
Biomarkers of therapeutic response with immune checkpoint inhibitors
Introduction
The past two decades have seen significant advances in the therapeutic utilization of immune checkpoint inhibitors (ICPIs) for a wide range of malignancies. Currently available immunotherapeutic drugs include monoclonal antibodies directed against programmed cell death protein (PD-1), programmed cell death protein ligand (PD-L1) and cytotoxic T-lymphocyte antigen 4 (CTLA-4) (1,2). Since the approval of ipilimumab in 2011 for management of advanced, unresectable melanoma (3), ICPIs continue to revolutionize the therapeutic landscape of cancer and were named the top cancer advance of the year by American Society of Clinical Oncology in 2016 (4). The Food and Drug administration (FDA) has approved several agents for a number of unresectable, advanced and refractory cancers including lung cancer, melanoma, renal cell cancer (RCC), head and neck cancers, Hodgkin lymphoma and several others. Response in some patients treated with single agent immunotherapy can be quite dramatic; however, only a small subset of patients across cancer indications derive significant benefit (5). Combination of ICPIs with other immunotherapeutic or chemotherapeutic agents improves response rates but this comes with a cost in the form of clinical and financial toxicities (6,7). The current array of predictive biomarkers for ICPI therapy are limited in predictive accuracy due to significant variability with tumor histology, tumor heterogeneity, lack of standardization of pre analytical techniques and subsequent challenges with clinical interpretation. Thus, development of biomarkers that are broadly applicable and well standardized for ICPI therapy across cancer subtypes is of critical interest to current clinical and investigative efforts.
An approach to organize current and emerging biomarkers of response to ICPIs is to consider categorization based on four broad elements of cancer immunity that govern the anticancer response (Figure 1) (7). The first category of cancer immunity is the inflamed tumor, as it is now well known that tumor inflammation is one of the hallmarks of cancer. Our understanding of the immune mediated cancer elimination is based on the widely accepted process of the “cancer immunity cycle” (8). Innate and adaptive immunity work together to eliminate evolving tumors. Signs of an inflammatory tumor microenvironment (TME) that can serve as biomarkers for response to ICPIs include PD-L1 expression, the presence of tumor infiltrating lymphocytes (TILs), and mRNA profiles with increased expression of genes associated with inflammation (9).
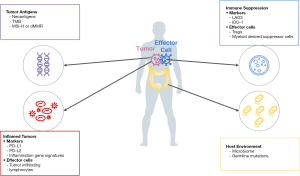
The second category of cancer immunity is tumor antigens. Tumor antigens are molecules presented on the surface of tumor cells that make tumor recognizable by the immune system. Presentation of tumor antigens can be affected by genetic or epigenetic alterations (10,11). States of higher tumor antigenicity may be represented by high tumor mutational burden (TMB) or microsatellite instability (MSI) which is usually caused by deficient mismatch repair (MMR) proteins.
The third category of tumor immunity is immune suppression where tumor proliferation is related to progressive immune dysfunction and tolerance leading to a loss of immune surveillance, thereby generating tumor immunity (12,13). PD-L1 expression bridges the first and third category because PD-L1 expression can be triggered by inflammation and the presence of PD-L1 on the tumor and immune cells causes immune suppression. Other proteins that affect immune evasion, such as lymphocyte activation gene 3 (LAG-3) and indoleamine 2,3-dioxygenase (IDO), are being investigated as the next generation of cancer immunotherapy predictive biomarkers.
The fourth category of tumor immunity is the overall tumor host environment. Factors like the host microbiome and germline mutations can influence not only the development of cancer but also response to chemotherapy and immunotherapy (14).
Here, we will review the key clinically relevant biomarkers of response to ICPIs and explore the limitations in their broader applicability in the treatment of cancer.
The inflamed tumor
PD-L1
Over the last decade, PD-L1 emerged as a companion and complementary diagnostic biomarker in parallel with the advent of anti-PD-1 and anti-PD-L1 immunotherapy. A majority of TILs express PD-1 after activation and it binds to two specific ligands, PD-L1 and PD-L2. Engagement of PD-1 with its ligand results in elimination of activated T cells upon completion of their effector functions, thereby serving as an immune checkpoint (8,15). Tumor cells can engage this immune inhibitory pathway by expressing PD-L1 as an ICPI on their cell surface. Increased PD-L1 expression (8) is achieved either by development of adaptive immune resistance or by certain genomic alterations (16-19). PD-L1 adaptive immune resistance refers to the expression of PD-L1 on the tumor surface in response to T cell recognition of target antigens on the tumor surface and release of interferon gamma. PD-L1 expression and interaction with PD-1 allows the tumor cells to evade immune-mediated destruction. Therapeutic inhibition of the PD-1 and PD-L1 immune inhibitory axis (for example by ICPIs) can lead to persistent activation of T cells and anti-tumor activity.
One of the first clinical indications of the potential for PD-L1 expression to serve as a predictive biomarker for response to ICPIs came from a large pivotal study by Topalian et al. that evaluated anti-PD-1 therapy with nivolumab in a variety of tumor types (20). In this study, 60% of tested tumors were positive for PD-L1 expression (defined as >5% PD-L1 expression using immunohistochemistry (IHC) assay IHC-5H1). Amongst the PD-L1 positive tumors, an overall response rate (ORR) of 36% was observed compared to 0% ORR in patients with PD-L1 negative tumors. Anti-tumor activity of single agent PD-L1 therapy was seen in melanoma, RCC and non-small cell lung cancer (NSCLC) but no activity was seen in metastatic castrate resistant prostate cancer and metastatic colon cancer regardless of PD-L1 status. Overtime, multiple studies have established that patients who have tumors that express PD-L1 correlate with higher response rates to ICPIs across multiple classes of cancer, including NSCLC, RCC, urothelial cancer, triple-negative breast cancer, among others (20-22). The PD-L1 expression may also have a role in identifying patients who may benefit from ICPI maintenance therapy. This is supported by an unplanned post hoc analysis of the PACIFIC trial that did not demonstrate an improvement in overall survival with maintenance durvalumab post chemoradiation in patients with unresectable stage III NSCLC whose tumors had PD-L1 expression less than 1% (23).
Multiple assays for PD-L1 expression have been approved as companion and complementary diagnostics for use with PD-1/PD-L1 directed FDA approved drugs in different disease indications. These assays utilize different antibodies and PD-L1 expression cutoffs to predict response to ICPIs as established by various trials and agents used. Furthermore, differences in scoring systems such as the tumor proportion scoring system (TPS, total number of PD-L1 positive tumor cells) or combined positive scoring system (CPS, divides number of PD-L1 positive tumor cells, lymphocytes, macrophages by the number of tumor cells) prevent standardization across assays (24). McLaughlin et al. demonstrated poor concordance between two PD-L1 rabbit monoclonal antibodies (E1L3N and SP142) with approximately 25% discordance in expression of PD-L1 in tested samples from patients with NSCLC (25). The predictive value of PD-L1 expression for response to immunotherapy can also vary among the ICPIs agents used or disease setting. For instance, Balar et al. demonstrated correlation between increased PD-L1 expression in advanced urothelial cancer with response to first line pembrolizumab in cisplatin-ineligible patients (26). However, a study by Sharma et al. in recurrent, advanced urothelial cancer after platinum-based chemotherapy did not demonstrate a correlation between PD-L1 expression and response to nivolumab (27). There is also discordance between expected responses when using the same assays for same tumor histology based on the treatment setting (first line versus subsequent lines of treatment). For instance, atezolizumab showed a higher response with increased levels of PD-L1 expression in immune cells in patients with locally advanced and metastatic urothelial cancer who had progressed following treatment with platinum-based chemotherapy (objective response rate, ORR 26% vs. 10% in patients with PD-L1 ≥5% compared to PD-L1 1–4%) (21). This study utilized the SP142 antibody assay for assessment of PD-L1 expression and, interestingly, patients with no expression (PD-L1 <1%) also demonstrated an appreciable response rate of 8%. When the same PD-L1 assay with the SP142 antibody was used to evaluate the response to atezolizumab in treatment naïve, platinum-ineligible patients with advanced urothelial cancer, the degree of PD-L1 expression was not predictive of the response to atezolizumab (ORR 21% with PD-L1 <1%, ORR 21% with PD-L1 ≥1% but <5% and 28% in patients with PD-L1 ≥5%) (28). This variability in the predictive power of PD-L1 may be due to differences in tissue selection and processing, differences in techniques of performing (antigen retrieval, fixation) staining, and/or differences in interpretation of the results which all pose challenges with standardized testing across different laboratories.
