
Identification of hub genes and compounds controlling ovarian cancer stem cell characteristics via stemness indices analysis
Introduction
Globally, there are approximately 239,000 new cases of ovarian cancer (OC) each year, accounting for 3.6% of all cancer diagnoses, along with 152,000 OC deaths, accounting for 4.3% of all annual cancer deaths (1). Compared with other female cancers, OC has the worst 5-year survival rate (47% in the United States and Canada), which has improved only slightly over the past three decades. This dilemma is largely attributed to its late-stage diagnosis and recurrent relapse. Serous epithelial carcinomas, which account for a substantial proportion of OC cases, are often diagnosed at advanced stages (III–IV, 80%) and lack effective treatments to significantly improve survival. Meanwhile, the upfront treatment of OC mainly relies on primary debulking surgery to achieve no residual disease (R0) and platinum-based chemotherapy. However, it is quite difficult to achieve R0 in bulking surgery due to common widespread disseminated metastases surrounding abdominal organs (2). Furthermore, although the majority of patients respond very well to initial chemotherapy, chemotherapy resistance followed by recurrent disease remains common in OC.
Cancer stem cells (CSCs), which possess the capacity for long-term self-renewal and abnormal differentiation residing at the apex, have been postulated to be responsible for tumor initiation (3-5). A variety of solid tumors have been reported to contain CSCs, including breast (6), pancreatic (7), colorectal (8), and ovarian (9) cancers. Furthermore, CSCs have been shown to contribute to metastases and chemotherapy resistance in OC. Bapat et al. reported on the stem- and progenitor-like cells derived from multilayered spheroids in the ascites of OC patients for the first time in 2005 (10). Using an in vivo OC xenograft model, Liao et al. demonstrated that the CSCs from ascites spheroids promoted metastasis of the disease to the peritoneum and omentum (11). In addition, although the initial chemotherapy could efficiently eliminate the bulk of the tumor mass, chemoresistant residual tumor cells (which include CSCs) would survive. It has been reported that ascites cells collected from chemoresistant patients exhibited enhanced expression of genes associated with CSCs compared to those collected from chemonaive patients (12). Moreover, numerous studies have demonstrated that residual tumor is enriched in CSC-like cells following chemotherapy (12-14). Thus far, several biological pathways have been shown to be pivotal for the chemoresistance of OC stem cells, including the Wnt (15), Notch (16,17), and Hedgehog (18,19) pathways. Given that CSCs contribute greatly to OC metastases and chemoresistance, it has been acknowledged that anti-CSC therapy increases the chemotherapeutic response and improves the prognosis of OC (20).
Although there are currently numerous markers that can be used to isolate and characterize CSCs in OC, including cluster of differentiation (CD)44, CD117, aldehyde dehydrogenase (ALDH), CD133, and CD24 (9,21,22), it remains challenging to identify the overall extent of the stemness of a given tumor. Recently, Malta et al. evaluated the novel stemness indices of almost 12,000 samples involving 33 tumor types using a deep learning algorithm (23). Briefly, they initially used a one-class logistic regression (OCLR) deep learning algorithm to construct a signature in order to quantify stemness based on transcriptomic and epigenomic molecular profile sequencing from stem cells that vary in stemness degree. Next, the OCLR-based signatures were applied to available datasets of The Cancer Genome Atlas (TCGA) to compute the messenger RNA expression-based stemness index (mRNAsi) and corresponding DNA methylation-based stemness index (mDNAsi). The former reflected gene expression traits, while the latter reflected epigenetic traits. The mRNAsi and mDNAsi indices were shown to indicate the degree of oncogenic dedifferentiation of the samples and the extent of stemness. The authors found that high mRNAsi and mDNAsi indices values were correlated with biological processes active in CSCs, along with deeper cancer cell dedifferentiation. Moreover, metastatic tumor cells were found to have higher index values and were more dedifferentiated phenotypically. Recently, this methodology has been applied to individual tumors in order to extract stemness-involved genes, including lung carcinoma (24), breast cancer (25), bladder cancer (26), and medulloblastoma (27).
