary antibody at 4 °C overnight. Following this, the membrane was incubated with a secondary antibody for 2 h at room temperature. Antibodies were listed as follows: FN (Proteintech, Rosemont, IL, USA; Cat:15613-1-AP); Col-I (Proteintech, Cat:14695-1-AP); α-SMA (Abcam Cat:ab5694); Tublin (Proteintech, Cat:11224-1-AP); Myocilin (Proteintech, Cat No. 14238-1-AP). Corresponding secondary antibodies, rabbit secondary antibody (CST, No. 7074), and mouse secondary antibody (CST, No. 7076), were selected according to the manufacturer’s instructions.
Statistical analysis
The statistical results and analysis were performed using Excel software, and the experiment results were finally analyzed with SPSS statistical software (version 18.0, IBM, Armonk, NY, USA). The statistical description of data was expressed as mean ± standard error of the mean (SEM) or standard deviation (SD), and each experiment data had at least 3 replicates. A P value <0.05 was considered significant.
Results
Identification of significant differential genes
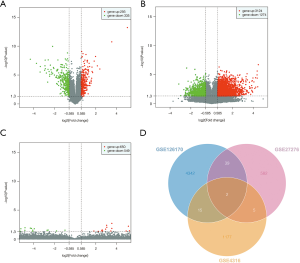
Only two genes intersected all three tested datasets. We used these intersection results for further statistical analysis. Among the tested mRNAs/genes, 293 and 335 were upregulated and downregulated, respectively, in the GSE27276 dataset (Figure 1A); there were 3,124 and 1,274 were upregulated and downregulated in GSE126170 (Figure 1B); and 650 and 549 upregulated and downregulated, respectively, in GSE4316 (Figure 1C). A total of 15 DEGs were identified between GSE4316 vs. GSE126170. More DEGs (39 genes) were identified between GSE126170 vs. GSE27276. Only five DEGs were identified between GSE4316 vs. GSE27276. We focused on the common DEGs between these datasets. The 61 common DEGs are listed in Table 1. In this study, two genes were identified, as shown in a Venn diagram (Figure 1D); ADORA3 and DNA damage-inducible transcript 4 protein (DDIT4) were significantly dysregulated in the POAG TM samples compared with the healthy controls in three datasets.

Full table
Dysregulated DEGs in datasets
The 61 intersecting genes were subjected to GO and KEGG enrichment analysis. The top 10 enrichment pathways in GO and KEGG, including the BPs, CCs, and MFs, are listed in Figure 2.
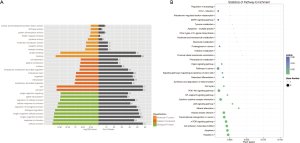
The GO functional enrichment analysis results suggested that the most highly regulated genes were significantly enriched in “cellular process” (ontology: BP), “binding” (ontology: MF), and “cell part” (ontology: CC). The GO results are listed in Figure 2A. The results showed that the majority of BPs focused on “cellular process”, “response to stimulus”, and “single-organism process”. The “cell part”, “cell” and “membrane-bounded organelle” accounted for most CC terms. Furthermore, most enrichment MFs were reported as “binding”, “protein binding”, and “receptor binding”. The top10 DEGs in the MF pathways were associated with “apoptosis”, “cell adhesion molecules (CAMs)”, and “mTOR signaling pathway”.
Several genes were tightly related to several pathways, such as BIRC3, BCL2A1, CLDN1, CSF1R, GADD45A, FZD10, IFNA5, and MAOB. The gene BCL2A1 was enriched in the “mTOR signaling pathway”, “herpes simplex infection”, “hepatitis C”, and “cytokine-cytokine receptor interaction”. Furthermore, the top 10 DEGs were enriched in “apoptosis”, “transcriptional misregulation in cancer”, “mTOR signaling pathway”, and “PI3K-Akt signaling pathway” (Figure 2B). Interestingly, the enrichment of apoptosis and the PI3K-Akt signaling pathway were associated with TM injury.
Cluster analysis, signaling pathways, and PPI network
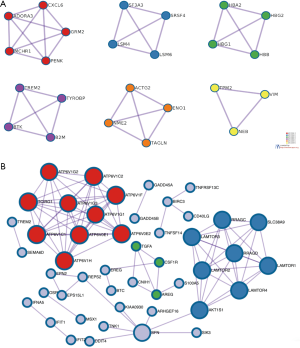
To analyze the activation pathways of pathophysiological changes in TM during POAG progression, we used the 61 intersecting genes to build the PPI network and cluster by MCODE. These results revealed that MCODE 1 was driven by ADORA3, CCXCL6, MCHR1, GRM2, and PENK (Figure 3A). The PPI network was built using STRING to demonstrate the inter-relationship of the 61 significantly expressed genes (Figure 3B). This analysis created a PPI network with 69 nodes and 187 edges. The relevance score cut-off was set as 0.8.