In addition to the technical challenges of PD-L1 testing mentioned above, intra-tumoral and inter-tumoral heterogeneity in PD-L1 expression also limits accurate assessment of PD-L1 expression (29,30). Moreover, it has been shown that PD-L1 expression is dynamic with chemotherapy, and that radiation therapy can induce PD-L1 expression (31,32). Thus, the response to ICPIs seen in PD-L1 negative tumors may be in the setting of tumor heterogeneity or temporally dynamic expression (33). Despite these limitations and caveats, PD-L1 is a clinically valuable predictive biomarker in certain tumor types like NSCLC where its use in determining first line treatment with ICPIs is now standard practice. Pembrolizumab is currently FDA approved as a single therapy for management of metastatic NSCLC with PD-L1 TPS ≥1% (PD-L1 IHC 22C3 assay), and in combination with chemotherapy regardless of the PD-L1 TPS (34). The FDA has designated PD-L1 as a companion diagnostic for certain indications and a complementary diagnostic marker for others based on emerging efficacy and safety data with frequent adaptations based on new findings. For instance, PD-L1 has recently emerged as a clinically relevant biomarker in urothelial cancer. In two ongoing trials, KEYNOTE-361 using PD-L1 IHC 22C3 PharmDx assay (NCT02853305) and IMvigor130 using Ventana PD-L1 SP142 assay (NCT02807636), patients with PD-L1 low status were found to have decreased survival with ICPI monotherapy compared to patients receiving platinum-based chemotherapy. Thus, FDA issued an alert after the initial review by the respective data monitoring committees leading to a revision in the trial protocols with enrollment of only PD-L1 high expression patients to the ICPI arm (35).
Tumor infiltrating lymphocytes and the tumor microenvironment
It has long been recognized that inflammatory cells including CD8+ cytotoxic T cells, natural killer cells, dendritic cells, macrophages and neutrophils mediate the cancer immunity cycle and their role as biomarkers is currently being explored (8). In 2011, TILs were identified as a prognostic marker in patients with breast cancer with the presence of CD8+ TILs associated with improved outcomes (36). Since TILs are a marker of inflammation in the TME, they have also been assessed as a potential predictive biomarker for response to ICPIs. In 2014, Tumeh et al. found that response to pembrolizumab in metastatic melanoma was associated with a higher CD8+ T cell density in the baseline biopsies (37). This finding of increased T cell density was also noted by Chen et al. in pretreatment tissue samples of responders treated with CTLA-4 blockade for melanoma (38). However, neither of these studies could establish a clear cut-off for T cell infiltration to differentiate between responders and non-responders. KEYNOTE-086 (39) and KEYNOTE-173 (40) also found increased TILs in pretreatment biopsy samples of patients with triple negative breast cancer who had a better response to treatment with single agent pembrolizumab and neoadjuvant pembrolizumab plus chemotherapy respectively compared to patients with lower TIL levels. Recent investigations have focused on characterizing the type, density and immune phenotype of intratumoral TILs as predictors of clinical outcomes to ICPIs (41). Studies of the transcriptomic profile of TILs have shown significant heterogeneity in the expression of molecules of T cell activation and presence of tissue resident memory cells that may predict better outcomes in patients with NSCLC and breast cancer (42,43).
In addition to assessing hematoxylin-eosin morphology and single analyte IHC, recent years have seen development of “molecular imaging” with IHC and immunofluorescence multiplexing methods (mIHC and mIF respectively) and technologies to allow a more comprehensive assessment of the markers of inflammation or lack thereof in the tumor-host microenvironment (44). In 2016, Zhang and Chen (45) introduced a classification system for the tumor immune microenvironment (TIME) based on the expression of PD-L1 and TILs as PD-L1 expression in tumors correlates with the presence of TILs (46). The four proposed classes of TME included T1 (PD-L1 and TIL negative), T2 (PD-L1 and TIL positive), T3 (PD-L1 negative and TIL positive) and T4 (PD-L1 positive and TIL negative). In this model, the T2 tumors are predicted to account for the most responses to anti-PD therapy while T1 tumors are predicted to be inherently resistant. In another study, Gettinger et al. used qualitative mIF to evaluate the location and immunophenotype of TILs in NSCLC samples prior to treatment with ICPIs. They found a population of “dormant” TIL phenotype with elevated CD3 expression and low T cell Ki67and granzyme B (indicators of T cell proliferation and activation) was associated with a favorable and durable response to ICPI therapy (47). These models need further clinical validation and perhaps incorporation of other characteristics of the tumor immune microenvironment to allow us to predict responses in patients treated with ICPI therapy reliably. One potential translation of evaluation of the complex immune interactions in the TME as a clinical biomarker is the development of the “Immunoscore” for many cancers, including colorectal cancer (48). The Immunoscore quantifies the lymphocyte populations at the tumor center and margins with higher scores correlating with longer patient survival. In some studies, the Immunoscore outperforms the classical TNM classification as a prognostic marker (49). In addition to serving as a prognostic factor, integrative analyses have also revealed that Immunoscore can predict disease specific recurrence and survival in patients with colorectal cancer (50,51). The considerable prognostic impact of combining stromal PD-L1 and PD-1 expression on TILs in a novel Immunoscore approach within each pathologic stage of NSCLC has also been demonstrated (52). Another novel noninvasive tool for detection of variations in whole body CD8+ T cells as a response to treatment with ICPIs has been developed and this immuno-PET imaging modality is currently being investigated for potential utility in clinical practice (53). The role of the TME in suppressing response to ICPIs is also an area of active research that is described in greater detail in the section on “immune suppression” in this manuscript.
Gene expression profiling
It has now been established that interferon-γ (IFN-γ) is a key cytokine in the TME that plays a vital role in the development of antitumor immune response (54). However, feedback inhibition of the IFN-γ signaling pathway can lead to the upregulation of PD-L1 and PD-L2 in the tumor and immune microenvironment compromising antitumor immunity (55,56). As discussed above, the assessment of PD-L1 expression can be used as a predictive marker for response to ICPIs, but PD-L1 expression likely only represents a small component of T cell biology in the TME. Newer technologies are now available that allow us to elucidate gene expression profiles and understand the complexity of tumor and immune cell interaction.
In the POPLAR trial, an 8-gene T-effector and IFN-γ gene signature was noted to be associated with improved overall survival in previously treated patients with advanced/metastatic NSCLC treated with atezolizumab (57). The T-cell inflamed gene expression profile (GEP) consisting of an 18 gene signature was validated and refined by Ayers et al. using baseline tumor samples of pembrolizumab-treated patients in different clinical studies. This 18 gene signature is currently being evaluated in several ongoing trials (58). Higgs et al. also demonstrated that IFN-γ mRNA signatures may be used to identify patients with NSCLC or urothelial cancer who may have improved outcomes with durvalumab regardless of the PD-L1 status (59). Cristescu et al. then evaluated GEP in over 300 patients with 22 tumor types from four KEYNOTE clinical trials and found that clinical responders had higher GEP scores compared to non-responders (60). Another analysis of 475 patients with 20 solid tumor types enrolled in KEYNOTE-028 also demonstrated that higher GEP scores were seen in clinical responders (61). Interestingly, GEP had low correlation with TMB in both of these studies, thereby suggesting that these biomarkers may capture distinct features of T cell activation and neoantigenicity.