Herein, we examined the stemness indices of 379 OC samples and 88 normal tissues, as well as the corresponding expression profiles to screen hub genes that are linked to OC stemness. We also explored the function of these hub genes and identified the underlying compounds that disturb them, which may be considered inhibitors of OC stem cells. We present the following article in accordance with the STREGA (Strengthening the Reporting of Genetic Association Studies) reporting checklist (available at http://dx.doi.org/10.21037/atm-20-3621).
Methods
The study complies with the ethical standards of the institution and/or the National Research Council, as well as the 1964 Helsinki Declaration and its subsequent amendments, or similar ethical standards.
Gene expression data collection and identification of differentially expressed genes (DEGs)
Messenger ribonucleic acid (mRNA) sequencing (mRNA-seq) data of 467 human samples, including 379 OC samples and 88 normal samples, were derived from TCGA and the Genotype Tissue Expression (GTEx) project using the University of California, Santa Cruz (UCSC) Xena browser (https://xena.ucsc.edu/) (28). Meanwhile, the corresponding clinical information of the OC samples was also collected. The R package “limma” was then used to identify DEGs in the OC and normal samples. The criteria for DEG screening were as follows: |log2 (fold change)| >2, and false discovery rate (FDR) <0.05.
Weighted correlation network analysis and the identification of hub genes
The R package “weighted gene co-expression network analysis” (WGCNA) was utilized to identify modules of highly correlated DEGs as previously described (29,30). Firstly, the DEG expression matrix was used to define gene co-expression similarity based on the Pearson’s correlation coefficient of paired genes, followed by an adjacency matrix transformed from co-expression similarity using the power function. Here, with a soft threshold β=4 and with the R2>0.9, the co-expression network distribution exhibited an essentially scale-free topology. The adjacency matrix was then transformed into a topological overlap matrix (TOM), and TOM-based dissimilarity was calculated by 1-TOM. The TOM-based dissimilarity subsequently led to distinct modules defined as clusters of densely interconnected genes (29).
Malta’s works provided a method to calculate the mRNAsi and epigenetically regulated mRNAsi (EREG-mRNAsi) indices, which reflect the stemness features of the samples (23). The R software was then used to evaluate the module-trait correlations with mRNAsi and EREG-mRNAsi. We selected two modules with the highest correlation of mRNAsi. Hub genes were defined as those with a gene significance (GS) score of >0.5 and a module membership (MM) score of >0.5, where the GS score represents the correlation between the gene and the mRNAsi index, and the MM score denotes the correlation between the gene and its module.
Gene co-expression analysis
The R package “corrplot” was used to calculate the Pearson’s correlations between the hub genes within a module based on the gene expression levels.
Protein-protein interaction (PPI) network
The online tool, STRING (Search Tool for the Retrieval of Interacting Genes/Proteins) version 11.0 (https://string-db.org/), was used to construct the hub genes’ PPI network (31). The minimum required interaction score threshold was set as 0.4. Meanwhile, a bar plot was drawn to visualize the counts of adjacent nodes of each gene in the PPI network.
Functional analysis of hub genes
The R packages “clusterProfiler” and “enrichplot” were used to perform the Gene Ontology (GO) project and the Kyoto Encyclopedia of Genes and Genomes (KEGG) functional pathway enrichment analyses. The terms with adjusted P values of <0.05 were considered statistically significant. The top ranked terms were shown as a bubble plot drawn by the R package “ggplot2”.
Prediction of anti-stemness compounds and identification of its three-dimensional (3D) conformers
The Connectivity Map (CMap, https://portals.broadinstitute.org/CMap/) is a collection of gene expression profiles from cultured human cell lineages treated with small molecule compounds. It helps scientists to discover functional connections between compounds and genes (32). Hub genes derived from brown modules (as up-regulated genes), and from green and red modules (as down-regulated genes) were imported into the CMap database. Subsequently, the enrichment score, which represents the similarities between the expression profiles of cells cultured with compounds and hub genes, was computed, and compounds listed by enrichment score were revealed. Using a P value of <0.01 and an enrichment score of <0, we identified the compounds that were significantly negatively correlated with hub genes. This indicated that these compounds may downregulate hub genes positively correlated to stemness and upregulate hub genes negatively correlated to stemness; thus, these compounds were considered to have anti-stemness properties. The PubChem (https://pubchem.ncbi.nlm.nih.gov/compound) provided 3D structures for the selected compounds (33).