ADORA3 was upregulated in POAG specimens and the bioinformatics analysis
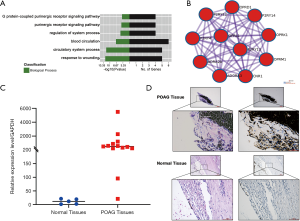
Dysregulated gene expression is considered a high-risk factor for diseases; therefore, ADORA3 was selected as our research target. The GO results showed that ADORA3-relative pathways were focused on the “adenosine receptor signaling pathway”, “G protein-coupled receptor signaling pathway”, “inflammatory response”, and “negative regulation of nuclear factor kappa light chain enhancer of activated B cells (NF-κB) transcription factor activity” in non-tumor pathways. In cancer pathways, ADORA3 showed a significantly negative regulation in “cell proliferation and migration” (Figure 4A). Additionally, we created a PPI network with ADORA3 as the independent factor, which incorporated 11 nodes and 55 edges (Figure 4B). We validated ADORA3 via RT-PCR based on the log2(FC) expression value using 10 samples of POAG TM tissue and 5 healthy controls. These results revealed significantly upregulated ADORA3 mRNA in the POAG TM specimens (100%) compared to healthy controls (Figure 4C). Furthermore, the HE results showed structural damage in the TM tissue in the POAG samples compared with the control tissue (Figure 4D). The IHC results confirmed the overexpression of ADORA3 in the HTM of POAG samples compared with healthy controls (Figure 4D).
ADORA3 leading to TM oxidative stress injury
To further investigate the functional role of ADORA3 in the pathogenesis of POAG, we used the three sources of HTMCs (hTM-6, N49TM3, and N55TM4) and established an oxidative stress injury model via treatment with H2O2. We used CCK-8 to find the maximal nonlethal dose of H2O2 to treat HTMCs and found that 300 μΜ was the optimal concentration (Figure 5A and Figure S1B). First, we observed that ADORA3 overexpression reduced cell viability in H2O2-treated vs. nontreated cells in all three HTMCs (Figure 5B). Previous studies have shown that high IOP glaucoma damages the TM and leads to collagen deposition, cellular conformational changes, and induces the overexpression of FN, Col-I, and α-SMA (1,24,25). The mRNA expression of these genes was upregulated in the H2O2-treated cells compared with the nontreated cells in all 3 HTMCs (Figure 5C). The WB analysis revealed increased overexpression of ADORA3 was associated with a significant increase of FN, Col-I, and α-SMA protein expression (Figure 5D). Moreover, in the ADORA3 group overexpressions, the expression of FN, Col-I, and α-SMA showed a more significantly upregulated tendency in the ADORA3-overexpressed H2O2 group. These results indicated that ADORA3 protein has a regulatory effect in the HTM oxidative injury of POAG.
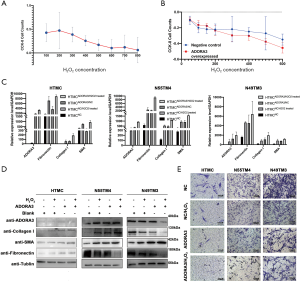
The migration and mobility of HTMCs can directly affect AH outflow and IOP; therefore, we analyzed the migration capability of HTMCs in the oxidative injury model via transwell migration assay. A significant reduction of cell migration was observed in the H2O2-treated cells compared with the untreated groups. Moreover, the untreated ADORA3 overexpressed cells also showed decreased cell migration, and the greatest reduction of migration capability was in the ADORA3-overexpressed H2O2 group (Figure 5E).
Discussion
High IOP is an important factor of POAG (2,26). Oxidative stress is a major pathogenic causative factor of malfunction in the TM (9,10). The dysfunction blocks the dynamic equilibrium of AH flow, leading to elevated IOP (13). Previous studies have shown that the TMCs in POAG have a noticeable variation in autophagy dysfunction and immune inflammation (6,27). Additionally, ROS and oxidative stress are associated with HTMC dysfunction (28). Collagen and elastic fiber degeneration in the TM induces increased deposition of the ECM (28). Moreover, other characteristics, including non-coding RNAs (28), pathological genes, and related mutations (29), contribute to this injury.