Recent efforts have also focused on development of immune-predictive scores as predictors of response to ICPIs. Auslander et al. built the immune-predictor score (IMPRES) as a predictor of response to ICPIs in patients with melanoma that encompasses 15 pairwise transcriptomics relations between immune checkpoint genes (62). In validation studies across multiple melanoma datasets, IMPRES was demonstrated to have a high predictive performance and further studies of its predictive performance in other cancer types are needed (62). Jiang et al. have also developed a predictive computational method, Tumor Immune Dysfunction and Exclusion (TIDE), that models the mechanisms of tumor immune evasion, i.e., prevention of cytotoxic T cell infiltration and induction of T cell dysfunction. These models and pre-treatment RNA sequencing GEP were used to predict clinical outcomes in patients with melanoma treated with ICPIs with results suggesting that TIDE may be a more accurate predictor of response than PD-L1 and TMB (63). There is significant interest in validating these immune predictive scores in clinical trials and it is highly likely that computational modeling will refine our understanding of the predictive biomarkers of ICPI responses.
Tumor antigens
Tumor neoantigens are generated by somatic alterations in the tumor cell genome as a result of point or nonsense mutations, chromosomal translocations, splicing variants or epigenetic alterations in antigen expression. The cancer immunity cycle is initiated when immunogenic neoantigen derived proteins are released into the TME after tumor cell apoptosis (14). These neoantigens are recognized as foreign and processed by the dendritic cells to activate tumor-specific cytotoxic T-cells. The cytotoxic T-cells in turn attack the neoantigen targets on tumor cells, thereby releasing more tumor neoantigens and renewing the cancer immunity cycle (33). MMR, MSI and TMB can be considered surrogate markers of these immunogenic neoantigens as a higher mutational burden within a tumor significantly increases the changes of generation of a neoantigen that would generate an effective adaptive T cell response (33).
MMR and MSI
The DNA MMR system edits and corrects the DNA mismatches that can occur during DNA replication and recombination repair (64). Deficient MMR (dMMR) occurs due to inactivation of one or more of the four main MMR proteins, i.e., MLH1, MSH2, PMS2 and MSH6. MLH1 and MSH2 account for 90% of cases of dMMR with the other two genes, PMS2 and MSH6 accounting for the remainder (65). dMMR can be caused by germline mutations (Lynch syndrome) or acquired somatic mutations leading to an increased rate of mismatch errors and alterations in the lengths of microsatellite regions referred to as MSI (66,67). Although initially identified in colorectal cancer, dMMR occurs in several tumor types including endometrial, gastrointestinal, biliary, and thyroid carcinomas among others (68,69). Microsatellites are short tandem repeats of DNA in the noncoding regions of the genome (70). dMMR leads to increased mismatched errors in the microsatellite regions resulting in MSI and neoantigen formation. The severity of this instability in tumors is described as MSI high, MSI low and microsatellite stable (71).
The potential of MMR to be utilized as a predictive biomarker was first demonstrated in a phase 2 trial evaluating pembrolizumab in colorectal cancer by Le et al. (72). The patients with dMMR had a significantly higher objective response rate of 40% compared to patients with proficient MMR (pMMR) (72). Le et al. subsequently performed a large trial evaluating the efficacy of pembrolizumab in patients with dMMR tumors across 12 tumor histologies (73). The patients with dMMR had a significant and durable response to PD-1 blockade. The study also demonstrated a rapid in vivo expansion of neoantigen directed T-cells in the responding patients (73). In 2017, these results led to the accelerated tumor agnostic FDA approval of pembrolizumab for patents with dMMR or MSI high tumors (74). This was the first FDA approval of an anti-cancer treatment based solely on a biomarker instead of a tumor type (74).
Defects in MMR are screened by two methods: IHC for the 4 MMR proteins (MLH1, MSH2, PMS2 and MSH6) (75) and polymerase chain reaction (PCR) testing for detection of MSI (76). Both tests have been shown to be equally sensitive with current recommendations in place for use of IHC-testing as the first screening tool due to its universal availability and cost effectiveness (77,78). Although the predictive value of defects in MMR for response to ICPI therapy is compelling, its clinical utilization is mainly limited to certain tumor types like colorectal and endometrial cancer. This may be due to a lower frequency of dMMR in other tumor types or due to the limitations of the assays as the microsatellite repeats currently evaluated in the PCR assays may not be representative for all cancer types (79). Thus, other assays to test and complement the defects in MMR including next generation sequencing and TMB are currently under development (80,81).
TMB
TMB is defined as the number of somatic mutations (non-synonymous single nucleotide variants) per megabase (33). These mutations that are acquired by the tumor cells result in the development of neoantigens that are expressed on the surface of tumor cells. These neoantigens can be recognized as foreign by T cells leading to activation of the cancer immunity cycle (14). Tumors then may develop mechanisms of immune evasion such as upregulation of immune checkpoints and thus may be susceptible to ICPI therapy (33). TMB is highly variable among different tumor types with lung cancer, melanoma and bladder cancer representing the cancers with generally higher TMB. The higher prevalence of mutations in these cancer types may reflect higher effects of environmental factors on tumorigenesis in these tumor types (82).
In 2014, Snyder et al. observed that a high TMB in patients with melanoma correlated with long term clinical benefit upon treatment with CTLA-4 blockade (83). Although this study suggested that the neoantigen signature might be more relevant than TMB in predicting response, this finding was not reproduced in subsequent studies using the currently available bioinformatics algorithms (84,85). Several subsequent studies demonstrated that mutational burden can be a predictor of response to ICPIs in other tumor types like lung cancer (86,87), head and neck cancer (88) and urothelial cancer (21). In 2017, Yarchoan et al. conducted a meta-analysis and found a significant correlation between TMB and objective response to PD-1 blockade in 27 different tumor types (89). In a large study of 151 patients, Goodman et al. also found that TMB is an independent predictor of response to ICPI therapy in over 20 different tumor types (90). It has also been demonstrated that other mutations and DNA repair defects like dMMR, DNA polymerase epsilon (POLE) and DNA polymerase delta 1 (POLD1) generate a high TMB and are associated with enhanced response to ICPI therapy (91,92).
Most of the studies demonstrating predictive potential of TMB and clinical benefit to ICPI therapy utilized whole-exome sequencing (WES) for assessment of TMB. Thus, access to testing and its high cost remains a major barrier to its practical application. More recently, studies have shown that targeted next generation sequencing is concordant with WES and may be applicable (93). Another limitation of TMB is the lack of a standardized cutoff predictive of clinical benefit. It has been postulated that different tumor types may have different TMB cutoffs to reliably predict response (94) with higher TMB cutoff, such as using a cut-off for 20% of cases with the highest TMB values for each tumor histology associated with better survival (95). Interestingly, it is now recognized that mutations are not equally immunogenic with evidence suggesting that frameshift mutations may create new open reading frames and generate a large number of mutagenic neoantigens thereby contributing to a highly immunogenic phenotype (96). Further, there remains a concern that TMB evaluation may underestimate spatial and intra-tumoral heterogeneity. Some of these challenges may be overcome by utilizing circulating cell free tumor DNA (ctDNA) shed into blood by tumor cells. A recent retrospective analysis of two randomized trials (OAK and POPLAR trials) showed that blood based TMB (bTMB) may serve as a promising alternate predictive biomarker (97). Subsequently, Aggarwal et al. (98) utilized a 500 gene NGS panel to assess bTMB in a study of 66 patients with metastatic NSCLC initiating treatment with first line pembrolizumab based therapy. In this study, a bTMB of >16 mutations/megabase pair was associated with improved progression free survival. It was also noted that concurrent mutations in STK11/KEAP1/PTEN and ERBB2 can be used as signals for patients with high bTMB who are not likely to respond (98). Based on this data, TMB has emerged as a promising predicting biomarker of response to ICPI therapy but the remaining complexities associated with TMB evaluation continue to challenge its clinical application. It is best used in a composite biomarker setting and further studies to establish tumor mutational signatures are needed.
Immune suppression
Studies have shown that both the tumor mutational signatures and their microenvironment contribute to high response rates in MSI-high tumors on treatment with ICPIs (99). MSI-high colorectal cancers tend to have a highly infiltrated population of CD8+ T cells (100). To counterbalance this highly immune phenotype, MSI-H colorectal cancers also express high levels of immune checkpoints, including PD-1, PD-L1, CTLA-4, lymphocyte activating 3 (LAG3) and IDO1 (99). Although initially thought to be an MHC II inhibitor, LAG3 is now known to cause down-modulation of lymphocyte response by widespread inhibition of several other ligands (101). Studies have shown a strong synergy between the PD-1 and LAG3 inhibitory pathways (102). Of the several LAG3 inhibitors under development, relatlimab is one of the most extensively studied. When used in combination with nivolumab, it has been shown to have a significant response rate (11.5%) and disease control rate (49%) in ICI-pretreated melanoma patients.103 These responses correlated with LAG3 expression (103).