Statistical analysis
All analysis was carried out in R version 3.6.2 and corresponding packages. For all data, *P<0.05, **P<0.01, and ***P<0.001 were considered as significant.
Results
Screening DEGs
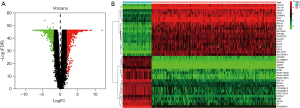
In total, we identified 2,253 DEGs in 379 OC samples and 88 normal samples using the “limma” package in R software. This included 1,017 upregulated genes and 1,236 downregulated genes in OC tissues. The volcano plot (Figure 1A) and heatmap (Figure 1B) of the DEGs were shown.
Identification of gene modules and hub genes correlated to mRNAsi and EREG-mRNAsi
We then utilized WGCNA to identify gene modules that were correlated to mRNAsi and EREG-mRNAsi based on the DEGs. The scale independence and mean connectivity [with various soft threshold (β) ranging from 1 to 20] are shown in Figure 2A,B, respectively. When β=4, the R^2 was >0.9 and the co-expression network distribution exhibited approximately scale-free topology. Altogether, five cohesive modules of highly correlated DEGs were obtained (Figure 2C). Next, we related the modules to sample traits (mRNAsi and EREG-mRNAsi indices) that represented their stemness levels. The green module was the most significantly correlated to mRNAsi (cor =−0.71), followed by the brown module (cor =0.64) and the red module (cor =−0.46) (Figure 2D). A minor relationship between the brown module and EREG-mRNAsi was also observed (cor =0.3). We then chose these three modules for the selection of highly connected intramodular genes (hub genes).
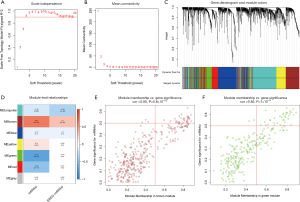
The threshold for identifying hub genes in modules correlated to mRNAsi indices was defined as an MM score of >0.5 and a GS score of >0.5. We obtained 31 hub genes in the brown modules (Figure 2E), 36 hub genes in the green modules (Figure 2F), and five hub genes in the red modules (Table 1). However, there were no genes left in the brown modules correlated to EREG-mRNAsi that satisfied the GS score >0.5 and MM score >0.5.
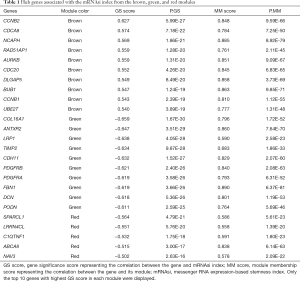
Full table
Hub gene expression in OC
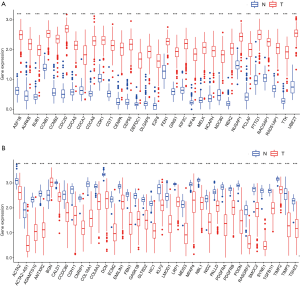
Since we had screened numerous hub genes that were either positively or negatively related to OC mRNAsi indices, we compared the expression levels of these genes between OC and normal samples. The hub genes derived from the brown module, which were positively correlated to mRNAsi indices, had higher expression levels in OC samples compared to normal samples (Figure 3A). In contrast, hub genes derived from the green module that were negatively correlated to mRNAsi indices were downregulated in OC (Figure 3B) and the similar result was found in the hub genes derived from the red module (data not shown). This result is consistent with the view that tumors originated from aberrant dedifferentiation cells and had a higher degree of stemness compared to the normal tissue, which was fully differentiated and resided at the apex.
