
FEN1 is a prognostic biomarker for ER+ breast cancer and associated with tamoxifen resistance through the ERα/cyclin D1/Rb axis
Introduction
Breast cancer is the most common malignant tumour occurring in women (1,2). Roughly 70–75% of breast cancers are oestrogen receptor-positive (ER+) and effective anti-ER endocrine therapy has brought benefits to reduce cancer-related mortality. Tamoxifen, an ER antagonist, remains an important choice for endocrine therapy in patients with ER+ breast cancer (3-5). Unfortunately, approximately 30% of women eventually relapses and dies due to the emergence of tamoxifen resistance (6,7). Previous studies have shown that the ERα pathway interacts with DNA damage responses and DNA repair reactive kinases, increasing genomic instability and causing failure of breast cancer treatment (8,9). Therefore, in-depth exploration of DNA damage repair systems and related mechanisms of tamoxifen resistance has important clinical value for overcoming tamoxifen resistance in patients with ER+ breast cancer.
Flap endonuclease-1 (FEN1) is a highly conserved structure-specific nuclease and possesses multiple activities including flap endonuclease, 5'-exonuclease and gap-endonuclease, which allow FEN1 to play an essential role in Okazaki fragment maturation, long-patch base excision repair, stalled replication fork rescue, telomere maintenance, and apoptotic DNA fragmentation (10-16): because the lack of the activity of FEN1 nuclease leads to the initiation of cancer, FEN1 is generally regarded as a tumour suppressor in maintaining the integrity of genomes (17,18). However, partially due to its essential role in DNA replication and repair, over-expression of FEN1 confers proliferation, migration, and drug resistance in cancer cells (10,19-26). A higher FEN1 expression level could be observed in multiple types of cancer, including breast cancer, which is related to poor differentiation and poor prognosis (22,27-31). In addition, our group also found that over-expression of FEN1 can promote breast cancer cells in terms of proliferation, migration, and drug resistance (21,24,32). Although functions of FEN1 in activating cancer progression are characterised extensively and FEN1 interactions with ERα have been studied (8,33), few researchers have investigated the function and molecular mechanisms of FEN1-mediated endocrine therapy resistance.
In this study, we present evidence suggesting that FEN1 is a prognostic biomarker for patients with ER+ breast cancer, especially in predicting disease recurrence and overall survival (OS) of these patients with adjuvant tamoxifen therapy through on-line database and IHC analysis from samples collected in our center. Then, we found that FEN1 rendered the ER+ breast cancer cells insensitive to the growth inhibitory effects of tamoxifen in vitro, which was associated with the activation of the ERα/cyclin D1/Rb axis. These findings provide better evidence as to how FEN1 contributes to tamoxifen resistance and serves as a critical regulator in activation of the ERα/cyclin D1/Rb axis.
We present the following article in accordance with the REMARK reporting checklist (available at http://dx.doi.org/10.21037/atm-20-3068).
Methods
The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013). The study was approved by institutional/regional/national ethics/committee/ethics board of the first hospital of China Medical University (No. [2016]120: the registration number of ethics board) and informed consent was taken from all the patients.
Kaplan-Meier (KM) plotter analysis
We used KM Plotter (http://kmplot.com/analysis/), a database that integrates gene expression data and clinical data, to obtain survival data for breast cancer, in relation to expression levels of genes of interest (34). Kaplan Meier plotter has information of 54,675 genes on survival, including 5,143 breast, 1816 ovarian, 2,437 lung and 1,065 gastric cancer patients with a mean follow-up of 69/40/49/33 months, respectively.
Brifly, the best specific probes (JetSet probes) for FEN1(Affy ID:204767_s_at) was entered to obtain KM plots. Information on relapse free survival (RFS) and overall patient survival (OS) was extracted. Furthermore, information on number of cases along with median values of mRNA expression levels, hazard ratios (HR) with 95% CIs and P values were extracted from the KM plotter webpage and considered significant having P values 0.05.
GEO datasets analysis, Microarray data analysis and Gene signature definition
The mRNA expression profiling of all the samples in this study were performed on the Human Affymetrix Human Genome U133 Plus 2.0 Array or Illumina Genome Analyzer II. GSE9195 was used to show the association between FEN1 expression and tamoxifen efficacy, and DFS was analysed (35). GSE25710 was used to obtain ERα ChIP-Seq data, and the map of ER binding at whole genome level was analysed (36).