Multiple non-lymphoid cells also inhabit the TME (104). In particular, the presence of tumor-associated macrophages has emerged as a negative prognostic biomarker (105). Enzymes and growth factor receptors on these non-lymphoid cells are being studied as possible biomarkers and targets for immune based therapies. IDO1 is an intracellular enzyme that initiates the breakdown of tryptophan that is crucial for survival of cytotoxic T cells. It was first shown to be a critical factor in development of maternal-fetal immune tolerance (106). It has been shown that most human tumors constitutively express IDO1 and resist immune rejection (107). Due to its well-defined molecular structure and functions, IDO1 has generated unparalleled interest among novel target directed drug development. Preliminary results from a study of combination indoximod and sipuleucel-T for patients with refractory metastatic prostate cancer showed promising results with improved PFS compared to sipuleucel-T alone (108). Although preliminary activity was also seen in other tumor types, the first large human phase 2/3 trial of an oral IDO1 inhibitor in combination with pembrolizumab for advanced melanoma showed no indication that IDO1 inhibition provided an increased benefit (109). This led to a significant scaling back on the trials for IDO inhibitors and its development as a biomarker (110). Preclinical studies have shown that lung cancer associated fibroblasts are also potentially immunosuppressive (111). Targeting these fibroblasts using nintedanib in combination with nivolumab and ipilimumab in advanced NSCLC is currently being investigated in an ongoing phase I/II study (NCT03377023).
Over 100 other molecular targeting multiple pathways are currently under investigation (110). There has been an exceptional growth of interest in targeting signaling pathways in addition to the core checkpoints, but better biomarker identification remains a challenge in this arena.
Host environment
Several host factors like germline polymorphisms or host microbiome may contribute to a patient’s sensitivity to ICPI therapy.
It is becoming increasingly clear that patients with alterations in the tumor DNA damage repair genes generate a higher neoantigen load and thus may have a favorable response when treated with ICPI (112). Recent investigations have shown that patients with germline alterations in the DNA double-strand break repair pathways exhibit unique immune signatures in their T-cells upon induction of DNA damage that may interact with the immune response (113,114). This suggests that a patient’s ability to repair DNA damage may regulate their response to ICPI therapy and further studies are warranted to understand this impact of germline mutations. Preclinical studies have suggested that human leukocyte antigen class I (HLA-I) molecules may act as tumor suppressor genes affecting the invasive potential of cancer cells (115,116). Heterozygosity in HLA-I loci has also been associated with improved survival in patients with advanced cancers with varied histologic types after treatment with ICPIs (117).
Interest in the role of the microbiome as a predictive biomarker of response to immunotherapy has grown in the recent years (118,119). Studies have shown that differences in the composition of the human gut microbiome are noted among responders and non-responders to ICPI therapy. In two studies (120,121) on patients with metastatic melanoma receiving PD-1 blockade therapy, responders had significantly higher gut microbiome diversity with enrichment of certain species (Akkermansia muciniphila in study by Matson et al. and Rumonicococcaceae family, Faecalibacterium genus in study by Gopalakrishnan et al). Enrichment for these species was also correlated with significant higher CD8+ T cells in these tumors suggesting enhanced immune response (121). Interestingly, when the stool from responders was transplanted into mice, they had a higher proportion of CD8+ T cells with a better response to ICPI therapy when compared to mice transplanted with stool from non-responders. Together, these studies provide gripping evidence that the gut microbiome may serve as a potential biomarker for response to immune checkpoint blockade.
Certain viral infections have also been associated with improved response to ICPIs with the hypothesis that viral proteins may lead to immune activation. For instance, Panda et al. reported a meaningful clinical response in a patient with Epstein-Barr virus (EBV) positive gastric cancer without high TMB or MSI (122). This observation was also supported by an analysis of patients with advanced gastric cancer who were treated with pembrolizumab as a salvage treatment with dramatic responses seen in patients with EBV positive tumors (123). A higher response rate to pembrolizumab was seen in patients with human papillomavirus (HPV) positive head and neck squamous cell cancers (HNSCC) compared to HPV negative HNSCC (124). However, these observations of improved response in virally driven cancers are not consistent across cancer types or different ICPI agents (125,126).
Recently, accumulating evidence also suggests that combining ICPIs with therapies targeting regulatory pathways governing host immune response to ICPIs may provide a synergistic effect. Small molecule inhibitors targeting histone deacytelases (HDACs) have been shown to have complex effects on immune cell function with preclinical (127,128) and clinical studies (129,130) showing potential therapeutic benefit. Combination immunotherapy targeting both tumor vasculature and immune cells has also emerged as a promising approach especially in the management of RCC (131,132), endometrial cancer (133) and hepatocellular carcinoma (HCC) (134). Further, combinations of fibroblast growth factor receptor inhibitors and immunotherapy are also being investigated in NSCLC (135). These combinations can be associated with an increase in toxicities compared to ICPI monotherapy and the next critical step would be to identify a biomarker that helps predict which patients are most likely to benefit with the combination regimens.
Conclusions
Despite significant advances in the era of immunotherapy, our ability to accurately identify the minority of patients who derive benefit from immunotherapy remains very limited. Thus, there is a critical need to develop and validate biomarkers that better predict response. So far, only PD-L1 and MMR/MSI testing are FDA-approved as validated biomarkers for anti-PD-1/PD-L1 immunotherapy. This is partly due to the complex interaction between the multiple tumors, host, environmental and immune mechanisms that drive response or lack thereof to immune based therapies. Another challenge in the development and broader application of predictive biomarkers is the significant variability in standardization and interpretation of these biomarkers as described in this review. Furthermore, the targets of these ICPIs are dynamic and change over time and location owing to the complex changes in the TME and immune milieu. The biomarkers reviewed here represent a small fraction of the currently ongoing investigations into the mechanisms underlying immune destruction of cancer. The development of an effective predictive biomarker for checkpoint inhibitor-based immunotherapy will likely integrate multimodal approaches with advanced analyses of host, tumor, immune and environmental factors.
Acknowledgments
Funding: None.
Footnote
Provenance and Peer Review: This article was commissioned by the Guest Editors (Umang Swami and Mohammed M. Milhem) for the series “Cancer Immunotherapy: Recent Advances and Challenges” published in Annals of Translational Medicine. The article has undergone external peer review.