Correlations between intramodular hub genes at the transcription level
To identify the mutual correlations between intramodular hub genes at the transcription level, we analyzed the Pearson’s correlation coefficients between intramodular hub genes. As shown in Figure 4, there was a statistically significant relationship between 31 hub genes derived from the brown module, and between 36 hub genes derived from the green module. In the brown module, the most robust correlation existed between cell division cycle 20 (CDC20) and kinesin family member 2C (KIF2C). In the green module, the strongest correlation existed between elastin microfibril interfacer 1 (EMILIN1) and platelet derived growth factor receptor alpha (PDGFRA).
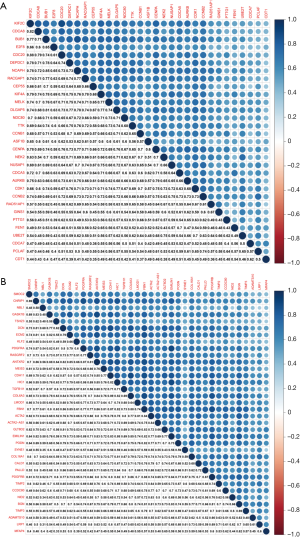
Correlations between intramodular hub genes at the protein level
We subsequently analyzed the correlations between intramodular hub genes at the protein level by constructing a PPI network (using the STRING online tools). The PPI network demonstrated a strong and wide-ranging relationship between the hub genes. Also, the edge number of each node gene in the PPI network in the brown module was almost equal (Figure 5A,B), while the edge number in the green module was not (Figure 5C,D), which indicated that hub genes derived from the brown module composed a more dense interaction network.
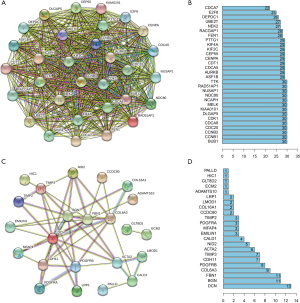
Functional analysis of hub genes
The hub genes we found proved to be highly interconnected. We examined GO annotation and the KEGG pathway to elaborate the functions of these hub genes. The top ranked GO terms and KEGG pathways based on the combined hub genes derived from modules in the brown, green, and red are displayed in Figure 6. The results demonstrated that hub genes primarily participated in biological processes associated with cell division, including organelle fission, sister chromatid segregation, nuclear chromosome segregation, and others.
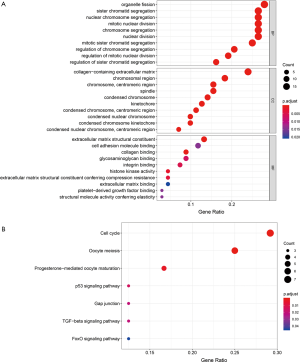
Prediction of anti-stemness compounds
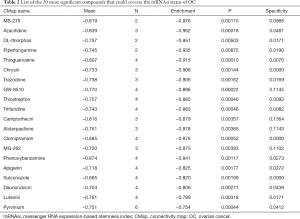
The CMap can be used to predict compounds that may induce or reverse a given gene expression signature. We employed CMap to predict which compounds could disturb hub genes that were correlated to OC mRNAsi indices. The top 20 ranked compounds are displayed in Table 2. Most of these compounds have been reported to be anti-stemness or anti-cancer drugs, while 11 compounds (MS-275, DL-thiorphan, piperlongumine, thioguanosine, GW-8510, trifluridine, alsterpaullone, clomipramine, phenoxybenzamine, sulconazole, and pyrvinium) have never been studied in OC stemness. We used the PubChem online tool (33) to draw the 3D construction of these 11 compounds (Figure 7A,B,C,D,E,F,G,H,I,J,K).
Full table
Discussion
OC is the most lethal gynecological malignancy. The high-grade serous subtype contributes to the majority of OC deaths, mainly as a result of the advanced stage of patients upon initial diagnosis and the high likelihood of relapse after chemotherapy. Over the past decade, ovarian CSCs have been shown to play a critical role in tumor growth, metastasis, and chemoresistance. A deep understanding of the molecular mechanisms of CSCs in OC will help to improve outcomes once CSC targeted therapy is available. In this study, we performed a bioinformatics analysis to identify OC stem cell-associated genes and discover the underlying compounds that kill ovarian OC stem cells.
