
Narrative review of generative adversarial networks in medical and molecular imaging
Introduction
Recent years have witnessed the rapidly expanding use of artificial intelligence (AI) and machine learning (ML) in medical imaging. However, as a “data-hungry” technology, large datasets are indispensable to meet the increasing demand of high-quality images (1) as training sets, preferably addressing all aspects of radiology, including interventional, neuroradiology, and molecular imaging. As such, extensive imaging datasets must be pooled from various centers, reviewed, and appropriately annotated by experts. Given the increasing number of different imaging techniques applied to a wide variety of disease conditions, new approaches are needed to overcome the hurdles of traditional data collection and to provide an appropriate number of scans that can be utilized for training of AI methods.
In this regard, Goodfellow and coworkers have introduced generative adversarial networks (GANs), which generate synthetic data with similar characteristics as their “real” counterparts (2). Such artificially created images can be added to existing datasets and may provide a larger number of images to enhance the variety within a dataset and, ultimately, to improve ML algorithms. Further applications of GANs in medical imaging include augmenting datasets of patients afflicted with orphan diseases (3) or to duplicate rare presentations of more common diseases that would not be encountered to the extent that an effective ML algorithm could be trained from real images (4-8). Moreover, in laboratory animal research, replacement is seen as the ultimate goal to further reduce the use of live animals (9) enabling GANs to open doors for a practical implementation potentially simulating disease onset or progression.
Given the increasing number of applications for GANs in medical imaging, this narrative review aimed to provide a comprehensive overview of established methods for estimating generative models via an adversarial process (including data augmentation, domain translation, de-noising, super-resolution (SR), domain adaptation, and image generation with disease severity and radiogenomics) and to highlight future aspects in the highly innovative and rapidly developing field of AI. GANs for natural images has been proposed and its usefulness has been validated. However, to the best of our knowledge, there are no published review papers on GANs for medical and molecular imaging.
We present the following article in accordance with the Narrative Review reporting checklist (available at http://dx.doi.org/10.21037/atm-20-6325).
General techniques of deep learning with GANs
Convolutional neural network (CNN)
Based on Huber-Wiesel’s hierarchical hypothesis of visual information processing, a CNN consists of convolutional layers that extract the patterns in images and pooling layers to provide shift-invariance of the patterns. In a multi-layered CNN, local features can be extracted from shallow layers and global features from deep layers (10).
GAN and DCGAN
Proposed by Goodfellow et al. (2), GANs consist of two sub-networks, a generator and a discriminator. The generator takes a latent (random) vector as an input and generates an image that is very similar to the training data (real image). The discriminator takes the generated or real image as an input and determines whether the input is fake or real. By training the generator and discriminator to compete with each other, the GAN will be able to generate images that more closely resemble real data. GAN has become a remarkable technology because of its success in generating clearer images than the conventional method of variational auto-encoding.
Deep convolutional GAN (DCGAN) is a model that applies a CNN to a GAN to improve the quality of the generated images (11). In order to improve authenticity judgment, the discriminator uses convolution to extract features from the image. For discriminators, convolution technique to extract features of the image and the generator has a high resolution to generate the image, while for generators, a type of transposed convolution is employed for generating high-resolution images.
Conditional GAN (cGAN)
Aiming at generating images of a certain modality, disease condition, or specific region of the human body by exclusively using typical GANs or DCGANs, a separate network model for each condition has to be established and appropriately trained. The generator in the cGAN takes not only latent vectors but also conditional labels as input and thus, the cGAN is capable of learning the relationship between real and fake (12). As the training proceeds, the generator can create an image with a given conditional label corresponding to situations such as brain magnetic resonance images (MRI) after stroke, computed tomography (CT) images displaying lung nodules, or oncologic positron emission tomography (PET) images (3,13,14).
Pix2Pix
As an image-to-image translation, Pix2Pix is a GAN that uses images instead of labeled features, such as specific disease conditions (15). Using real image pairs and pairs of real and generated images, Pix2Pix learns the relationships between images and such a pair-wise learning approach allows the conversion of an image from one domain (modality) to another. For instance, in medical imaging, the Pix2Pix is utilized to convert MRI to CT images for the same subject and region (16).
CycleGAN
The Pix2Pix method requires a large amount of paired data to perform pair-wise learning. Paired training data, however, will not always be available and thus, recent developments of GANs allow for the translation of an image from a source domain to a target domain, even if paired datasets are not available (17).
For the two domain datasets A and B, CycleGAN provides two generators and two discriminators, that is, generators that converts an image of domain A (or B) into domain B (or A), and discriminators that determine whether an image for domain B (or A) is real or generated one from domain A (or B). For instance, CycleGAN could generate a CT image from an MR image of another subject without the need of additional imaging.