In this study, we applied systematic bioinformatics analysis to 3 individual RNA microarrays from the GEO database. We analyzed the significant DEGs in all 3 datasets to obtain the intersecting genes that were high-risk factors. We constructed a PPI network with 61 DEGs and analyzed the MCODE in the network from these data. The GSE4316 dataset contains genome-wide HTM tissue genes from three pairs of control and two pairs of POAG samples. The GSE27276 dataset contains samples from one patient with POAG with the myocilin mutation, limiting the statistical analysis for all DEGs; therefore, this dataset yielded the lowest results. The GSE126170 dataset mainly focused on the expression and role of expressed IncRNA; therefore, it provided the most DEGs in this analysis. Each dataset was analyzed using a different platform, which led to a significant difference in the number of DEGs extracted between datasets. To investigate the extracted intersecting genes’ function, we analyzed the network and found 6 cluster modules (Figure 3A). The top 1 MCODE, which was driven by ADORA3, was correlated with downstream pathways, including G alpha (I) signaling events, GPCR ligand binding, and Class A/1 (rhodopsin-like receptors). In our screening and intersection results, two significantly dysregulated genes (ADORA3 and DDIT4) showed the highest probability to affect the function of HTM in POAG. Therefore, we hypothesized that ADORA3 and DDIT4 might be TM regulators in POAG. The functioning of ADORA3 is related to adenosine receptor signaling, inflammatory response, and signal transduction (30-33). The function of DDIT4 is associated with hypoxia response, neuron migration, and apoptotic process.
The gene ADORA3 is an adenosine receptor subtype (34). Adenosine is a purine nucleoside that is ubiquitously present in extracellular spaces (34,35). A previous study assessing adenosine triphosphate ATP release mechanisms reported that the first step of AH dynamics in purinergic regulation is the cellular release of ATP. Also, ATP-derived adenosine affects the outflow pathway of IOP in purinergic regulation (36). Interestingly, adenosine agonists can reduce TM cell volume by targeting ATP-released adenosine (37), and purinergic receptors can influence the AH dynamics (38,39).
Moreover, hypoxia and mechanical stress increase the release of adenosine (35). The upregulation of ADORA3 plays a role in the pathogenesis of a variety of diseases, including cell apoptosis and ischemia-reperfusion injury in chronic heart failure (40,41), Wnt signaling pathway in cancer (42), apoptosis in hepatocellular carcinoma (43), and pro-inflammatory regulation in pulmonary ischemia-reperfusion injury (44). Inhibition of ADORA3 in a diabetic rat model’s kidneys serves as an upstream factor that influences downstream factors’ expression in the inflammatory response (19). Additionally, ADORA3 is expressed at a low level in the TM in Rhesus monkeys under normal conditions (45). The upregulation of ADORA3 has been observed in pseudoexfoliation syndrome, which can be a complication of POAG, suggesting that ADORA3 may participate in the pathogenies of HTM (46).
Moreover, elevated expression of ADORA3 may participate in the oxidative stress response (32). Our study used RT-PCR and IHC methods to reveal an upregulation of ADORA3 in POAG HTMC at the transcriptional and post-transcriptional levels compared with control samples. We hypothesized that this overexpression induces cellular injury and alterations of HTM tissue structure.
The transwell cell migration assay was used to evaluate HTMC injury. In pigmentary glaucoma, the migration of HTMCs was largely inhibited through the Rho signaling pathway (47). A decrease in cell migration influences the compliance of HTM, which may further decrease the outflow of AH and increase IOP (48). Moreover, oxidative stress could also damage the cell proliferation and migration function of HTMCs (23,48). In this study, upregulated ADORA3 in HTMCs inhibited cell migration. These data suggest that ADORA3 influences the migration of HTMCs and the outflow of AH, which may be plausible pathogenesis for POAG.
Fibrosis of HTM is a characteristic pathologic hallmark of POAG; therefore, we hypothesized that aberrant ADORA3 expression would lead to HTM fibrosis. A previous study reported that ADORA3 antagonism could reduce kidney fibrosis injury in diabetic rats (20). The deposition of ECM, including FN, Col-I, and α-SMA, may lead to intracellular and extracellular cell contractility changes, cytoskeleton collapse, collagen production, and deposition (1,28). Additionally, excessive ECM deposition increases AH outflow resistance and elevates IOP, which occurs in the POAG (49,50). Our results revealed that profibrotic ECM protein expression was upregulated in the POAG overexpressed HTMCs, indicating that ADORA3 participated in POAG.