Compared to other tumors, the CSCs in OC have not yet been clearly defined (34). However, certain stem cell markers, including CD133, receptor tyrosine kinase like orphan receptor 1 (ROR1), enhancer of zeste homolog 2 (EZH2), and GLI family zinc finger 2 (GLi2), have more frequently been found in recurrent/resistant OC and have not been identified in primary OC (35,36). As a novel index of stemness, the mRNAsi and EREG-mRNAsi indices were calculated using an OCLR machine learning algorithm, and were shown to reliably stratify tumors of given stemness phenotypes. Based on the expression profiles of OC and normal samples, along with the corresponding stemness indices, we identified five modules in the DEGs and screened hub genes in two of the five modules (with the most significant relationship with mRNAsi). As anticipated, hub genes that were positively and negatively related to stemness indices were upregulated and downregulated in OC, respectively. Furthermore, the correlation analysis showed that strong interactions between these genes were established at both the mRNA and protein levels. We also retrieved previous literature regarding these hub genes and found that most hub genes have been reported to be differentially expressed between stem cells and differentiated cells. For example, human embryonic stem (ES) cells had high expression levels of cell division cycle-associated 8 (CDCA8); however, differentiated human ES cell only had lowered levels of CDCA8 (37). Also, KIF2C was significantly more expressed in mouse neural stem cells compared to astrocytes (38). Furthermore, hypermethylated in cancer 1 (HIC1), which is negatively associated with stemness indices, was reported to be a marker of tissue-resident mesenchymal progenitors in skeletal muscle (39), and deletion of HIC1 stimulated the hyperplasia of mesenchymal progenitors. The hub genes identified in our study primarily participate in biological processes associated with cell division, including cell cycle regulation, alignment, and segregation of chromosomes (Figure 6). This was further verified by our GO annotation and KEGG pathway enrichment analysis, and signifies that OC stem cells are active in cell division and proliferation, which is consistent with previous studies that focused on other types of carcinomas (24,26).
Although the majority of patients with OC respond well to initial platinum-based chemotherapy, patients with advanced disease eventually experience recurrence and acquire resistance to chemotherapy. During the past decade, bevacizumab (antiangiogenic agent) has been approved for platinum-resistant disease as it helps to significantly improve progression-free survival (PFS) (40), with some evidence indicating that poly (ADP-ribose) polymerase inhibitors (PARPi) may be efficacious in patients with resistant disease (41,42). Also, identification of vulnerabilities in chemoresistant OC cells will help to develop new therapeutics for recurrent/resistant disease. The crucial role of OC stem cells in metastasis and chemoresistance has increased the enthusiasm of researchers to explore the targets of CSCs as a potential therapeutic strategy. Anti-CSCs therapies are generally classified into three main strategies: (I) targeting the markers of CSCs, (II) agents that induce the differentiation of CSCs, and (III) the targeting of vital pathways for CSC survival (43-45).
In this study we evaluated the overall degree of stemness in OC samples using mRNAsi indices and screened genes that were associated with mRNAsi. We then predicted compounds that could be considered underlying therapeutic agents for OC with enriched stem cells. This yielded complex components, including two epigenetic modification drugs (MS-275 and azacitidine), two cyclin dependent kinase (CDK) inhibitors (GW-8510 and alsterpaullone), two anti-depressant agents (clomipramine and trazodone), and various other types of drugs. Nevertheless, many of the top 20 ranked compounds have been investigated in stem cells. For example, MS-275, an inhibitor of histone deacetylases (HDACi), has been reported to induce the differentiation of human adipose tissue-extracted mesenchymal stem cells into neuronal tissue via the Wnt signaling pathway (46). Also, daunorubicin, an anthracycline aminoglycoside antineoplastic, has been extensively studied in leukemia and other neoplasm CSCs (47,48). Ultimately, we identified 11 compounds that have never been reported in OC stem cells. As the findings of the present study were obtained from the analyses of databases, the functions of 11 compounds in OC stem cells can be further examined in future studies.