Microarray technology was utilized to investigate changes in mRNA profiles with FEN1 siRNA versus control group in MCF-7 cells. Total RNA was extracted using TRIzol® reagent (Invitrogen; Thermo Fisher Scientific, Inc.) and the RNA was purified, amplified, labeled, and hybridized according to the manufacturer’s protocol (Genechem Company, Shanghai, China). Further data analysis was performed with the R software package, such as limma package. FEN1 with an expression fold change > |1.5| was considered to be statistically significantly differentially expressed. Based on the differentially expressed genes (DEGs), Gene Set Enrichment Analysis (GSEA) was performed with molecular signatures database (MSigDB) for pathways analysis (37,38).
Unsupervised hierarchical cluster analysis of genes that were changed when MCF-7 was treated with E2 (E2 vs. Veh.) or E2 plus 4-OHT (E2+ 4-OHT vs. E2) using GSE25316 dataset compared to genes that were differentially expressed upon FEN1. Genes that were shared between ERα-dependent core genes and FEN1-regulated ones were identified as the signature genes that are controlled by both FEN1 and ERα signaling. The gene signatures were determined to coregulate by FEN1 and ERα signaling according to the ERα-dependent core genes, which were defined by changed upon E2 stimulation and transcription start site of ERα binding peak (39).
Patient tissue specimens and immunohistochemistry (IHC)
This study retrospectively analyzed 65 patients with ER+ breast cancer. These patients were admitted to the First Affiliated Hospital of China Medical University from 2002 to 2008, and had the end of five-year tamoxifen treatment or developed relapse under regular adjuvant hormone therapy (tamoxifen 20 mg/d). Clinical pathological data of the cohorts are shown in Table 1. Patient cohorts for IHC staining, tumor specimen collection, survey data, and all clinical and pathologic information were reviewed and approved by the Ethics Committee of China Medical University. The current study includes follow-up data available as of October 31, 2019, the median follow-up time was 152 months. The relapse-free survival (RFS) was set on the period from the date of surgery to recurrence. The overall survival (OS) was set on the period from the date of surgery to death or to the most recent clinic visit. Antibody used for IHC: Mouse Anti-Human FEN1 (working concentrations were 1:200) were purchased from Santa Cruz Biotechnology (CA, USA). The results of IHC were assessed with double-blind method, the staining results were reviewed and approved by two specialists in Department of Pathology in the first hospital of China Medical University. The positive staining of FEN1 was defined as those showing nuclei or cytoplasmic staining of tumor cells. Briefly, the scoring method that takes both staining intensity and proportion of stained cells into account. The staining intensity was classified into four categories according to the color of immune reactions: negative, 0, no staining; weak, 1, light brown; moderate, 2, brown in color; and strong, 3, with dark brown staining. The proportion of positively stained cells was reported as: 0–25%, 1; 26–50%, 2; 51–75%, 3; and 76–100%, 4. The overall expression level of FEN1 was obtained by the staining intensity and the proportion of positively stained cells. A median expression score of 6 was taken as the cut-off value, samples with scores of 0–4 were considered as low expressing, others with scores of 6–12 were defined as high expressing.
Full table
Cell culture, small interfering RNA (siRNA) transfection and lentiviral transfection
The human ER+ breast cancer cell line MCF-7 and T47D were obtained from the Type Culture Collection of the Chinese Academy of Sciences (Shanghai, China). both cell lines were cultured in DMEM (GIBCO BRL, Grand Island, NY) containing 10% fetal bovine serum (GIBCO BRL), 100 U/mL penicillin and 100 µg/mL streptomycin in a humidified incubator at 37 °C with an atmosphere of 5% CO2.
Small interfering RNAs (siRNAs) for FEN1 ordered from RiboBio Company (Guangzhou, China). The target sequence of FEN1 was 5'-GGGTCAAGAGGCTGAGTAA-3' (sense), 5'-UUACUCAGCCUCUUGACCCdTdT-3' (anti-sense), and negative control: 5'-UUCUCCGAACGUGUCACGUtt-3' (sense), 5'-ACGUGACACGUUCGGAGAAtt-3' (anti-sense). The siRNAs (100 nM) were transfected into cells using Lipofectamine 2000 (Invitrogen) according to the manufacturer’s instructions. Seventy-two hours after transfection, cells were harvested for the subsequent experiments.