Peer Review File: Available at http://dx.doi.org/10.21037/atm-20-6396
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/atm-20-6396). The series “Cancer Immunotherapy: Recent Advances and Challenges” was commissioned by the editorial office without any funding or sponsorship. JEG reports grants and personal fees from AstraZeneca, personal fees from Blueprint Medicines, grants from Boehringer Ingelheim, grants and personal fees from Bristol-Myers Squibb, personal fees from EMD Serono - Merck KGaA, grants from Genentech, grants from G 1 Therapeutics, personal fees from Inivata, grants and personal fees from Merck, grants and personal fees from Novartis, grants from Pfizer, grants from Ludwig Institute of Cancer Research, outside the submitted work. SP reports that she is Advisory Board of AstraZeneca, Consultant of G1 therapeutics, and reports Research funding from 5 for the Fight. Dr. TAB reports grants from Bristol Meyers Squibb (BMS), outside the submitted work. The authors have no other conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Ribas A, Wolchok JD. Cancer immunotherapy using checkpoint blockade. Science 2018;359:1350-5. [Crossref] [PubMed]
- Arora S, Velichinskii R, Lesh RW, et al. Existing and Emerging Biomarkers for Immune Checkpoint Immunotherapy in Solid Tumors. Adv Ther 2019;36:2638-78. [Crossref] [PubMed]
- Cameron F, Whiteside G, Perry C. Ipilimumab: first global approval. Drugs 2011;71:1093-104. [Crossref] [PubMed]
- Dizon DS, Krilov L, Cohen E, et al. Clinical Cancer Advances 2016: Annual Report on Progress Against Cancer From the American Society of Clinical Oncology. J Clin Oncol 2016;34:987-1011. [Crossref] [PubMed]
- Carretero-González A, Lora D, Ghanem I, et al. Analysis of response rate with ANTI PD1/PD-L1 monoclonal antibodies in advanced solid tumors: a meta-analysis of randomized clinical trials. Oncotarget 2018;9:8706-15. [Crossref] [PubMed]
- Postow MA, Chesney J, Pavlick AC, et al. Nivolumab and ipilimumab versus ipilimumab in untreated melanoma. N Engl J Med 2015;372:2006-17. [Crossref] [PubMed]
- Chen Y, Zhou Y, Tang L, et al. Immune-Checkpoint Inhibitors as the First Line Treatment of Advanced Non-Small Cell Lung Cancer: A Meta-Analysis of Randomized Controlled Trials. J Cancer 2019;10:6261-8. [Crossref] [PubMed]
- Chen DS, Mellman I. Oncology meets immunology: the cancer-immunity cycle. Immunity 2013;39:1-10. [Crossref] [PubMed]
- Schreiber RD, Old LJ, Smyth MJ. Cancer immunoediting: integrating immunity’s roles in cancer suppression and promotion. Science 2011;331:1565-70. [Crossref] [PubMed]
- Sjöblom T, Jones S, Wood LD, et al. The consensus coding sequences of human breast and colorectal cancers. Science 2006;314:268-74. [Crossref] [PubMed]
- Segal NH, Parsons DW, Peggs KS, et al. Epitope landscape in breast and colorectal cancer. Cancer Res 2008;68:889-92. [Crossref] [PubMed]
- Dunn GP, Bruce AT, Ikeda H, Old LJ, Schreiber RD. Cancer immunoediting: from immunosurveillance to tumor escape. Nat Immunol 2002;3:991-8. [Crossref] [PubMed]
- Miller JFAP, Sadelain M. The journey from discoveries in fundamental immunology to cancer immunotherapy. Cancer Cell 2015;27:439-49. [Crossref] [PubMed]
- Chen DS, Mellman I. Elements of cancer immunity and the cancer-immune set point. Nature 2017;541:321-30. [Crossref] [PubMed]
- Ahmadzadeh M, Johnson LA, Heemskerk B, et al. Tumor antigen-specific CD8 T cells infiltrating the tumor express high levels of PD-1 and are functionally impaired. Blood 2009;114:1537-44. [Crossref] [PubMed]
- Parsa AT, Waldron JS, Panner A, et al. Loss of tumor suppressor PTEN function increases B7-H1 expression and immunoresistance in glioma. Nat Med 2007;13:84-8. [Crossref] [PubMed]
- Ota K, Azuma K, Kawahara A, et al. Induction of PD-L1 Expression by the EML4-ALK Oncoprotein and Downstream Signaling Pathways in Non-Small Cell Lung Cancer. Clin Cancer Res 2015;21:4014-21. [Crossref] [PubMed]
- Roemer MGM, Advani RH, Ligon AH, et al. PD-L1 and PD-L2 Genetic Alterations Define Classical Hodgkin Lymphoma and Predict Outcome. J Clin Oncol 2016;34:2690-7. [Crossref] [PubMed]
- Goodman AM, Piccioni D, Kato S, et al. Prevalence of PDL1 Amplification and Preliminary Response to Immune Checkpoint Blockade in Solid Tumors. JAMA Oncol 2018;4:1237-44. [Crossref] [PubMed]
- Topalian SL, Hodi FS, Brahmer JR, et al. Safety, activity, and immune correlates of anti-PD-1 antibody in cancer. N Engl J Med 2012;366:2443-54. [Crossref] [PubMed]
- Rosenberg JE, Hoffman-Censits J, Powles T, et al. Atezolizumab in patients with locally advanced and metastatic urothelial carcinoma who have progressed following treatment with platinum-based chemotherapy: a single-arm, multicentre, phase 2 trial. Lancet 2016;387:1909-20. [Crossref] [PubMed]
- Schmid P, Adams S, Rugo HS, et al. Atezolizumab and Nab-Paclitaxel in Advanced Triple-Negative Breast Cancer. N Engl J Med 2018;379:2108-21. [Crossref] [PubMed]
- Gray JE, Villegas A, Daniel D, et al. Three-Year Overall Survival with Durvalumab after Chemoradiotherapy in Stage III NSCLC—Update from PACIFIC. Journal of Thoracic Oncology 2020;15:288-93. [Crossref] [PubMed]
- Udall M, Rizzo M, Kenny J, et al. PD-L1 diagnostic tests: a systematic literature review of scoring algorithms and test-validation metrics. Diagn Pathol 2018;13:12. [Crossref] [PubMed]
- McLaughlin J, Han G, Schalper KA, et al. Quantitative Assessment of the Heterogeneity of PD-L1 Expression in Non-Small-Cell Lung Cancer. JAMA Oncol 2016;2:46-54. [Crossref] [PubMed]
- Balar AV, Castellano D, O’Donnell PH, et al. First-line pembrolizumab in cisplatin-ineligible patients with locally advanced and unresectable or metastatic urothelial cancer (KEYNOTE-052): a multicentre, single-arm, phase 2 study. Lancet Oncol 2017;18:1483-92. [Crossref] [PubMed]
- Sharma P, Callahan MK, Bono P, et al. Nivolumab monotherapy in recurrent metastatic urothelial carcinoma (CheckMate 032): a multicentre, open-label, two-stage, multi-arm, phase 1/2 trial. Lancet Oncol 2016;17:1590-8. [Crossref] [PubMed]
- Balar AV, Galsky MD, Rosenberg JE, et al. Atezolizumab as first-line treatment in cisplatin-ineligible patients with locally advanced and metastatic urothelial carcinoma: a single-arm, multicentre, phase 2 trial. Lancet 2017;389:67-76. [Crossref] [PubMed]
- Haragan A, Field JK, Davies MPA, Escriu C, Gruver A, Gosney JR. Heterogeneity of PD-L1 expression in non-small cell lung cancer: Implications for specimen sampling in predicting treatment response. Lung Cancer 2019;134:79-84. [Crossref] [PubMed]
- Liu Y, Dong Z, Jiang T, et al. Heterogeneity of PD-L1 Expression Among the Different Histological Components and Metastatic Lymph Nodes in Patients With Resected Lung Adenosquamous Carcinoma. Clin Lung Cancer 2018;19:e421-30. [Crossref] [PubMed]
- Leduc C, Adam J, Louvet E, et al. TPF induction chemotherapy increases PD-L1 expression in tumour cells and immune cells in head and neck squamous cell carcinoma. ESMO Open 2018;3:e000257 [Crossref] [PubMed]
- Lim YJ, Koh J, Kim S, et al. Chemoradiation-Induced Alteration of Programmed Death-Ligand 1 and CD8+ Tumor-Infiltrating Lymphocytes Identified Patients With Poor Prognosis in Rectal Cancer: A Matched Comparison Analysis. Int J Radiat Oncol Biol Phys 2017;99:1216-24. [Crossref] [PubMed]
- Walk EE, Yohe SL, Beckman A, et al. The Cancer Immunotherapy Biomarker Testing Landscape. Arch Pathol Lab Med 2020;144:706-24. [Crossref] [PubMed]
- Gandhi L, Rodríguez-Abreu D, Gadgeel S, et al. Pembrolizumab plus Chemotherapy in Metastatic Non-Small-Cell Lung Cancer. N Engl J Med 2018;378:2078-92. [Crossref] [PubMed]
- FDA. Alerts Health Care Professionals and Oncology Clinical Investigators about an Efficacy Issue Identified in Clinical Trials for Some Patients Taking Keytruda (pembrolizumab) or Tecentriq (atezolizumab) as Monotherapy to Treat Urothelial Cancer with Low Expression of PD-L1. FDA [Internet] 2018 [cited 2020 Jul 27]. Available online: https://www.fda.gov/drugs/drug-safety-and-availability/fda-alerts-health-care-professionals-and-oncology-clinical-investigators-about-efficacy-issue
- Mahmoud SMA, Paish EC, Powe DG, et al. Tumor-infiltrating CD8+ lymphocytes predict clinical outcome in breast cancer. J Clin Oncol 2011;29:1949-55. [Crossref] [PubMed]
- Tumeh PC, Harview CL, Yearley JH, et al. PD-1 blockade induces responses by inhibiting adaptive immune resistance. Nature 2014;515:568-71. [Crossref] [PubMed]
- Chen PL, Roh W, Reuben A, et al. Analysis of Immune Signatures in Longitudinal Tumor Samples Yields Insight into Biomarkers of Response and Mechanisms of Resistance to Immune Checkpoint Blockade. Cancer Discov 2016;6:827-37. [Crossref] [PubMed]
- Loi S, Adams S, Schmid P, et al. Relationship between tumor infiltration lymphocyte (TIL) levels and response to pembrolizumab in metastatic triple-negative breast cancer (mTNBC): results from KEYNOTE-086. Ann Oncol 2019;30:iii2. [Crossref]
- Loi S, Schmid P, Aktan G, et al. Relationship between tumor infiltrating lymphocytes (TILs) and response to pembrolizumab (Pembro)+chemotherapy (Chemo) as neoadjuvant treatment (NAT) for triple-negative breast cancer (TNBC): phase Ib KEYNOTE-173 trial. Ann Oncol 2019;30:mdz095.003.