In our study, the experimental in vitro POAG model we established used an H2O2 oxidative stress injury model in HTMC, which has been previously reported in TM injury studies (23). Moreover, we performed further experiments on HTMCs using the H2O2-treated model and untreated controls. We overexpressed ADORA3 via plasmid transfection in normal and H2O2-treated cells, which was confirmed via an increase in ADORA3 mRNA and protein expression. Next, we explored the dysregulation of ADORA3 and the relative injury factors in POAG and TM injury’s inflammatory response progression. The RT-PCR and WB results showed an increase in mRNA and protein expression of FN, Col-I, and α-SMA, which was associated with ADORA3 overexpression in each group.
Conclusions
In conclusion, we analyzed three datasets to identify the intersecting regulators of the pathological process of POAG. We extracted the DEGs and found that upregulated ADORA3 showed an inflammatory response function during TM oxidative stress injury. Our results provide a viable molecular target of the underlying pathological mechanism of TM damage in POAG.
Acknowledgments
Funding: This study was supported by the Key Research and Development Program of Hunan Province (2018SK2090); the National Key Research and Development Program of China (2016YFC1201800); the National Natural Science Foundation of China (81671225, 81871089); and the Project of Graduate Independent Exploration and Innovation Plan of Central South University (2020zzts218).
Footnote
Reporting Checklist: The authors have completed the MDAR reporting checklist. Available at http://dx.doi.org/10.21037/atm-20-6154
Data Sharing Statement: Available at http://dx.doi.org/10.21037/atm-20-6154
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/atm-20-6154). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. All the tissue samples in our study were collected according to the Declaration of Helsinki (as revised in 2013) on biomedical research involving human subjects. This study protocol was reviewed and approved by the Center for Medical Ethics Committees of the Third Xiangya Hospital of Central South University (Scientific Research Project No. 2016-S147). Written informed consent was provided by all donors and their guardians.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Wang M, Zheng Y. Oxidative stress and antioxidants in the trabecular meshwork. PeerJ 2019;7:e8121 [Crossref] [PubMed]
- Wang N, Chintala SK, Fini ME, et al. Activation of a tissue-specific stress response in the aqueous outflow pathway of the eye defines the glaucoma disease phenotype. Nat Med 2001;7:304-9. [Crossref] [PubMed]
- Fechtner RD, Weinreb RN. Mechanisms of optic nerve damage in primary open angle glaucoma. Surv Ophthalmol 1994;39:23-42. [Crossref] [PubMed]
- Zhavoronkov A, Izumchenko E, Kanherkar RR, et al. Pro-fibrotic pathway activation in trabecular meshwork and lamina cribrosa is the main driving force of glaucoma. Cell Cycle 2016;15:1643-52. [Crossref] [PubMed]
- Boland MV, Ervin AM, Friedman DS, et al. Comparative effectiveness of treatments for open-angle glaucoma: a systematic review for the US Preventive Services Task Force. Ann Intern Med 2013;158:271-9. [Crossref] [PubMed]
- Baleriola J, Garcia-Feijoo J, Martinez-de-la-Casa JM, et al. Apoptosis in the trabecular meshwork of glaucomatous patients. Mol Vis 2008;14:1513-6. [PubMed]
- Wang M, Wan H, Wang S, et al. RSK3 mediates necroptosis by regulating phosphorylation of RIP3 in rat retinal ganglion cells. J Anat 2020;237:29-47. [Crossref] [PubMed]
- Ammar DA, Hamweyah KM, Kahook MY. Antioxidants protect trabecular meshwork cells from hydrogen peroxide-induced cell death. Transl Vis Sci Technol 2012;1:4. [Crossref] [PubMed]
- Abu-Amero KK, Kondkar AA, Mousa A, et al. Decreased total antioxidants status in the plasma of patients with pseudoexfoliation glaucoma. Mol Vis 2011;17:2769-75. [PubMed]