FEN1 overexpressing and FEN1 knockdown lentivirus were purchased from Genechem Company (Shanghai, China). The processes of lentivirus transfection were performed as described previously (21). In brief, the lentiviral vectors LV-GFP-FEN1-RNAi, LV-GFP-FEN1-3FLAG and empty vector controls were synthesized (Genechem Company). The target sequence of FEN1 was the same as siRNA. To estimate transfection efficiency, our experiments utilize LV-GFP-FEN1-RNAi, LV-GFP-FEN1-3FLAG and LV-GFP-NC in which GFP is expressed as a fusion. The percent of GFP-positive cells was determined by florescent microscopy (BX61, Olympus, Japan) 120 hours after transfection. Poor transfection can result in low translocation efficiency. Western blot analysis was performed to detect the knock-down and overexpression efficiency.
Cell viability assay and colony formation experiments
MTT assay was used to measure the cell viability after using tamoxifen. Cells were inoculated in the 96-well plate, 5,000 cells per well. Incubation for 24 hours to make sure all cells were attached. After 96 h treatment, MTT was added to incubate for 4 h, and then dimethyl sulfoxide (DMSO) was added. OD value of the survival cells were determined under 570 nm wavelength using microplate reader (Bio-Tek, GA, USA). The percentage of cell viability was calculated. 4-OHT was purchased from Sigma-Aldrich (Merck, China).
As for colony formation assay, 1,000 cells were inoculated in 24-well plates, then cells were treated with 1 uM 4-OHT and maintained in an incubator of 5% CO2 at 37 °C for 14 days. The culture medium was changed every three days. At the end of the experiments, cells were washed with PBS and fixed with 75% ethanol for 5 min at room temperature and then stained with Giemsa for 30 min at room temperature. Colonies with more than 50 cells were counted under an inverted microscope.
Western blot analysis
For western blot, the process was described previously (40). The membrane was incubated with the indicated primary and secondary antibodies, and the proteins were visualized by an enhanced ECL kit (Beyotime, China). Antibody: FEN1 (Genetex), ERɑ (Santa Cruz Biotechnology), phosphorylated(p)-ER (Cell Signaling Technology, Danvers, MA, USA), Cyclin D1 (Santa Cruz Biotechnology), Rb (Santa Cruz Biotechnology), p-Rb (Santa Cruz Biotechnology), E2F (Santa Cruz Biotechnology), Cyclin B (Santa Cruz Biotechnology), Cyclin E (Santa Cruz Biotechnology) and GAPDH (Cell Signaling Technology, Danvers, MA, USA). Imaging Densitometer with Molecular Analyst Software (Bio-Rad) and expressed as the ratios to the density of GAPDH bands.
Statistical analysis
Each experiment was repeated at least 3 times unless otherwise specified. Group data comparisons were conducted by χ2 tests. The results were expressed as mean ± standard deviation (SD) in this study. Associations between FEN1 expression and clinical parameters were evaluated using Chi-square test analysis. The KM method, two-tailed log-rank test, and Cox proportional hazard model were used for survival analysis. P<0.05 is considered to be statistically significant. All statistical tests were performed on SPSS 20.0 software.
Results
High expression of FEN1 correlated with worse prognosis in ER+ breast cancer patients receiving tamoxifen treatment
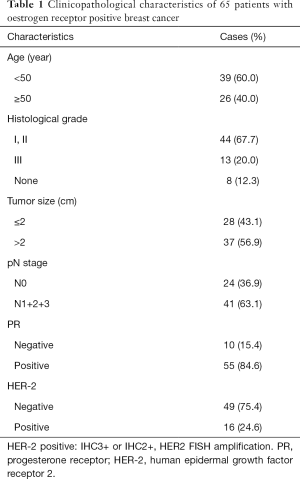
To investigate the clinical relevance of FEN1 in ER+ breast cancer, we first correlated the mRNA expression levels of FEN1 with the RFS and OS using a KM plotter. The results showed that the high expression of FEN1 was associated with a shorter RFS for patients with ER+ breast cancer [Figure 1A, Hazard ratio (HR) = 1.64 (1.39–1.93); logrank P=4.8×10−9]. However, FEN1 expression was not prognostic in ER– breast cancers [Figure 1B; HR = 1.05 (0.84–1.32); logrank P=0.66]. Further, the correlation of FEN1 expression in tamoxifen-treated ER+ patients was assessed. A high expression of FEN1 correlated with a shorter RFS in the ER+ breast cancer patients who received tamoxifen [Figure 1C; HR = 1.62 (1.19–2.21); logrank P=0.0019]. The results of OS were consistent with the RFS (Figure 1D,E,F). Taken together, the above results suggest that FEN1 may play an important role in predicting the prognosis of patients with ER+ breast cancer and patients who received tamoxifen treatment.