- Galon J, Costes A, Sanchez-Cabo F, et al. Type, Density, and Location of Immune Cells Within Human Colorectal Tumors Predict Clinical Outcome. Science 2006;313:1960-4. [Crossref] [PubMed]
- Ganesan AP, Clarke J, Wood O, et al. Tissue-resident memory features are linked to the magnitude of cytotoxic T cell responses in human lung cancer. Nat Immunol 2017;18:940-50. [Crossref] [PubMed]
- Savas P, Virassamy B, Ye C, et al. Single-cell profiling of breast cancer T cells reveals a tissue-resident memory subset associated with improved prognosis. Nat Med 2018;24:986-93. [Crossref] [PubMed]
- Wickenhauser C, Bethmann D, Feng Z, et al. Multispectral Fluorescence Imaging Allows for Distinctive Topographic Assessment and Subclassification of Tumor-Infiltrating and Surrounding Immune Cells. Methods Mol Biol 2019;1913:13-31. [Crossref] [PubMed]
- Zhang Y, Chen L. Classification of Advanced Human Cancers Based on Tumor Immunity in the MicroEnvironment (TIME) for Cancer Immunotherapy. JAMA Oncol 2016;2:1403-4. [Crossref] [PubMed]
- Taube JM, Anders RA, Young GD, et al. Colocalization of inflammatory response with B7-h1 expression in human melanocytic lesions supports an adaptive resistance mechanism of immune escape. Sci Transl Med 2012;4:127ra37 [Crossref] [PubMed]
- Gettinger SN, Choi J, Mani N, et al. A dormant TIL phenotype defines non-small cell lung carcinomas sensitive to immune checkpoint blockers. Nat Commun 2018;9:3196. [Crossref] [PubMed]
- Angell H, Galon J. From the immune contexture to the Immunoscore: the role of prognostic and predictive immune markers in cancer. Curr Opin Immunol 2013;25:261-7. [Crossref] [PubMed]
- Mlecnik B, Tosolini M, Kirilovsky A, et al. Histopathologic-based prognostic factors of colorectal cancers are associated with the state of the local immune reaction. J Clin Oncol 2011;29:610-8. [Crossref] [PubMed]
- Mlecnik B, Bindea G, Angell HK, et al. Integrative Analyses of Colorectal Cancer Show Immunoscore Is a Stronger Predictor of Patient Survival Than Microsatellite Instability. Immunity 2016;44:698-711. [Crossref] [PubMed]
- Pagès F, Mlecnik B, Marliot F, et al. International validation of the consensus Immunoscore for the classification of colon cancer: a prognostic and accuracy study. Lancet 2018;391:2128-39. [Crossref] [PubMed]
- Paulsen EE, Kilvaer TK, Khanehkenari MR, et al. Assessing PDL-1 and PD-1 in Non-Small Cell Lung Cancer: A Novel Immunoscore Approach. Clinical Lung Cancer 2017;18:220-33.e8. [Crossref] [PubMed]
- Tavaré R, Escuin-Ordinas H, Mok S, et al. An Effective Immuno-PET Imaging Method to Monitor CD8-Dependent Responses to Immunotherapy. Cancer Res 2016;76:73-82. [Crossref] [PubMed]
- Ikeda H, Old LJ, Schreiber RD. The roles of IFN gamma in protection against tumor development and cancer immunoediting. Cytokine Growth Factor Rev 2002;13:95-109. [Crossref] [PubMed]
- Bald T, Landsberg J, Lopez-Ramos D, et al. Immune cell-poor melanomas benefit from PD-1 blockade after targeted type I IFN activation. Cancer Discov 2014;4:674-87. [Crossref] [PubMed]
- Bellucci R, Martin A, Bommarito D, et al. Interferon-γ-induced activation of JAK1 and JAK2 suppresses tumor cell susceptibility to NK cells through upregulation of PD-L1 expression. Oncoimmunology 2015;4:e1008824 [Crossref] [PubMed]
- Fehrenbacher L, Spira A, Ballinger M, et al. Atezolizumab versus docetaxel for patients with previously treated non-small-cell lung cancer (POPLAR): a multicentre, open-label, phase 2 randomised controlled trial. Lancet 2016;387:1837-46. [Crossref] [PubMed]
- Ayers M, Lunceford J, Nebozhyn M, et al. IFN-γ-related mRNA profile predicts clinical response to PD-1 blockade. J Clin Invest 2017;127:2930-40. [Crossref] [PubMed]
- Higgs BW, Morehouse CA, Streicher K, et al. Interferon Gamma Messenger RNA Signature in Tumor Biopsies Predicts Outcomes in Patients with Non-Small Cell Lung Carcinoma or Urothelial Cancer Treated with Durvalumab. Clin Cancer Res 2018;24:3857-66. [Crossref] [PubMed]
- Cristescu R, Mogg R, Ayers M, et al. Pan-tumor genomic biomarkers for PD-1 checkpoint blockade-based immunotherapy. Science 2018;362:eaar3593 Erratum in: Science 2019;363(6430). [Crossref] [PubMed]
- Ott PA, Bang YJ, Piha-Paul SA, et al. T-Cell-Inflamed Gene-Expression Profile, Programmed Death Ligand 1 Expression, and Tumor Mutational Burden Predict Efficacy in Patients Treated With Pembrolizumab Across 20 Cancers: KEYNOTE-028. J Clin Oncol 2019;37:318-27. [Crossref] [PubMed]
- Auslander N, Zhang G, Lee JS, et al. Robust prediction of response to immune checkpoint blockade therapy in metastatic melanoma. Nat Med 2018;24:1545-9. [Crossref] [PubMed]
- Jiang P, Gu S, Pan D, et al. Signatures of T cell dysfunction and exclusion predict cancer immunotherapy response. Nat Med 2018;24:1550-8. [Crossref] [PubMed]
- Iyer RR, Pluciennik A, Burdett V, et al. DNA mismatch repair: functions and mechanisms. Chem Rev 2006;106:302-23. [Crossref] [PubMed]
- Buza N, Ziai J, Hui P. Mismatch repair deficiency testing in clinical practice. Expert Rev Mol Diagn 2016;16:591-604. [Crossref] [PubMed]
- Lynch HT, Jascur T, Lanspa S, et al. Making sense of missense in Lynch syndrome: the clinical perspective. Cancer Prev Res (Phila) 2010;3:1371-4. [Crossref] [PubMed]
- Hewish M, Lord CJ, Martin SA, et al. Mismatch repair deficient colorectal cancer in the era of personalized treatment. Nat Rev Clin Oncol 2010;7:197-208. [Crossref] [PubMed]
- Cortes-Ciriano I, Lee S, Park WY, et al. A molecular portrait of microsatellite instability across multiple cancers. Nat Commun 2017;8:15180. [Crossref] [PubMed]
- Bonneville R, Krook MA, Kautto EA, et al. Landscape of Microsatellite Instability Across 39 Cancer Types. JCO Precis Oncol 2017;2017:PO.17.00073.