Progressive growing GAN (PGGAN)
As a major drawback, high-resolution images cannot be generated by using GANs and DCGANs. PGGANs, however, start with generation of low resolution images (overall features), and gradually increase the resolution of the generated images (detailed features) (18). In addition to continuous training to reach higher resolution of the duplicated images, the so-called minibatch standard deviation was also developed to guarantee diversity of generated images.
Super-resolution GAN (SRGAN)
SRGAN enables SR processing, generating high-resolution images, also described as photo-realistic natural images for 4x upscaling factors (19). Such a deep residual network allows for recovery of photo-realistic textures, even from heavily down sampled images (19).
GANs in medical and molecular imaging
PubMed, IEEE, Google Scholar and arXiv were searched for relevant literature from January 2017 to May 2020 using with the following keywords: “artificial intelligence”, “neural network”, “deep learning”, “generative adversarial networks”, “GAN”, “GANs”, “medical image”, “medical imaging”, “molecular imaging”, “nuclear medicine”, “MRI”, “CT”, “PET”, “SPECT”, “X-ray”, “data augmentation”, “modality conversion”, “de-noising”, “image reconstruction”, “super-resolution”, “domain adaptation”, “image generation”. The selected language is English. Therefore, database and language biases are limitations of this review articles. Although literatures from arXiv were preprints, those were cited in order to keep up with the lasted trends of GANs. Thirty-eight literature was reviewed in the following sections. Twenty-one of them reported that publicly available datasets were utilized as training data.
Image synthesis
Some of the aforementioned techniques have already been applied to different imaging modalities and diseases. Given the relatively static and invariable anatomy of the brain compared to other body parts such as the lower abdomen, the vast majority of these studies have been applied to brain disorders.
2D-sectional images—GANs applied to MRI
First, data augmentation techniques have been proposed for 2D sectional images derived from MRI. For instance, a DCGAN was utilized to synthesize artificial MR images using T1W images of healthy subjects and patients afflicted with recent stroke. The likelihood that images were DCGAN-created versus acquired was evaluated by a mix of neuroradiologists and radiologists from other subspecialties in a binary fashion to identify real vs. created images. In this quality control study, DCGAN-created brain MR images were able to convince neuroradiologists that they were viewing true images instead of artificial brain MR images. Thus, DCGAN-derived brain MRI may be nearing readiness to be implemented for synthetic data augmentation for data-hungry technologies, such as supervised ML, which in turn can pave the way to incorporate these techniques in even highly complex medical imaging cases (3).
In a separate study, a GAN-based network model was trained with brain MR images from T1-weighted (T1), post-contrast T1-weighted (T1Gd), T2-weighted (T2), and T2 fluid attenuated inversion recovery (FLAIR) sequences (20) demonstrating semantic segmentation of enhancing tumor, peritumoral edema, and necrosis (non-enhancing tumor core) regions on gliomas. Supervised data including generated images improved the segmentation performance, again potentially addressing the scarcity of readily available medical images (20).
CycleGAN is one of the standard methods for learning the relationships between different datasets. Ideally, when an image IB is generated for dataset B using an image IA in dataset A, the image IA is the same as I’A generated using the image IB. However, it is not guaranteed that the mapping between the two image datasets is unique. A so-called one2one CycleGAN was proposed to realize the mapping uniqueness (21) and the results showed the superiority of the opposed method over the baseline CycleGAN. Using a small sample size dataset in training GANs could result in synthesizing low diversity and/or low-quality images. A network model was developed for tumor image generation with even a limited sample dataset as the training data (22). That model consisted of components with clear roles generating shape and texture of tumors, and produced tumor images by merging the generated tumor into background tumor-free images. Experiments on FLAIR images with 19/20 co-gain absent/present (control/mutated) classes showed that the proposed method generated high quality images compared to conventional network models, and captured the characteristics of the two tumor classes.
Reflecting clinical reality, datasets consisting of multiple diseases have recently been investigated, but resulting inter-class imbalances in sample sizes have to be addressed. As such, a recently introduced GAN was conditioned on global information such as acquisition configuration (scanner type, protocol, etc.) or lesion type in addition to local information from segmentation masks on the lesions. The learning of the developed GAN was performed with brain glioma MR images (FLAIR, T2, and T1) and dermoscopic images of skin lesions (melanoma, seborrheic keratosis, and nevus), respectively. The GAN succeeded in synthesizing realistic images for both MR imaging and dermoscopic imaging (23).
2D-sectional images—GANs applied to chest X-ray, mammography, and PET
Apart from MRI, data augmentation of 2D images by GAN has also been used in conventional X-ray. For instance, a combination of real and artificial images to train a deep convolutional neural network (DCNN) to detect pathology across five classes of chest X-rays (cardiomegaly, normal, pleural effusion, pulmonary edema, and pneumothorax) was used. Augmenting the original dataset with GAN-generated images significantly improved performance of chest pathology classification when compared to the accuracy of the original dataset (24).