Patients with high expression of FEN1 showed tamoxifen resistance
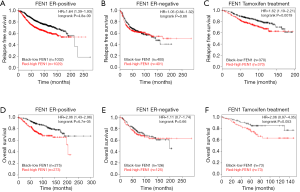
We re-analysed ER+ breast cancer patients from GSE9195 dataset, in which the patients received tamoxifen as an adjuvant treatment. There was a significant increase of FEN1 mRNA level in patients that were resistant to tamoxifen compared to the sensitive group (Figure 2A; P=0.0062).
Among the 65 cancer specimens in the current study, 32 patients (49.23%) demonstrated high FEN1 expression. High expression of FEN1 was significantly correlated with lymph node positivity (P=0.013), but not with age (P=0.265), histological grade (P=0.431) and tumor size (P=0.162) (Table 2). The high expression staining of FEN1 was 33.3% in disease-free patients, 87.5% in less than 2-year recurrence patients and 73.3% in more than 2-year recurrence patients (Figure 2B,C). IHC staining showed that the expression of FEN1 was significantly increased in the less than 2-year recurrent tamoxifen-resistant tumours than that in tamoxifen-sensitive tumours (recurrence-free and more than 2-year recurrence); high FEN1 expression was associated with poor prognosis in breast cancer patients receiving tamoxifen therapy (DFS, Figure 2D, P<0.001; OS, Figure 2E, P<0.001).
Full table
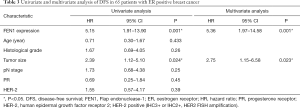
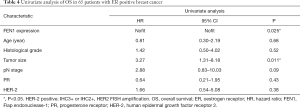
Univariate and multivariate analysis for PFS and OS
Furthermore, the univariate and a Cox multivariable proportional hazard model were constructed to evaluate independent prognostic significance of FEN1 expression and some clinicopathological characteristics. Univariate regression analysis showed that FEN1 expression (P=0.001) and tumor size (P=0.024) were significantly associated with DFS in 65 patients with ER positive BC (Table 3). These variables with P<0.10 were included in multivariate regression analysis using a forward step-wise method. The results showed that FEN1 expression (P=0.001) and tumor size (P=0.023) were independent factors for DFS. We next assessed whether these variables had the prognostic impacts on OS as well. Multivariable analysis of outcomes for the entire cohort showed that FEN1 expression (P=0.025) and tumor size (P=0.011) were significantly associated with worse survival (Table 4). Since equation does not converge, HR and CIs was not available.
Full table
Full table
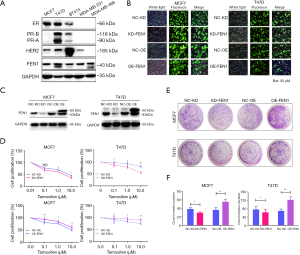
FEN1 rendered tamoxifen resistance in ER-positive breast cancer cell lines
To explore further the actual function of FEN1 in tamoxifen resistance, we over-expressed or knocked-down FEN1 in MCF-7 and T47D cell lines (Figure 3A,B,C), and then measured cellular response to increasing concentrations of 4-hydroxytamoxifen (4-OHT), the active metabolite of tamoxifen. After over-expression of FEN1, MCF-7 and T47D demonstrated a decreased sensitivity to tamoxifen, but we observed an inhibitory effect of tamoxifen on MCF-7 and T47D cell proliferation after deletion of FEN1 (Figure 3D,E,F). Combining the above, these results indicated that FEN1 plays an essential role in driving tamoxifen resistance and may act as a promising therapeutic target.