Conclusions
We compared the available expression profiles of OC and normal tissues and identified hub genes associated with stemness characteristics. We also explored the functions of these hub genes and the underlying compounds that disturb them, which may be considered underlying targets for inhibiting OC stem cells.
Acknowledgments
Funding: This study was supported by the National Key Research and Development Program (No. 2019YFC1005200 and 2019YFC1005202) and the Hubei Province Health and Family Planning Scientific Research Project (No. WJ 2019M127).
Footnote
Reporting Checklist: The authors have completed the STREGA reporting checklist. Available at http://dx.doi.org/10.21037/atm-20-3621
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/atm-20-3621). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. It also complies with the ethical standards of the institution and/or the National Research Council, as well as the 1964 Helsinki Declaration and its subsequent amendments, or similar ethical standards.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Torre LA, Trabert B, DeSantis CE, et al. Ovarian cancer statistics, 2018. CA Cancer J Clin 2018;68:284-96. [Crossref] [PubMed]
- Lengyel E. Ovarian Cancer Development and Metastasis. Am J Pathol 2010;177:1053-64. [Crossref] [PubMed]
- Medema JP. Cancer stem cells: The challenges ahead. Nat Cell Biol 2013;15:338-44. [Crossref] [PubMed]
- Vermeulen L. The developing cancer stem-cell model: clinical challenges and opportunities. Lancet Oncol 2012;13:e83-9. [Crossref] [PubMed]
- Vermeulen L, Sprick MR, Kemper K, et al. Cancer stem cells - old concepts, new insights. Cell Death Differ 2008;15:947-58. [Crossref] [PubMed]
- Al-Hajj M, Wicha MS, Benito-Hernandez A, et al. Prospective identification of tumorigenic breast cancer cells. Proc Natl Acad Sci 2003;100:3983-8. [Crossref] [PubMed]
- Li C, Heidt DG, Dalerba P, et al. Identification of Pancreatic Cancer Stem Cells. Cancer Res 2007;67:1030-7. [Crossref] [PubMed]
- Dalerba P, Dylla SJ, Park IK, et al. Phenotypic characterization of human colorectal cancer stem cells. Proc Natl Acad Sci 2007;104:10158-63. [Crossref] [PubMed]
- Zhang S, Balch C, Chan MW, et al. Identification and Characterization of Ovarian Cancer-Initiating Cells from Primary Human Tumors. Cancer Res 2008;68:4311-20. [Crossref] [PubMed]
- Bapat SA, Mali AM, Koppikar CB, et al. Stem and Progenitor-Like Cells Contribute to the Aggressive Behavior of Human Epithelial Ovarian Cancer. Cancer Res 2005;65:3025-9. [Crossref] [PubMed]
- Liao J, Qian F, Tchabo N, et al. Ovarian Cancer Spheroid Cells with Stem Cell-Like Properties Contribute to Tumor Generation, Metastasis and Chemotherapy Resistance through Hypoxia-Resistant Metabolism. PLoS One 2014;9:e84941. [Crossref] [PubMed]
- Latifi A, Luwor RB, Bilandzic M, et al. Isolation and Characterization of Tumor Cells from the Ascites of Ovarian Cancer Patients: Molecular Phenotype of Chemoresistant Ovarian Tumors. PLoS One 2012;7:e46858. [Crossref] [PubMed]
- Ahmed N, Abubaker K, Findlay J, et al. Cancerous ovarian stem cells: Obscure targets for therapy but relevant to chemoresistance. J Cell Biochem 2013;114:21-34. [Crossref] [PubMed]