Regarding other modalities, for mammography, generating synthetic full field digital mammograms of 1280×1024 pixels was accomplished with a progressive growing GAN (25). For PET imaging, a cGAN and DCGAN-based model (RADIOGAN) succeeded in generating maximum intensity projection images of normal, head & neck cancer, esophageal cancer, lung cancer, and lymphoma with a small sample size for each class of lesion (14).
3D images—GANs applied to MRI
Over time, volumetric imaging has become increasingly important in modern medical imaging. It has been reported that the generation of 3D brain MR images was possible with a small set of training data from a network model with a combination of a variational auto-encoder (VAE) and a GAN (26). The model outperformed baseline models in both quantitative and qualitative measurements for generating normal brain images, and also successfully generated FLAIR, T1W images for tumor and T2W images for stroke as shown in Figure 1.
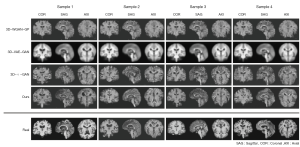
Other groups have used similar techniques for other imaging modalities. For example, with FDG-PET images, a 3D cGAN model with 3D U-Net like generator was developed to convert low injected dose images to high dose images (27). After training with full-dose (an average of 203 MBq) and low-dose (about a quarter of full-dose) images, the proposed model outperformed the state-of-the art methods in both qualitative and quantitative measures.
A GAN-based technique was utilized to produce smooth CT interpolations with high quality and accuracy of human organ structures in order to fill the gaps between adjacent CT slices (28). The experiments that were performed showed great improvement on two difference matrix sizes (256×256 and 512×256) when compared quantitatively and qualitatively with conventional methods based on optimizing mean squared error or a perceptual loss between ground truth and interpolated slices.
Modality conversion (domain translation, image-to-image)
Generation of CT images from MR images
Methods for conversion of MR images to CT images or vice versa are an area that is being actively studied. A number of GAN-based methods have been proposed, and in this section, we will discuss the most recent advances.
MR to CT conversion is a particularly salient example of where modality conversion could be utilized in routine clinical practice. Potential applications include bone segmentation in MR images and estimation of attenuation maps in PET imaging (i.e., improved attenuation correction for PET/MR). For example, Hiasa et al. performed a conversion in 2D from MR to CT images by applying a CycleGAN technique trained with T1W MR and CT images of lower abdominal regions including the hip joints (29).
Again, emphasizing the importance of volumetric imaging, conversion in 3D has also been accomplished. A GAN based on a 3D fully CNN was used (30). The method significantly outperformed state-of-the-art methods in prediction of brain and pelvis CT images with measurement of mean absolute error and peak signal-to-noise ratio.
With the growing installed base of PET/MR scanners, the need for accurate methods to perform attenuation correction from MR is acute. Estimation of attenuation maps from non-attenuation corrected data have been investigated. Briefly, the generator received a non-attenuation corrected PET (NAC PET) image and synthesized the pseudo-CT image (31). Training data consisted of 50 paired CT and NAC PET images and validation data of 20 paired CT and NAC PET images. Minimal underestimation was present (<5%) for standardized uptake values (SUVs) in all brain regions on the NAC PET images corrected for attenuation with the synthesized CT images when compared to the attenuation-corrected PET images.
Another GAN succeeded in generation of attenuation-corrected PET from non-attenuation-corrected PET images for whole-body 18F-FDG PET imaging (32). Training data were 25 pairs of whole body 18F-FDG images with and without attenuation correction. In comparing deep-learning-based attenuation-corrected PET with original attenuation-corrected PET, average mean error and normalized mean square error of the whole-body were 0.62%±1.26% and 0.72%±0.34%.
Other application in image-to-image translation
A GAN trained with 17 PET/CT pairs for liver regions was used to synthesize PET images from CT images (33). Using a test set consisting of 8 CT scans with a total of 26 liver tumors, 92.3% of all tumors were detected in the synthesized PET images compared to the real PET images.
Image-to-Image translation has also been applied to correct voxel intensity non-uniformity of MRI images (34). In order to estimate a corrected MR image from an uncorrected one, a GAN was trained using pairs of uncorrected and corrected MR images. Experiments showed higher accuracy and better tissue uniformity compared to a clinically established algorithm.
Furthermore, instead of individual network modes for each task of image-to-image translation, a comprehensive framework, named MedGAN has been proposed (35). For three different tasks of PET/CT translation, correction of MR motion artefacts, and PET image denoising, the MedGAN outperformed other existing approaches in regards to perceptual analysis by 5 experienced radiologists, as well as quantitative evaluation.