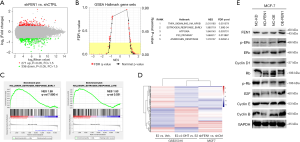
FEN1 over-expression elicited an Endo-R gene signature and ER/Cyclin D/Rb axis
The better to understand the effects of FEN1 in tamoxifen resistance, the RNA-seq analysis revealed a total of 271 up-regulated genes and 336 down-regulated genes (FC >1.5, P<0.05) in si-FEN1 MCF-7 cells compared to the control group (Figure 4A, Table S1). Functional annotation of these differential genes in the GSEA to interrogate the oncogenic gene signatures from the Molecular Signatures Database (MSigDB) showed that the “OESTROGEN_RESPONSE_EARLY” pathway (P=7.89×10−4) was mostly annotated according to the adjusted P value (Figure 4B). Interestingly, within the down-regulated genes, the most enriched term was “OESTROGEN RESPONSE EARLY” (P=7.89×10−4) and “OESTROGEN RESPONSE LATE” (P=0.031), suggesting that over-expression of FEN1 enhanced ER-related downstream signalling (Figure 4C). In addition, the FEN1-induced mRNA profile was in agree with the Endo-R Gene Signature that was up-regulated in the E2-stimulatedcases and ran contrary to tamoxifen inhibition (Figure 4D). These data suggested that the high expression of FEN1 potentially drives a transcriptional programme associated with high ER signalling that contributes to endocrine resistance. To confirm the above analysis, we knocked-down and over-expressed FEN1 respectively. As shown in Figure 4E, p-ERα, cyclin D1, and p-Rb were up-regulated after FEN1 over-expression, and vice versa. We may thus infer that over-expression of FEN1 activates the ERα/cyclin D1/Rb axis.
Discussion
For more than four decades, tamoxifen has been used to treat ER+ breast cancer as a classic medicine for endocrine therapy; however, a proportion of patients with ER+ breast cancer that received tamoxifen treatment eventually acquired resistance thereto (6,7). The major challenge is identifying new therapeutic targets or specific biomarkers that are predictive of the therapeutic responses to endocrine therapy to achieve successful treatment. A study has highlighted a correlation between FEN1 over-expression and poor prognosis in breast cancer (31). In this study, our results showed that elevated FEN1 mRNA expressions were correlated with shorter RFS, DMFS, and OS in breast cancer patients, and more significant in ER-positive breast cancer and tamoxifen treatment failure subtype. Next, the IHC analysis of FEN1 protein levels in 55 ER+ breast cancer patients also demonstrated that high expression of FEN1 protein was highly significantly associated with shorter DFS and OS in tamoxifen treatment. Our results showed that the FEN1 is an important prognostic biomarker of breast cancer patients, especially in the tamoxifen treatment failure group. The above results showed that FEN1 may be a biomarker of tamoxifen resistance. The in-depth study of the function and mechanism of FEN1 participation in tamoxifen resistance plays an important role in screening the benefit to patients of tamoxifen and reversing drug resistance.
In recent years, studies have reported that the high expression of FEN1 is involved in drug resistance processes such as chemotherapies, radiation treatment, and targeted therapy (24-26). Through inhibiting expression of FEN1 or application of FEN1 small molecule inhibitors, it can reverse drug resistance and synergistic chemotherapy/radiotherapy sensitivity (26,41-47). These results indicated that FEN1 is a key molecule associated with the resistance to anti-cancer therapy, however, the precise functions of FEN1 in tamoxifen-resistance remain unknown. The further to confirm this ability, we established FEN1 over-expressed and knocked-down ER+ breast cancer cell lines. Next, both MTT experiments and colony formation experiments showed that tamoxifen-sensitive breast cancer cell lines MCF-7 and T47D were less sensitive to tamoxifen after FEN1 was over-expressed, and the inhibitory effect of tamoxifen on the growth of breast cancer cells was enhanced by knocking out the FEN1 gene. These results indicated that breast cancer cell over-expressed FEN1 is resistant to tamoxifen and knocking out FEN1 would enhance the inhibitory effect of tamoxifen on cells. A high expression of FEN1 was related to drug resistance, and it was significant to reversing drug resistance, especially tamoxifen resistance caused by over-expression of FEN1.