- Aydın Yaz Y, Yıldırım N, Yaz Y, et al. Role of oxidative stress in pseudoexfoliation syndrome and pseudoexfoliation glaucoma. Turk J Ophthalmol 2019;49:61-7. [Crossref] [PubMed]
- Gabelt BT, Kaufman PL. Changes in aqueous humor dynamics with age and glaucoma. Prog Retin Eye Res 2005;24:612-37. [Crossref] [PubMed]
- Zhou L, Li Y, Yue BY. Oxidative stress affects cytoskeletal structure and cell-matrix interactions in cells from an ocular tissue: the trabecular meshwork. J Cell Physiol 1999;180:182-9. [Crossref] [PubMed]
- Tripathi RC. Mechanism of the aqueous outflow across the trabecular wall of Schlemm's canal. Exp Eye Res 1971;11:116-21. [Crossref] [PubMed]
- Nettesheim A, Shim MS, Hirt J, et al. Transcriptome analysis reveals autophagy as regulator of TGFbeta/Smad-induced fibrogenesis in trabecular meshwork cells. Sci Rep 2019;9:16092. [Crossref] [PubMed]
- Saccà SC, Izzotti A, Rossi P, et al. Glaucomatous outflow pathway and oxidative stress. Exp Eye Res 2007;84:389-99. [Crossref] [PubMed]
- Vranka JA, Kelley MJ, Acott TS, et al. Extracellular matrix in the trabecular meshwork: intraocular pressure regulation and dysregulation in glaucoma. Exp Eye Res 2015;133:112-25. [Crossref] [PubMed]
- Thomson BR, Carota IA, Souma T, et al. Targeting the vascular-specific phosphatase PTPRB protects against retinal ganglion cell loss in a pre-clinical model of glaucoma. Elife 2019;8:e48474 [Crossref] [PubMed]
- He HR, Li YJ, He GH, et al. The polymorphism in ADORA3 decreases transcriptional activity and influences the chronic heart failure risk in the Chinese. Biomed Res Int 2018;2018:4969385 [Crossref] [PubMed]
- Garrido W, Jara C, Torres A, et al. Blockade of the adenosine A3 receptor attenuates caspase 1 activation in renal tubule epithelial cells and decreases interleukins IL-1beta and IL-18 in diabetic rats. Int J Mol Sci 2019;20:4531. [Crossref] [PubMed]
- Antonioli L, Blandizzi C, Csoka B, et al. Adenosine signalling in diabetes mellitus--pathophysiology and therapeutic considerations. Nat Rev Endocrinol 2015;11:228-41. [Crossref] [PubMed]
- Fredholm BB, Chen JF, Masino SA, et al. Actions of adenosine at its receptors in the CNS: insights from knockouts and drugs. Annu Rev Pharmacol Toxicol 2005;45:385-412. [Crossref] [PubMed]
- Keller KE, Bhattacharya SK, Borrás T, et al. Consensus recommendations for trabecular meshwork cell isolation, characterization and culture. Exp Eye Res 2018;171:164-73. [Crossref] [PubMed]
- Xu L, Zhang Y, Guo R, et al. HES1 promotes extracellular matrix protein expression and inhibits proliferation and migration in human trabecular meshwork cells under oxidative stress. Oncotarget 2017;8:21818-33. [Crossref] [PubMed]
- Filla MS, Faralli JA, Desikan H, et al. Activation of αvβ3 integrin alters fibronectin fibril formation in human trabecular meshwork cells in a ROCK-independent manner. Invest Ophthalmol Vis Sci 2019;60:3897-913. [Crossref] [PubMed]
- Patel GC, Millar JC, Clark AF. Glucocorticoid receptor transactivation is required for glucocorticoid-induced ocular hypertension and glaucoma. Invest Ophthalmol Vis Sci 2019;60:1967-78. [Crossref] [PubMed]
- Weinreb RN, Aung T, Medeiros FA. The pathophysiology and treatment of glaucoma: a review. JAMA 2014;311:1901-11. [Crossref] [PubMed]
- Grant WM. Experimental aqueous perfusion in enucleated human eyes. Arch Ophthalmol 1963;69:783-801. [Crossref] [PubMed]
- Welge-Lüssen U, Birke K. Oxidative stress in the trabecular meshwork of POAG. Klin Monbl Augenheilkd 2010;227:99-107. [PubMed]
- Guo JH, Su C, Jiang SY, et al. MicroRNA-1 regulates fibronectin expression in human trabecular meshwork cells under oxidative stress. Zhonghua Yan Ke Za Zhi 2019;55:355-60. [PubMed]
- Iyer SV, Ranjan A, Elias HK, et al. Genome-wide RNAi screening identifies TMIGD3 isoform1 as a suppressor of NF-κB and osteosarcoma progression. Nat Commun 2016;7:13561. [Crossref] [PubMed]