De-noising
In CT and nuclear imaging, from the perspective of reducing the exposure dose and/or shortening acquisition time, GANs have been applied to noise reduction on images scanned under low-dose conditions. For single-photon emission computed tomography (SPECT) imaging, de-noising techniques based on GANs have been explored (36). When an image of high noise level is given, the generator estimates an image from the noisy image that is equivalent to the real low noise image in terms of signal-to-noise ratio. From an experiment on abdominal simulated images with low (128 MBq) and high (987 MBq) injected dose using an XCAT phantom (37), region-of-interest analysis of noise level demonstrated that the GAN-based method has the potential to decrease the noise level of SPECT images.
For CT images that are expected to provide anatomic information, it is important to remove noise while preserving the shape and contrast of the organs. In order to achieve this requirement, GANs that introduce perceptual loss and sharpness loss have been developed. The former class of GANs introduces perceptual loss using a pre-trained network known as a VGG, and was evaluated on abdominal CT images with normal dose and simulated quatre-dose (38). The generator has the role of denoising the low-dose CT images. The discriminator judges whether the input is a normal dose image or a denoised one. In practice, that approach solved the over-smoothing problem and denoised the images with increased contrast for lesion detection.
In the latter type of GAN, an additional sharpness detection network was introduced to measure the sharpness of the denoised image (39). The networks were trained with pairs of high and low dose CT images, in which effective dose for high and low were 14.14, 7.07, and 0.71 mSv, to generate noise-reduced versions of the low-dose images. Figure 2 demonstrated the denoised images comparable with high-dose CT images in term of peak signal-to-noise ratio and structured similarity index.
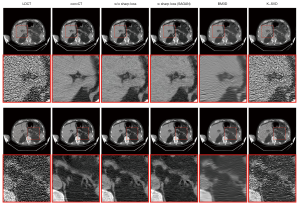
In regards to EKG-gated cardiac CT scans without contrast enhancement for coronary calcium scoring, de-noising in low dose CT images is useful to avoid excessive radiation dose for the patient (40). Jelmer and colleagues trained networks with pairs of low- and routine-dose CT images to generate noise-reduced images based off of the low-dose images. The GAN-based noise reduction allowed accurate quantification of coronary artery calcification from low dose cardiac CT images.
Image reconstruction
In addition to conventional analytical and iterative image reconstruction methods, GANs may be utilized for this task. In this section, we discuss GANs for reconstruction of MR images with compressed sensing (CS-MRI), which enables a reduce acquisition time while maintaining image quality.
A model referred to as RefineGAN provided faithful interpolation in k-space and outperformed the state-of-the-art CS-MRI methods in terms of both running time and image quality for open-source MRI databases of brain, chest, and knee. That was true even for low sampling rate, i.e., 10% of fully acquired data (41).
Another GAN-based framework, GANCS, was developed which was applicable to reconstruction of CS-MRI (42). In a study on a contrast-enhanced MR dataset of pediatric patients, expert radiologists rated images by the GANCS as high quality in regards to improvement of fine texture details when compared to the conventional CS methods. Processing times of reconstruction were faster than the current state-of-the-art CS-MRI schemes by two orders of magnitude.
As a means to improve fidelity between the CS reconstructed image and the fully sampled image, a GAN with an adaptation algorithm of trained generative model parameters to the complete data was created (43). The use of the GAN allowed the reconstruction of high fidelity MR images of knee from noisy and/or incomplete measurement data.
At times in diagnostic imaging, only one region of the image is important. In the case of cardiac MRI, the cardiac region is more important to radiologists than are other regions. From that perspective, a network called Recon-GLGAN utilized region-of-interest features from full-sampling reconstructed images as prior information to improve quality in the CS-MRI reconstructions (44) as shown in Figure 3. A study with cardiac MR images showed better reconstruction quality in terms of peak signal-to-noise ratio, structural similarity index, and normalized mean square error metrics as compared to those of conventional methods. Segmentation tests of heart regions from the whole image showed similar results to fully sampled images.
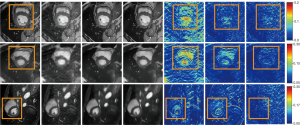
SR
SR is a technique for producing a high-resolution image from a low-resolution image. While the main processing in the conventional method is to increase the image size by interpolation and emphasize the edges by filtering, noise in the image is also emphasized. GAN-based approaches, on the other hand, learn patterns in the same region of paired low- and high-resolution training images to improve resolution in the low-resolution images. In this field, there is a lot of research on MR images that provide anatomic information. Basically, the generator receives a low-resolution image as input and generates a SR image. The discriminator receives the generated image or the true high-resolution image and determines its authenticity.