Previous findings have indicated that FEN1 may active EGFR signalling, and promote epithelial-mesenchymal transition (EMT) and anti-apoptosis, which are the classic mechanism of endocrine resistance (21,48,49). Schultz-Norton et al. found that oestrogen promotes the binding of FEN1 to multiple domains of ERα, including the DNA domain, C-terminal, and carboxy-terminal domains, thereby enhancing ERα-mediated oestrogen-responsive gene transcription (33). Moreover, the study from our team has reported that FEN1 may mediate trastuzumab resistance via enhancing ERα-target gene transcription (24). The abnormal activation of ERα-target gene transcription was another mechanism of tamoxifen resistance (50). These previous studies have provided several underlying molecular mechanisms of tamoxifen resistance caused by FEN1. To find the reason, we performed GSEA using the MSigDB hallmark gene sets, which indicated two hallmark gene sets (“oestrogen response early” and “oestrogen response late”) were positively correlated with FEN1 expression. Further verification through western blot assay showed that FEN1 over-expression could significantly increase the level of p-ERα, cyclin D1, p-RbSer807/811, and E2F to initiate transcription of downstream target genes such as cyclin E and cyclin B, suggesting that the promotion of Rb phosphorylation was probably involved in FEN1-induced tamoxifen resistance. This was consistent with earlier studies reporting that FEN1 enhances ERα-mediated oestrogen-responsive gene transcription (33). From these results, we proposed that FEN1 stimulated the activation of the ERα/cyclinD1/Rb axis to promote tamoxifen resistance. However, this study had the some limitations of retrospective studies conducted at a single-center and smaller sample size. Therefore, the findings require validation with large-scale, multi-center clinical studies.
Conclusions
In summary, our study confirmed that the high expression of FEN1 was related to poor survival in ER+ breast cancer patients. We found that breast cancer cells were less sensitive to tamoxifen after FEN1 was over-expressed and became sensitive to tamoxifen when FEN1 was knocked out. For the first time we found that FEN1 may participate in tamoxifen resistance via the ERα/Cyclin D1/Rb axis, which provided evidence that may improve precision treatment with tamoxifen. In future, whether inhibition of FEN1 may reverse tamoxifen resistance (or not) warrants further investigation.
Acknowledgments
Funding: This work was supported by National Natural Science Foundation of China (No. 81672605); National Science and Technology Major Project of the Ministry of Science and Technology of China (No. 2017ZX09304025); The Key Research and Development Program of Liaoning Province(2018225060); Special Project of Liaoning Province of China (2019020176-JH1/103); Science and Technology Plan Project of Liaoning Province (No. 2013225585); Technological The general project of Liaoning province department of education (No. L2015588); Science and Technology Plan Project of Liaoning Province (No. 2015020458); National Key R&D Program of China (Grant #2018YFC1311600).
Footnote
Reporting Checklist: The authors have completed the REMARK reporting checklist. Available at http://dx.doi.org/10.21037/atm-20-3068
Peer Review File: Available at http://dx.doi.org/10.21037/atm-20-3068
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/atm-20-3068). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013). The study was approved by institutional/regional/national ethics/committee/ethics board of the first hospital of China Medical University (No. [2016]120: the registration number of ethics board) and informed consent was taken from all the patients.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Bray F, Ferlay J, Soerjomataram I, et al. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin 2018;68:394-424. [Crossref] [PubMed]