- Palmer TM, Harris CA, Coote J, et al. Induction of multiple effects on adenylyl cyclase regulation by chronic activation of the human A3 adenosine receptor. Mol Pharmacol 1997;52:632-40. [Crossref] [PubMed]
- Bouma MG, Jeunhomme TM, Boyle DL, et al. Adenosine inhibits neutrophil degranulation in activated human whole blood: involvement of adenosine A2 and A3 receptors. J Immunol 1997;158:5400-8. [PubMed]
- Dougherty C, Barucha J, Schofield PR, et al. Cardiac myocytes rendered ischemia resistant by expressing the human adenosine A1 or A3 receptor. FASEB J 1998;12:1785-92. [Crossref] [PubMed]
- Guzman-Aranguez A, Gasull X, Diebold Y, et al. Purinergic receptors in ocular inflammation. Mediators Inflamm 2014;2014:320906 [Crossref] [PubMed]
- Agarwal R, Agarwal P. Newer targets for modulation of intraocular pressure: focus on adenosine receptor signaling pathways. Expert Opin Ther Targets 2014;18:527-39. [Crossref] [PubMed]
- Li A, Leung CT, Peterson-Yantorno K, et al. Mechanisms of ATP release by human trabecular meshwork cells, the enabling step in purinergic regulation of aqueous humor outflow. J Cell Physiol 2012;227:172-82. [Crossref] [PubMed]
- Fleischhauer JC, Mitchell CH, Stamer WD, et al. Common actions of adenosine receptor agonists in modulating human trabecular meshwork cell transport. J Membr Biol 2003;193:121-36. [Crossref] [PubMed]
- Burnstock G, Vaughn B, Robson SC. Purinergic signalling in the liver in health and disease. Purinergic Signal 2014;10:51-70. [Crossref] [PubMed]
- Fonseca B, Martínez-Águila A, Pérez de Lara MJ, et al. Changes in P2Y purinergic receptor expression in the ciliary body in a murine model of glaucoma. Front Pharmacol 2017;8:719. [Crossref] [PubMed]
- Hannon JP, Pfannkuche HJ, Fozard JR. A role for mast cells in adenosine A3 receptor-mediated hypotension in the rat. Br J Pharmacol 1995;115:945-52. [Crossref] [PubMed]
- Boros D, Thompson J, Larson DF. Adenosine regulation of the immune response initiated by ischemia reperfusion injury. Perfusion 2016;31:103-10. [Crossref] [PubMed]
- Fishman P, Bar-Yehuda S, Madi L, et al. A3 adenosine receptor as a target for cancer therapy. Anticancer Drugs 2002;13:437-43. [Crossref] [PubMed]
- Ayoub BM, Attia YM, Ahmed MS. Structural re-positioning, in silico molecular modelling, oxidative degradation, and biological screening of linagliptin as adenosine 3 receptor (ADORA3) modulators targeting hepatocellular carcinoma. J Enzyme Inhib Med Chem 2018;33:858-66. [Crossref] [PubMed]
- Zhou Y, Murthy JN, Zeng D, et al. Alterations in adenosine metabolism and signaling in patients with chronic obstructive pulmonary disease and idiopathic pulmonary fibrosis. PLoS One 2010;5:e9224 [Crossref] [PubMed]
- Beach KM, Hung LF, Arumugam B, et al. Adenosine receptor distribution in Rhesus monkey ocular tissue. Exp Eye Res 2018;174:40-50. [Crossref] [PubMed]
- Schlötzer-Schrehardt U, Zenkel M, Decking U, et al. Selective upregulation of the A3 adenosine receptor in eyes with pseudoexfoliation syndrome and glaucoma. Invest Ophthalmol Vis Sci 2005;46:2023-34. [Crossref] [PubMed]
- Koga T, Koga T, Awai M, et al. Rho-associated protein kinase inhibitor, Y-27632, induces alterations in adhesion, contraction and motility in cultured human trabecular meshwork cells. Exp Eye Res 2006;82:362-70. [Crossref] [PubMed]
- Last JA, Pan T, Ding Y, et al. Elastic modulus determination of normal and glaucomatous human trabecular meshwork. Invest Ophthalmol Vis Sci 2011;52:2147-52. [Crossref] [PubMed]
- Acott TS, Kelley MJ. Extracellular matrix in the trabecular meshwork. Experimental Eye Research 2008;86:543-61. [Crossref] [PubMed]
- Vranka JA, Bradley JM, Yong-Feng Y, et al. Mapping molecular differences and extracellular matrix gene expression in segmental outflow pathways of the human ocular trabecular meshwork. PLoS One 2015;10:e0122483 [Crossref] [PubMed]