Meta-SRGAN, which is a network model that utilizes SRGAN and a Meta-Upscale Module. This architecture produces SR for 2D MR images with arbitrary scale and high fidelity (45). Meta-SRGAN outperformed traditional interpolation methods and achieved competitive results with the state-of-the-art method with low memory usage.
Instead of a single GAN, an ensemble learning approach was applied for SR of MR images of knees by training multiple GANs and combining their outputs to final output (46). The ensemble approach outperformed some state-of-the-art methods based on deep learning in terms of structural similarity index and peak signal-to-noise ratio. This method was also able to suppress artifacts and keep more image details.
Extending SR technology from 2D to 3D would be expected to increase computation time and memory consumption, the so-called “curse of dimensionality”. Along with techniques to suppress the computational cost as well as patch-wise (small region) learning, SR technique for 3D images has been developed. A network model based on SRGAN with improved upsampling techniques succeeded in generating realistic images from normal control T1W images down-sampled by a factor of 4 when compared to results by classical interpolation (47).
To generate high resolution 2D and 3D images, a multi-scale GAN with patch-wise learning was developed (48). Starting from a low-resolution scale of the image, the training was repeated and conditioned on the previous scale to produce a higher resolution. The GAN suppressed the artifacts that occur in patch-wise training and produced 3D chest CT and chest X-ray images with matrix sizes of 512×512×512 and 2,048×2,048, respectively. In addition to SR, the GAN was also used for medical image domain translation, such as low dose to high dose chest CT images and T1W to T2W 3D brain MR images.
In another approach, a network referred to as mDCSRN-GAN realized SR for brain MR images of 3D volume and preserved continuous structures by estimating multi-level structure information and adding it on low resolution images of inputs (49,50). For images down-sampled by a factor of 4, the method recovered local image textures and details more accurately as shown in Figure 4, and quickly than current state-of-the-art deep learning approaches by a factor of 6.
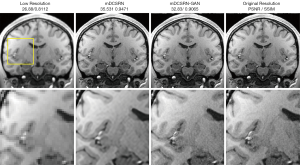
Domain adaptation
Supervised learning for medical image analysis requires appropriate labels and annotation by medical experts, which takes an enormous amount of time and effort. Domain adaptation is a method for constructing high-performance neural networks in a target domain by adapting knowledge such as supervised labels and annotations from a source domain to the target domain with no or insufficient knowledge.
Unsupervised domain adaptation using adversarial training of 3D multi-scale CNNs was demonstrated for the segmentation of traumatic brain injuries (TBI) on brain MR images (51). Their network model leaned features of TBI lesions on gradient-echo with MPRAGE, FLAIR, T2W, and proton density (PD) images without binary labels. The features were adapted to detect TBI lesions on images of MPRAGE, FLAIR, T2W, and PD for other patients. Figure 5 showed similar accuracy to results by supervised learning using binary labels for TBI lesions.
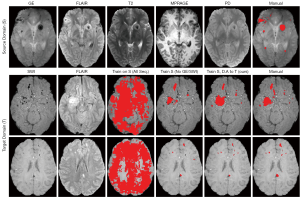
An unsupervised domain adaptation method based on Cycle-GAN has also been developed (52). Using the network model and tissue region labels of gray matter, white matter, and cerebrospinal fluid in a brain MRI dataset, the Alzheimer’s Disease Neuroimaging Initiative dataset was adopted to another brain MRI dataset, BaTS. The adopted tissue labels contributed to improved accuracy of brain tumor segmentation significantly in the BaTS dataset.
An unsupervised domain adaptation method based on Cycle-GAN was investigated for image registration independent from training dataset (53). The networks trained with chest X-ray images were applied to brain MR or multimodal retinal image registration. The investigated method outperformed conventional methods without domain adaptation in registration performance.
Image generation with disease severity and radiomics
Finally, it is appropriate to discuss some examples of the future of the GAN in medical and molecular imaging. Visualization of the progression of chronic obstructive pulmonary disease (COPD) in X-ray chest images were studied with VR-GAN, which learned features of X-ray chest images with quantitative COPD severity based on forced expiratory volume/forced vital capacity (54). Inputs to the generator are an X-ray image with two conditions of real severity for the image and desired severity. The generator makes a disease effect map between the desired severity and the baseline (top row in Figure 6). Final output image is obtained by summing the effect map to the baseline image (bottom row in Figure 6). The generated images agreed with radiologists’ expectations and produced realistic images.