- Li YC, Korol AB, Fahima T, et al. Microsatellites within genes: structure, function, and evolution. Mol Biol Evol 2004;21:991-1007. [Crossref] [PubMed]
- Imai K, Yamamoto H. Carcinogenesis and microsatellite instability: the interrelationship between genetics and epigenetics. Carcinogenesis 2008;29:673-80. [Crossref] [PubMed]
- Le DT, Uram JN, Wang H, et al. PD-1 Blockade in Tumors with Mismatch-Repair Deficiency. N Engl J Med 2015;372:2509-20. [Crossref] [PubMed]
- Le DT, Durham JN, Smith KN, et al. Mismatch-repair deficiency predicts response of solid tumors to PD-1 blockade. Science 2017;357:409-13. [Crossref] [PubMed]
- Prasad V, Kaestner V, Mailankody S. Cancer Drugs Approved Based on Biomarkers and Not Tumor Type-FDA Approval of Pembrolizumab for Mismatch Repair-Deficient Solid Cancers. JAMA Oncol 2018;4:157-8. [Crossref] [PubMed]
- Giardiello FM, Allen JI, Axilbund JE, et al. Guidelines on genetic evaluation and management of Lynch syndrome: a consensus statement by the US Multi-society Task Force on colorectal cancer. Am J Gastroenterol 2014;109:1159-79. [Crossref] [PubMed]
- Boland CR, Thibodeau SN, Hamilton SR, et al. A National Cancer Institute Workshop on Microsatellite Instability for cancer detection and familial predisposition: development of international criteria for the determination of microsatellite instability in colorectal cancer. Cancer Res 1998;58:5248-57. [PubMed]
- Shia J. Immunohistochemistry versus microsatellite instability testing for screening colorectal cancer patients at risk for hereditary nonpolyposis colorectal cancer syndrome. Part I. The utility of immunohistochemistry. J Mol Diagn 2008;10:293-300. [Crossref] [PubMed]
- Zhang L. Immunohistochemistry versus microsatellite instability testing for screening colorectal cancer patients at risk for hereditary nonpolyposis colorectal cancer syndrome. Part II. The utility of microsatellite instability testing. J Mol Diagn 2008;10:301-7. [Crossref] [PubMed]
- Hause RJ, Pritchard CC, Shendure J, et al. Classification and characterization of microsatellite instability across 18 cancer types. Nat Med 2016;22:1342-50. [Crossref] [PubMed]
- Middha S, Zhang L, Nafa K, et al. Reliable Pan-Cancer Microsatellite Instability Assessment by Using Targeted Next-Generation Sequencing Data. JCO Precis Oncol 2017;2017:PO.17.00084.
- Stadler ZK, Battaglin F, Middha S, et al. Reliable Detection of Mismatch Repair Deficiency in Colorectal Cancers Using Mutational Load in Next-Generation Sequencing Panels. J Clin Oncol 2016;34:2141-7. [Crossref] [PubMed]
- Alexandrov LB, Nik-Zainal S, Wedge DC, et al. Signatures of mutational processes in human cancer. Nature 2013;500:415-21. Erratum in: Nature 2013;502:258. [Crossref] [PubMed]
- Snyder A, Makarov V, Merghoub T, et al. Genetic basis for clinical response to CTLA-4 blockade in melanoma. N Engl J Med 2014;371:2189-99. [Crossref] [PubMed]
- Van Allen EM, Miao D, Schilling B, et al. Genomic correlates of response to CTLA-4 blockade in metastatic melanoma. Science 2015;350:207-11. [Crossref] [PubMed]
- Hellmann MD, Callahan MK, Awad MM, et al. Tumor Mutational Burden and Efficacy of Nivolumab Monotherapy and in Combination with Ipilimumab in Small-Cell Lung Cancer. Cancer Cell 2018;33:853-61.e4. [Crossref] [PubMed]
- Rizvi NA, Hellmann MD, Snyder A, et al. Cancer immunology. Mutational landscape determines sensitivity to PD-1 blockade in non-small cell lung cancer. Science 2015;348:124-8. [Crossref] [PubMed]
- Hellmann MD, Ciuleanu TE, Pluzanski A, et al. Nivolumab plus Ipilimumab in Lung Cancer with a High Tumor Mutational Burden. N Engl J Med 2018;378:2093-104. [Crossref] [PubMed]
- Hanna GJ, Lizotte P, Cavanaugh M, et al. Frameshift events predict anti-PD-1/L1 response in head and neck cancer. JCI Insight 2018;3:e98811 [Crossref] [PubMed]
- Yarchoan M, Hopkins A, Jaffee EM. Tumor Mutational Burden and Response Rate to PD-1 Inhibition. N Engl J Med 2017;377:2500-1. [Crossref] [PubMed]
- Goodman AM, Kato S, Bazhenova L, et al. Tumor Mutational Burden as an Independent Predictor of Response to Immunotherapy in Diverse Cancers. Mol Cancer Ther 2017;16:2598-608. [Crossref] [PubMed]
- Campbell BB, Light N, Fabrizio D, et al. Comprehensive Analysis of Hypermutation in Human Cancer. Cell 2017;171:1042-56.e10. [Crossref] [PubMed]
- Nicolas E, Golemis EA, Arora S. POLD1: Central mediator of DNA replication and repair, and implication in cancer and other pathologies. Gene 2016;590:128-41. [Crossref] [PubMed]
- Rizvi H, Sanchez-Vega F, La K, et al. Molecular Determinants of Response to Anti-Programmed Cell Death (PD)-1 and Anti-Programmed Death-Ligand 1 (PD-L1) Blockade in Patients With Non-Small-Cell Lung Cancer Profiled With Targeted Next-Generation Sequencing. J Clin Oncol 2018;36:633-41. [Crossref] [PubMed]
- Panda A, Betigeri A, Subramanian K, et al. Identifying a Clinically Applicable Mutational Burden Threshold as a Potential Biomarker of Response to Immune Checkpoint Therapy in Solid Tumors. JCO Precis Oncol 2017;2017:PO.17.00146.
- Samstein RM, Lee C-H, Shoushtari AN, et al. Tumor mutational load predicts survival after immunotherapy across multiple cancer types. Nat Genet 2019;51:202-6. [Crossref] [PubMed]
- Turajlic S, Litchfield K, Xu H, et al. Insertion-and-deletion-derived tumour-specific neoantigens and the immunogenic phenotype: a pan-cancer analysis. Lancet Oncol 2017;18:1009-21. [Crossref] [PubMed]
- Gandara DR, Paul SM, Kowanetz M, et al. Blood-based tumor mutational burden as a predictor of clinical benefit in non-small-cell lung cancer patients treated with atezolizumab. Nat Med 2018;24:1441-8. [Crossref] [PubMed]
- Aggarwal C, Thompson JC, Chien AL, et al. Baseline Plasma Tumor Mutation Burden Predicts Response to Pembrolizumab-based Therapy in Patients with Metastatic Non-Small Cell Lung Cancer. Clin Cancer Res 2020;26:2354-61. [Crossref] [PubMed]
- Llosa NJ, Cruise M, Tam A, et al. The vigorous immune microenvironment of microsatellite instable colon cancer is balanced by multiple counter-inhibitory checkpoints. Cancer Discov 2015;5:43-51. [Crossref] [PubMed]
- Drescher KM, Sharma P, Watson P, et al. Lymphocyte recruitment into the tumor site is altered in patients with MSI-H colon cancer. Fam Cancer 2009;8:231-9. [Crossref] [PubMed]
- Xu F, Liu J, Liu D, et al. LSECtin expressed on melanoma cells promotes tumor progression by inhibiting antitumor T-cell responses. Cancer Res 2014;74:3418-28. [Crossref] [PubMed]
- Woo SR, Turnis ME, Goldberg MV, et al. Immune inhibitory molecules LAG-3 and PD-1 synergistically regulate T-cell function to promote tumoral immune escape. Cancer Res 2012;72:917-27. [Crossref] [PubMed]
- Ascierto PA, Bono P, Bhatia S, et al. Efficacy of BMS-986016, a monoclonal antibody that targets lymphocyte activation gene-3 (LAG-3), in combination with nivolumab in pts with melanoma who progressed during prior anti-PD-1/PD-L1 therapy (mel prior IO) in all-comer and biomarker-enriched. Ann Oncol 2017;28:v605-49. [Crossref]
- Belli C, Trapani D, Viale G, et al. Targeting the microenvironment in solid tumors. Cancer Treat Rev 2018;65:22-32. [Crossref] [PubMed]
- Zhang QW, Liu L, Gong CY, et al. Prognostic significance of tumor-associated macrophages in solid tumor: a meta-analysis of the literature. PLoS One 2012;7:e50946 [Crossref] [PubMed]
- Munn DH, Zhou M, Attwood JT, et al. Prevention of allogeneic fetal rejection by tryptophan catabolism. Science 1998;281:1191-3. [Crossref] [PubMed]
- Uyttenhove C, Pilotte L, Théate I, et al. Evidence for a tumoral immune resistance mechanism based on tryptophan degradation by indoleamine 2,3-dioxygenase. Nat Med 2003;9:1269-74. [Crossref] [PubMed]
- Jha GG, Gupta S, Tagawa ST, et al. A phase II randomized, double-blind study of sipuleucel-T followed by Ido pathway inhibitor, indoximod, or placebo in the treatment of patients with metastatic castration resistant prostate cancer (mCRPC). J Clin Oncol 2017;35:3066. [Crossref]
- Zakharia Y, McWilliams R, Shaheen M, et al. Interim analysis of the Phase 2 clinical trial of the IDO pathway inhibitor indoximod in combination with pembrolizumab for patients with advanced melanoma. Cancer Research 77:Abstract nr CT117.