- Harbeck N, Gnant M. Breast cancer. Lancet 2017;389:1134-50. [Crossref] [PubMed]
- Dobovišek L, Krstanović F, Borštnar S, et al. Cannabinoids and Hormone Receptor-Positive Breast Cancer Treatment. Cancers (Basel) 2020;12:525. [Crossref] [PubMed]
- Waks AG, Winer EP. Breast Cancer Treatment: A Review. JAMA 2019;321:288-300. [Crossref] [PubMed]
- Ruddy KJ, Ganz PA. Treatment of Nonmetastatic Breast Cancer. JAMA 2019;321:1716-7. [Crossref] [PubMed]
- Musgrove EA, Sutherland RL. Biological determinants of endocrine resistance in breast cancer. Nat Rev Cancer 2009;9:631-43. [Crossref] [PubMed]
- Jordan VC, O'Malley BW. Selective estrogen-receptor modulators and antihormonal resistance in breast cancer. J Clin Oncol 2007;25:5815-24. [Crossref] [PubMed]
- Schultz-Norton JR, Ziegler YS, Nardulli AM. ERalpha-associated protein networks. Trends Endocrinol Metab 2011;22:124-9. [Crossref] [PubMed]
- Ratanaphan A. A DNA repair BRCA1 estrogen receptor and targeted therapy in breast cancer. Int J Mol Sci 2012;13:14898-916. [Crossref] [PubMed]
- Zheng L, Jia J, Finger LD, et al. Functional regulation of FEN1 nuclease and its link to cancer. Nucleic Acids Res 2011;39:781-94. [Crossref] [PubMed]
- Singh P, Zheng L, Chavez V, et al. Concerted action of exonuclease and Gap-dependent endonuclease activities of FEN-1 contributes to the resolution of triplet repeat sequences (CTG)n- and (GAA)n-derived secondary structures formed during maturation of Okazaki fragments. J Biol Chem 2007;282:3465-77. [Crossref] [PubMed]
- Liu P, Qian L, Sung JS, et al. Removal of oxidative DNA damage via FEN1-dependent long-patch base excision repair in human cell mitochondria. Mol Cell Biol 2008;28:4975-87. [Crossref] [PubMed]
- Saharia A, Stewart SA. FEN1 contributes to telomere stability in ALT-positive tumor cells. Oncogene 2009;28:1162-7. [Crossref] [PubMed]
- Sampathi S, Bhusari A, Shen B, et al. Human flap endonuclease I is in complex with telomerase and is required for telomerase-mediated telomere maintenance. J Biol Chem 2009;284:3682-90. [Crossref] [PubMed]
- Zheng L, Zhou M, Chai Q, et al. Novel function of the flap endonuclease 1 complex in processing stalled DNA replication forks. EMBO Rep 2005;6:83-9. [Crossref] [PubMed]
- Parrish JZ, Yang C, Shen B, et al. CRN-1, a Caenorhabditis elegans FEN-1 homologue, cooperates with CPS-6/EndoG to promote apoptotic DNA degradation. EMBO J 2003;22:3451-60. [Crossref] [PubMed]
- Zheng L, Dai H, Zhou M, et al. Fen1 mutations result in autoimmunity, chronic inflammation and cancers. Nat Med 2007;13:812-9. [Crossref] [PubMed]
- Larsen E, Gran C, Saether BE, et al. Proliferation failure and gamma radiation sensitivity of Fen1 null mutant mice at the blastocyst stage. Mol Cell Biol 2003;23:5346-53. [Crossref] [PubMed]
- Illuzzi JL, Wilson DM 3rd. Base excision repair: contribution to tumorigenesis and target in anticancer treatment paradigms. Curr Med Chem 2012;19:3922-36. [Crossref] [PubMed]
- Kucherlapati M, Yang K, Kuraguchi M, et al. Haploinsufficiency of Flap endonuclease (Fen1) leads to rapid tumor progression. Proc Natl Acad Sci U S A 2002;99:9924-9. [Crossref] [PubMed]
- Zeng X, Qu X, Zhao C, et al. FEN1 mediates miR-200a methylation and promotes breast cancer cell growth via MET and EGFR signaling. FASEB J 2019;33:10717-30. [Crossref] [PubMed]
- Zhang K, Keymeulen S, Nelson R, et al. Overexpression of Flap Endonuclease 1 Correlates with Enhanced Proliferation and Poor Prognosis of Non-Small-Cell Lung Cancer. Am J Pathol 2018;188:242-51. [Crossref] [PubMed]
- He L, Zhang Y, Sun H, et al. Targeting DNA Flap Endonuclease 1 to Impede Breast Cancer Progression. EBioMedicine 2016;14:32-43. [Crossref] [PubMed]