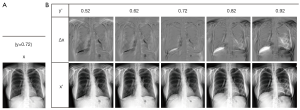
A GAN was developed for elucidation of the relationship between gene expression and CT image for lung nodules (55). When background image without nodules and the gene expression profile are given, the GAN can generate a nodule image with characteristics such as size and shape reflected by the genomic information. That model can separate genetic code to clusters remarkably with effective reducing data of the code from 5,172 to 128 dimensions. As shown in Figure 7, the generated images corresponding to representative three clusters showed the features such as nodule shape and boundary smoothness for each cluster.

Conclusions
As introduced above, the many different GAN architectures were developed as powerful and promising tools for medical and molecular imaging. GANs have realized image synthesis, modality conversion, and SR for volumetric imaging. The achievement of low-dose imaging and shortening acquisition time while maintaining image quality were considered to be important clinically. Domain adaptation that utilizes existing expertise is expected to be a rapid solution to emerging problems with less work. Further improvements in computational power and network models will enable new applications for higher dimensional images, such as volumetric and temporal imaging. As reviewed in the last topic, not only images but also various types of data as inputs could be one of the future directions of GANs in medical and molecular imaging. Use of public/open datasets for high verifiability and evaluation in large-scale studies will make the role of GANs more important.
Acknowledgments
Funding: This work was supported by Grant-in-Aid for Scientific Research (C) (Grant Number JP18K12073) of the Japan Society for the Promotion of Science (JSPS), the German Research Foundation (DFG; Clinician Scientist Program ME3696/3-1), the REBIRTH – Research Center for Translational Regenerative Medicine and the Prostate Cancer Foundation Young Investigator Award and National Institutes of Health grants CA134675, CA183031, CA184228, and EB024495.
Footnote
Provenance and Peer Review: This article was commissioned by the editorial office, Annals of Translational Medicine for the series “Artificial Intelligence in Molecular Imaging”. The article has undergone external peer review.
Reporting Checklist: The authors have completed the Narrative Review reporting checklist. Available at http://dx.doi.org/10.21037/atm-20-6325
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/atm-20-6325). The series “Artificial Intelligence in Molecular Imaging” was commissioned by the editorial office without any funding or sponsorship. SPR served as the unpaid Guest Editor of the series and serves as an unpaid editorial board member of Annals of Translational Medicine from Mar 2020 to Feb 2022. RAB reports non-financial support from Mediso Medical Imaging (Budapest, Hungary), personal fees from Bayer Healthcare (Leverkusen, Germany), personal fees from Eisai GmbH (Frankfurt, Germany), outside the submitted work. The authors have no other conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Chartrand G, Cheng PM, Vorontsov E, et al. Deep learning: A primer for radiologists. Radiographics 2017;37:2113-31. [Crossref] [PubMed]
- Goodfellow I, Pouget-Abadie J, Mirza M, et al. Generative adversarial nets. Advances in neural information processing systems. 2014. Available online: https://papers.nips.cc/paper/2014
- Kazuhiro K, Werner RA, Toriumi F, et al. Generative adversarial networks for the creation of realistic artificial brain magnetic resonance images. Tomography 2018;4:159-63. [Crossref] [PubMed]
- Rowe SP, Pienta KJ, Pomper MG, et al. Proposal for a structured reporting system for prostate-specific membrane antigen-targeted PET imaging: PSMA-RADS version 1.0. J Nucl Med 2018;59:479-85. [Crossref] [PubMed]
- Rowe SP, Pienta KJ, Pomper MG, et al. PSMA-RADS version 1.0: A step towards standardizing the interpretation and reporting of PSMA-Targeted PET imaging studies. Eur Urol 2018;73:485-7. [Crossref] [PubMed]
- Shenderov E, Gorin MA, Kim S, et al. Diagnosing small bowel carcinoid tumor in a patient with oligometastatic prostate cancer imaged with PSMA-Targeted [(18)F]DCFPyL PET/CT: Value of the PSMA-RADS-3D Designation. Urol Case Rep 2018;17:22-5. [Crossref] [PubMed]
- Werner RA, Solnes LB, Javadi MS, et al. SSTR-RADS version 1.0 as a reporting system for SSTR PET imaging and selection of potential PRRT candidates: a proposed standardization framework. J Nucl Med 2018;59:1085-91. [Crossref] [PubMed]
- Werner RA, Bundschuh RA, Bundschuh L, et al. Molecular imaging reporting and data systems (MI-RADS): a generalizable framework for targeted radiotracers with theranostic implications. Ann Nucl Med 2018;32:512-22. [Crossref] [PubMed]
- Russel WMS, Burch RL. The principles of humane experimental technique. London: Methuen, 1959.