- Abubaker K, Latifi A, Luwor R, et al. Short-term single treatment of chemotherapy results in the enrichment of ovarian cancer stem cell-like cells leading to an increased tumor burden. Mol Cancer 2013;12:24. [Crossref] [PubMed]
- Chau WK, Ip CK, Mak ASC, et al. c-Kit mediates chemoresistance and tumor-initiating capacity of ovarian cancer cells through activation of Wnt/β-catenin-ATP-binding cassette G2 signaling. Oncogene 2013;32:2767-81. [Crossref] [PubMed]
- Park JT, Chen X, Tropè CG, et al. Notch3 Overexpression Is Related to the Recurrence of Ovarian Cancer and Confers Resistance to Carboplatin. Am J Pathol 2010;177:1087-94. [Crossref] [PubMed]
- McAuliffe SM, Morgan SL, Wyant GA, et al. Targeting Notch, a key pathway for ovarian cancer stem cells, sensitizes tumors to platinum therapy. Proc Natl Acad Sci 2012;109:E2939-48. [Crossref] [PubMed]
- Ray A, Meng E, Reed E, et al. Hedgehog signaling pathway regulates the growth of ovarian cancer spheroid forming cells. Int J Oncol 2011;39:797-804. [PubMed]
- Song X, Yan L, Lu C, et al. Activation of hedgehog signaling and its association with cisplatin resistance in ovarian epithelial tumors. Oncol Lett 2018;15:5569-76. [Crossref] [PubMed]
- Roy L, Cowden Dahl K. Can Stemness and Chemoresistance Be Therapeutically Targeted via Signaling Pathways in Ovarian Cancer? Cancers (Basel) 2018;10:241. [Crossref] [PubMed]
- Szotek PP, Pieretti-Vanmarcke R, Masiakos PT, et al. Ovarian cancer side population defines cells with stem cell-like characteristics and Mullerian Inhibiting Substance responsiveness. Proc Natl Acad Sci 2006;103:11154-9. [Crossref] [PubMed]
- Choi YJ, Ingram PN, Yang K, et al. Identifying an ovarian cancer cell hierarchy regulated by bone morphogenetic protein 2. Proc Natl Acad Sci 2015;112:E6882-8. [Crossref] [PubMed]
- Malta TM, Sokolov A, Gentles AJ, et al. Machine Learning Identifies Stemness Features Associated with Oncogenic Dedifferentiation. Cell 2018;173:338-354.e15. [Crossref] [PubMed]
- Zhang Y, Tseng JTC, Lien IC, et al. mRNAsi Index: Machine Learning in Mining Lung Adenocarcinoma Stem Cell Biomarkers. Genes (Basel) 2020;11:257. [Crossref] [PubMed]
- Pei J, Wang Y, Li Y. Identification of key genes controlling breast cancer stem cell characteristics via stemness indices analysis. J Transl Med 2020;18:74. [Crossref] [PubMed]
- Pan S, Zhan Y, Chen X, et al. Identification of Biomarkers for Controlling Cancer Stem Cell Characteristics in Bladder Cancer by Network Analysis of Transcriptome Data Stemness Indices. Front Oncol 2019;9:613. [Crossref] [PubMed]
- Lian H, Han Y, Zhang Y, et al. Integrative analysis of gene expression and DNA methylation through one-class logistic regression machine learning identifies stemness features in medulloblastoma. Mol Oncol 2019;13:2227-45. [Crossref] [PubMed]
- Haeussler M, Zweig AS, Tyner C, et al. The UCSC Genome Browser database: 2019 update. Nucleic Acids Res 2019;47:D853-8. [Crossref] [PubMed]
- Zhang B, Horvath S. A general framework for weighted gene co-expression network analysis. Stat Appl Genet Mol Biol 2005;4:Article17.