- Mazzarella L, Duso BA, Trapani D, et al. The evolving landscape of ‘next-generation’ immune checkpoint inhibitors: A review. Eur J Cancer 2019;117:14-31. [Crossref] [PubMed]
- Cruz-Bermúdez A, Laza-Briviesca R, Vicente-Blanco RJ, et al. Cancer-associated fibroblasts modify lung cancer metabolism involving ROS and TGF-β signaling. Free Radic Biol Med 2019;130:163-73. [Crossref] [PubMed]
- Mouw KW, Goldberg MS, Konstantinopoulos PA, et al. DNA Damage and Repair Biomarkers of Immunotherapy Response. Cancer Discov 2017;7:675-93. [Crossref] [PubMed]
- Arora S, Yan H, Cho I, et al. Genetic Variants That Predispose to DNA Double-Strand Breaks in Lymphocytes From a Subset of Patients With Familial Colorectal Carcinomas. Gastroenterology 2015;149:1872-1883.e9. [Crossref] [PubMed]
- Nicolas E, Arora S, Zhou Y, et al. Systematic evaluation of underlying defects in DNA repair as an approach to case-only assessment of familial prostate cancer. Oncotarget 2015;6:39614-33. [Crossref] [PubMed]
- Garrido C, Paco L, Romero I, et al. MHC class I molecules act as tumor suppressor genes regulating the cell cycle gene expression, invasion and intrinsic tumorigenicity of melanoma cells. Carcinogenesis 2012;33:687-93. [Crossref] [PubMed]
- Garrido F, Ruiz-Cabello F, Aptsiauri N. Rejection versus escape: the tumor MHC dilemma. Cancer Immunol Immunother 2017;66:259-71. [Crossref] [PubMed]
- Chowell D, Morris LGT, Grigg CM, et al. Patient HLA class I genotype influences cancer response to checkpoint blockade immunotherapy. Science 2018;359:582-7. [Crossref] [PubMed]
- Gong J, Chehrazi-Raffle A, Placencio-Hickok V, Guan M, Hendifar A, Salgia R. The gut microbiome and response to immune checkpoint inhibitors: preclinical and clinical strategies. Clin Transl Med 2019;8:9. [Crossref] [PubMed]
- Zitvogel L, Ma Y, Raoult D, et al. The microbiome in cancer immunotherapy: Diagnostic tools and therapeutic strategies. Science 2018;359:1366-70. [Crossref] [PubMed]
- Matson V, Fessler J, Bao R, et al. The commensal microbiome is associated with anti-PD-1 efficacy in metastatic melanoma patients. Science 2018;359:104-8. [Crossref] [PubMed]
- Gopalakrishnan V, Spencer CN, Nezi L, et al. Gut microbiome modulates response to anti-PD-1 immunotherapy in melanoma patients. Science 2018;359:97-103. [Crossref] [PubMed]
- Panda A, Mehnert JM, Hirshfield KM, et al. Immune Activation and Benefit From Avelumab in EBV-Positive Gastric Cancer. J Natl Cancer Inst 2018;110:316-20. [Crossref] [PubMed]
- Kim ST, Cristescu R, Bass AJ, et al. Comprehensive molecular characterization of clinical responses to PD-1 inhibition in metastatic gastric cancer. Nat Med 2018;24:1449-58. [Crossref] [PubMed]
- Seiwert TY, Burtness B, Mehra R, et al. Safety and clinical activity of pembrolizumab for treatment of recurrent or metastatic squamous cell carcinoma of the head and neck (KEYNOTE-012): an open-label, multicentre, phase 1b trial. Lancet Oncol 2016;17:956-65. [Crossref] [PubMed]
- Hsu C, Lee SH, Ejadi S, et al. Safety and Antitumor Activity of Pembrolizumab in Patients With Programmed Death-Ligand 1-Positive Nasopharyngeal Carcinoma: Results of the KEYNOTE-028 Study. J Clin Oncol 2017;35:4050-6. [Crossref] [PubMed]
- Ferris RL, Blumenschein G, Fayette J, et al. Nivolumab for Recurrent Squamous-Cell Carcinoma of the Head and Neck. N Engl J Med 2016;375:1856-67. [Crossref] [PubMed]
- Christiansen AJ, West A, Banks KM, et al. Eradication of solid tumors using histone deacetylase inhibitors combined with immune-stimulating antibodies. Proc Natl Acad Sci U S A 2011;108:4141-6. [Crossref] [PubMed]
- Kroesen M, Gielen P, Brok IC, et al. HDAC inhibitors and immunotherapy; a double edged sword? Oncotarget 2014;5:6558-72. [Crossref] [PubMed]
- Pili R, Quinn DI, Albany C, et al. Immunomodulation by HDAC inhibition: Results from a phase Ib study with vorinostat and pembrolizumab in metastatic urothelial, renal, and prostate carcinoma patients. J Clin Oncol 2019;37:2572. [Crossref]
- Sullivan RJ, Moschos SJ, Johnson ML, et al. Efficacy and safety of entinostat (ENT) and pembrolizumab (PEMBRO) in patients with melanoma previously treated with anti-PD1 therapy. In: Proceedings of the 110th Annual Meeting of the American Association for Cancer Research; 2019 March 29 - April 3; Atlanta, GA. Philadelphia (PA): AACR; 2019. Abstract nr CT072.
- Rini BI, Plimack ER, Stus V, et al. Pembrolizumab plus Axitinib versus Sunitinib for Advanced Renal-Cell Carcinoma. N Engl J Med 2019;380:1116-27. [Crossref] [PubMed]
- Motzer RJ, Penkov K, Haanen J, et al. Avelumab plus Axitinib versus Sunitinib for Advanced Renal-Cell Carcinoma. N Engl J Med 2019;380:1103-15. [Crossref] [PubMed]
- Makker V, Rasco D, Vogelzang NJ, et al. Lenvatinib plus pembrolizumab in patients with advanced endometrial cancer: an interim analysis of a multicentre, open-label, single-arm, phase 2 trial. Lancet Oncol 2019;20:711-8. [Crossref] [PubMed]
- Cheng AL, Qin S, Ikeda M, et al. LBA3 - IMbrave150: Efficacy and safety results from a ph III study evaluating atezolizumab (atezo) + bevacizumab (bev) vs sorafenib (Sor) as first treatment (tx) for patients (pts) with unresectable hepatocellular carcinoma (HCC). Annals of Oncology 2019;30:ix186-7. [Crossref]
- Puri S, Niyongere S, Chatwal MS, et al. Phase I/II study of nivolumab and ipilimumab combined with nintedanib in advanced NSCLC. J Clin Oncol 2018;36:TPS9112 [Crossref]