- Lecun Y, Bottou L, Bengio Y, et al. Gradient-based learning applied to document recognition. Proc IEEE 1998;86:2278-324. [Crossref]
Radford A Metz L Chintala S Unsupervised representation learning with deep convolutional generative adversarial networks. 2015 . Available online: https://arxiv.org/abs/1511.06434Mirza M Osindero S Conditional generative adversarial nets. 2014 . Available online: https://arxiv.org/abs/1411.1784- Jin D, Xu Z, Tang Y, et al. (editors). CT-realistic lung nodule simulation from 3D conditional generative adversarial networks for robust lung segmentation. Medical Image Computing and Computer Assisted Intervention – MICCAI 2018. Granada, Spain: Springer International Publishing, 2018.
Amyar A Ruan S Vera P RADIOGAN: Deep convolutional conditional generative adversarial network to generate PET images. 2020 . Available online: https://arxiv.org/abs/2003.08663Isola P Zhu JY Zhou T Image-to-image translation with conditional adversarial networks. 2016 . Available online: https://arxiv.org/abs/2003.08663Kaiser B Albarqouni S. MRI to CT translation with GANs. 2019 . Available online: https://arxiv.org/abs/1901.05259Zhu JY Park T Isola P Unpaired image-to-image translation using cycle-consistent adversarial networks. 2017 . Available online: https://arxiv.org/abs/1703.1059310.1109/ICCV.2017.244 Karras T Aila T Laine S Progressive growing of GANs for improved quality, stability, and variation. 2017 . Available online: https://arxiv.org/abs/1710.10196- Ledig C, Theis L, Huszár F, et al. Photo-realistic single image super-resolution using a generative adversarial network. Honolulu, HI, USA: 2017 IEEE Conference on Computer Vision and Pattern Recognition (CVPR), 2017:105-14.
Carver E Dai Z Liang E Improvement of multiparametric MR image segmentation by augmenting the data with generative adversarial networks for glioma patients. 2019 . Available online: https://arxiv.org/abs/1910.00696Shen Z Zhou SK Chen Y One-to-one mapping for unpaired image-to-image translation. 2019 . Available online: https://arxiv.org/abs/1909.04110- Jonnalagedda P, Weinberg B, Allen J, et al. SAGE: Sequential attribute generator for analyzing glioblastomas using limited dataset. 2020. Available online: https://arxiv.org/abs/2005.07225
Qasim AB Ezhov I Shit S Red-GAN: Attacking class imbalance via conditioned generation. Yet another medical imaging perspective. 2020 . Available online: https://arxiv.org/abs/2004.10734- Salehinejad H, Valaee S, Dowdell T, et al. Generalization of deep neural networks for chest pathology classification in X-rays using generative adversarial networks. Calgary, AB, Canada: 2018 IEEE International Conference on Acoustics, Speech and Signal Processing (ICASSP), 2018:990-4.
Korkinof D Rijken T O'Neill M High-resolution mammogram synthesis using progressive generative adversarial networks. 2018 . Available online: https://arxiv.org/abs/1807.03401Kwon G Han C Kim DS Generation of 3D brain MRI using auto-encoding generative adversarial networks. 2019 . Available online: https://arxiv.org/abs/1908.0249810.1007/978-3-030-32248-9_14 - Wang Y, Yu B, Wang L, et al. 3D conditional generative adversarial networks for high-quality PET image estimation at low dose. NeuroImage 2018;174:550-62. [Crossref] [PubMed]
Li J Koh JC Lee WS Alternative supervision network for high-resolution CT image interpolation. 2020 . Available online: https://arxiv.org/abs/2002.0445510.1109/ICIP40778.2020.9191060 - Hiasa Y, Otake Y, Takao M, et al. (editors). Cross-modality image synthesis from unpaired data using CycleGAN. Simulation and Synthesis in Medical Imaging. SASHIMI 2018. Granada, Spain: Springer, Cham, 2018.
- Nie D, Trullo R, Lian J, et al., editors. Medical image synthesis with context-aware generative adversarial networks. Medical Image Computing and Computer Assisted Intervention − MICCAI 2017. Quebec City, QC, Canada: Springer International Publishing, 2017.
- Armanious K, Küstner T, Reimold M, et al. Independent brain (18)F-FDG PET attenuation correction using a deep learning approach with generative adversarial networks. Hell J Nucl Med 2019;22:179-86. [PubMed]
- Dong X, Lei Y, Wang T, et al. Deep learning-based attenuation correction in the absence of structural information for whole-body positron emission tomography imaging. Phys Med Biol 2020;65:055011 [Crossref] [PubMed]
Ben-Cohen A Klang E Raskin SP Virtual PET images from CT data using deep convolutional networks: Initial results. 2017 . Available online: https://arxiv.org/abs/1707.0958510.1007/978-3-319-68127-6_6 - Lei Y, Fu Y, Mao H, et al. Multi-modality MRI arbitrary transformation using unified generative adversarial networks. SPIE Medical Imaging: SPIE, 2020.