- Langfelder P, Horvath S. WGCNA: an R package for weighted correlation network analysis. BMC Bioinformatics 2008;9:559. [Crossref] [PubMed]
- Szklarczyk D, Gable AL, Lyon D, et al. STRING v11: protein-protein association networks with increased coverage, supporting functional discovery in genome-wide experimental datasets. Nucleic Acids Res 2019;47:D607-13. [Crossref] [PubMed]
- Lamb J. The Connectivity Map: Using Gene-Expression Signatures to Connect Small Molecules, Genes, and Disease. Science 2006;313:1929-35. [Crossref] [PubMed]
- Kim S, Chen J, Cheng T, et al. PubChem 2019 update: improved access to chemical data. Nucleic Acids Res 2019;47:D1102-9. [Crossref] [PubMed]
- Pujade-Lauraine E, Banerjee S, Pignata S. Management of Platinum-Resistant, Relapsed Epithelial Ovarian Cancer and New Drug Perspectives. J Clin Oncol 2019;37:2437-48. [Crossref] [PubMed]
- Tomao F, Papa A, Strudel M, et al. Investigating Molecular Profiles of Ovarian Cancer: An Update on Cancer Stem Cells. J Cancer 2014;5:301-10. [Crossref] [PubMed]
- Cojoc M, Mäbert K, Muders MH, et al. A role for cancer stem cells in therapy resistance: Cellular and molecular mechanisms. Semin Cancer Biol 2015;31:16-27. [Crossref] [PubMed]
- Dai C, Miao CX, Xu XM, et al. Transcriptional Activation of Human CDCA8 Gene Regulated by Transcription Factor NF-Y in Embryonic Stem Cells and Cancer Cells. J Biol Chem 2015;290:22423-34. [Crossref] [PubMed]
- Qin S, Huang X, Wang D, et al. Identification of characteristic genes distinguishing neural stem cells from astrocytes. Gene 2019;681:26-35. [Crossref] [PubMed]
- Scott RW, Arostegui M, Schweitzer R, et al. Hic1 Defines Quiescent Mesenchymal Progenitor Subpopulations with Distinct Functions and Fates in Skeletal Muscle Regeneration. Cell Stem Cell 2019;25:797-813.e9. [Crossref] [PubMed]
- Wilson MK, Pujade-Lauraine E, Aoki D, et al. Fifth Ovarian Cancer Consensus Conference of the Gynecologic Cancer InterGroup: recurrent disease. Ann Oncol 2017;28:727-32. [Crossref] [PubMed]
- Fong PC, Yap TA, Boss DS, et al. Poly(ADP)-ribose polymerase inhibition: frequent durable responses in BRCA carrier ovarian cancer correlating with platinum-free interval. J Clin Oncol 2010;28:2512-9. [Crossref] [PubMed]
- Kaufman B, Shapira-Frommer R, Schmutzler RK, et al. Olaparib monotherapy in patients with advanced cancer and a germline BRCA1/2 mutation. J Clin Oncol 2015;33:244-50. [Crossref] [PubMed]
- Kwon MJ, Shin Y. Regulation of Ovarian Cancer Stem Cells or Tumor-Initiating Cells. Int J Mol Sci 2013;14:6624-48. [Crossref] [PubMed]
- Ahmed N, Abubaker K, Findlay JK. Ovarian cancer stem cells: Molecular concepts and relevance as therapeutic targets. Mol Aspects Med 2014;39:110-25. [Crossref] [PubMed]
- Aguilar-Gallardo C, Rutledge EC, Martínez-Arroyo AM, et al. Overcoming Challenges of Ovarian Cancer Stem Cells: Novel Therapeutic Approaches. Stem Cell Rev Rep 2012;8:994-1010. [Crossref] [PubMed]
- Jang S, Jeong HS. Histone deacetylase inhibition-mediated neuronal differentiation via the Wnt signaling pathway in human adipose tissue-derived mesenchymal stem cells. Neurosci Lett 2018;668:24-30. [Crossref] [PubMed]
- Lin TY, Zhu Y, Li Y, et al. Daunorubicin-containing CLL1-targeting nanomicelles have anti-leukemia stem cell activity in acute myeloid leukemia. Nanomedicine Nanotechnology. Nanomedicine 2019;20:102004. [Crossref] [PubMed]
- Zhao WY, Zhang CX, Liu L, et al. Construction of Functional Targeting Daunorubicin Liposomes Used for Eliminating Brain Glioma and Glioma Stem Cells. J Biomed Nanotechnol 2016;12:1404-20. [Crossref] [PubMed]
(English Language Editors: A. Kassem and J. Gray)