- Armanious K, Jiang C, Fischer M, et al. MedGAN: Medical image translation using GANs. Comput Med Imaging Graph 2020;79:101684 [Crossref] [PubMed]
Zhang Q Sun J Mok GSP Low dose SPECT image denoising using a generative adversarial network. 2019 . Available online: https://arxiv.org/abs/1907.11944- Segars WP, Sturgeon G, Mendonca S, et al. 4D XCAT phantom for multimodality imaging research. Med Phys 2010;37:4902-15. [Crossref] [PubMed]
- Yang Q, Yan P, Zhang Y, et al. Low-dose CT image denoising using a generative adversarial network with Wasserstein distance and perceptual loss. IEEE Trans Med Imaging 2018;37:1348-57. [Crossref] [PubMed]
- Yi X, Babyn P. Sharpness-aware low-dose CT denoising using conditional generative adversarial network. J Digit Imaging 2018;31:655-69. [Crossref] [PubMed]
- Wolterink JM, Leiner T, Viergever MA, et al. Generative adversarial networks for noise reduction in low-dose CT. IEEE Trans Med Imaging 2017;36:2536-45. [Crossref] [PubMed]
- Quan TM, Nguyen-Duc T, Jeong W. Compressed sensing MRI reconstruction using a generative adversarial network with a cyclic loss. IEEE Trans Med Imaging 2018;37:1488-97. [Crossref] [PubMed]
- Mardani M, Gong E, Cheng JY, et al. Deep generative adversarial neural networks for compressive sensing MRI. IEEE Trans Med Imaging 2019;38:167-79. [Crossref] [PubMed]
- Bhadra S, Zhou W, Anastasio M. Medical image reconstruction with image-adaptive priors learned by use of generative adversarial networks. SPIE Medical Imaging: SPIE, 2020.
- Murugesan B, Vijaya Raghavan S, Sarveswaran K, et al., editors. Recon-GLGAN: A global-local context based generative adversarial network for MRI reconstruction. MLMIR: International Workshop on Machine Learning for Medical Image Reconstruction; 2019 Oct 17; Shenzhen, China: Springer International Publishing, 2019.
- Tan C, Zhu J, Lio’ P. Arbitrary Scale Super-Resolution for Brain MRI Images. Artificial Intelligence Applications and Innovations. Neos Marmaras, Greece: 16th IFIP WG 125 International Conference, AIAI 2020, Proceedings, Part I:165-76.
- Lyu Q, Shan H, Wang G. MRI super-resolution with ensemble learning and complementary priors. IEEE Trans Comput Imaging 2020;6:615-24. [Crossref]
Sanchez I Vilaplana V. Brain MRI super-resolution using 3D generative adversarial networks. 2018 . Available online: https://arxiv.org/abs/1812.11440Uzunova H Ehrhardt J Jacob F Multi-scale GANs for memory-efficient generation of high resolution medical images. 2019 . Available online: https://arxiv.org/abs/1907.0137610.1007/978-3-030-32226-7_13 Chen Y Shi F Christodoulou AG Efficient and accurate MRI super-resolution using a generative adversarial network and 3D multi-level densely connected network. 2018 . Available online: https://arxiv.org/abs/1803.0141710.1007/978-3-030-00928-1_11 Chen Y Christodoulou AG Zhou Z MRI super-resolution with GAN and 3D multi-level DenseNet: smaller, faster, and better. 2020 . Available online: https://arxiv.org/abs/2003.01217- Kamnitsas K, Baumgartner C, Ledig C, et al., editors. Unsupervised domain adaptation in brain lesion segmentation with adversarial networks. International Conference on Information Processing in Medical Imaging. Boone, NC, USA: Springer International Publishing, 2017.
- Tokuoka Y, Suzuki S, Sugawara Y. An inductive transfer learning approach using cycle-consistent adversarial domain adaptation with application to brain tumor segmentation. Proceedings of the 2019 6th International Conference on Biomedical and Bioinformatics Engineering; Shanghai, China: Association for Computing Machinery, 2019:44-8.
- Mahapatra D, Ge Z. Training data independent image registration using generative adversarial networks and domain adaptation. Pattern Recognition 2020;100:107109 [Crossref]
- Bigolin Lanfredi R, Schroeder JD, Vachet C, et al., editors. Adversarial regression training for visualizing the progression of chronic obstructive pulmonary disease with chest X-rays. Medical Image Computing and Computer Assisted Intervention – MICCAI 2019. Shenzhen, China: Springer International Publishing, 2019.
- Xu Z, Wang X, Shin HC, et al. Correlation via synthesis: end-to-end nodule image generation and radiogenomic map learning based on generative adversarial network. 2019. Available online: https://arxiv.org/abs/ 1907.03728