- Zeng X, Che X, Liu YP, et al. FEN1 knockdown improves trastuzumab sensitivity in human epidermal growth factor 2-positive breast cancer cells. Exp Ther Med 2017;14:3265-72. [Crossref] [PubMed]
- He L, Luo L, Zhu H, et al. FEN1 promotes tumor progression and confers cisplatin resistance in non-small-cell lung cancer. Mol Oncol 2017;11:640-54. [Crossref] [PubMed]
- Ward TA, McHugh PJ, Durant ST. Small molecule inhibitors uncover synthetic genetic interactions of human flap endonuclease 1 (FEN1) with DNA damage response genes. PLoS One 2017;12:e0179278. [Crossref] [PubMed]
- Lam JS, Seligson DB, Yu H, et al. Flap endonuclease 1 is overexpressed in prostate cancer and is associated with a high Gleason score. BJU Int 2006;98:445-51. [Crossref] [PubMed]
- Nikolova T, Christmann M, Kaina B. FEN1 is overexpressed in testis, lung and brain tumors. Anticancer Res 2009;29:2453-9. [PubMed]
- Singh P, Yang M, Dai H, et al. Overexpression and hypomethylation of flap endonuclease 1 gene in breast and other cancers. Mol Cancer Res 2008;6:1710-7. [Crossref] [PubMed]
- Sato M, Girard L, Sekine I, et al. Increased expression and no mutation of the Flap endonuclease (FEN1) gene in human lung cancer. Oncogene 2003;22:7243-6. [Crossref] [PubMed]
- Abdel-Fatah TM, Russell R, Albarakati N, et al. Genomic and protein expression analysis reveals flap endonuclease 1 (FEN1) as a key biomarker in breast and ovarian cancer. Mol Oncol 2014;8:1326-38. [Crossref] [PubMed]
- Wang J, Zhou L, Li Z, et al. YY1 suppresses FEN1 over-expression and drug resistance in breast cancer. BMC Cancer 2015;15:50. [Crossref] [PubMed]
- Schultz-Norton JR, Walt KA, Ziegler YS, et al. The deoxyribonucleic acid repair protein flap endonuclease-1 modulates estrogen-responsive gene expression. Mol Endocrinol 2007;21:1569-80. [Crossref] [PubMed]
- Györffy B, Lanczky A, Eklund AC, et al. An online survival analysis tool to rapidly assess the effect of 22,277 genes on breast cancer prognosis using microarray data of 1,809 patients. Breast Cancer Res Treat 2010;123:725-31. [Crossref] [PubMed]
- Loi S, Haibe-Kains B, Desmedt C, et al. Predicting prognosis using molecular profiling in estrogen receptor-positive breast cancer treated with tamoxifen. BMC Genomics 2008;9:239. [Crossref] [PubMed]
- Hurtado A, Holmes KA, Ross-Innes CS, et al. FOXA1 is a key determinant of estrogen receptor function and endocrine response. Nat Genet 2011;43:27-33. [Crossref] [PubMed]
- Mootha VK, Lindgren CM, Eriksson KF, et al. PGC-1alpha-responsive genes involved in oxidative phosphorylation are coordinately downregulated in human diabetes. Nat Genet 2003;34:267-73. [Crossref] [PubMed]
- Subramanian A, Tamayo P, Mootha VK, et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci U S A 2005;102:15545-50. [Crossref] [PubMed]
- Zhang Y, Liu T, Meyer CA, et al. Model-based analysis of ChIP-Seq (MACS). Genome Biol 2008;9:R137. [Crossref] [PubMed]
- Li H, Xu L, Li C, et al. Ubiquitin ligase Cbl-b represses IGF-I-induced epithelial mesenchymal transition via ZEB2 and microRNA-200c regulation in gastric cancer cells. Mol Cancer 2014;13:136. [Crossref] [PubMed]
- Mileo AM, Di Venere D, Mardente S, et al. Artichoke Polyphenols Sensitize Human Breast Cancer Cells to Chemotherapeutic Drugs via a ROS-Mediated Downregulation of Flap Endonuclease 1. Oxid Med Cell Longev 2020;2020:7965435. [Crossref] [PubMed]
- Dong S, Xiao Y, Ma X, et al. miR-193b Increases the Chemosensitivity of Osteosarcoma Cells by Promoting FEN1-Mediated Autophagy. Onco Targets Ther 2019;12:10089-98. [Crossref] [PubMed]
- Li JL, Wang JP, Chang H, et al. FEN1 inhibitor increases sensitivity of radiotherapy in cervical cancer cells. Cancer Med 2019;8:7774-80. [Crossref] [PubMed]
- Zhu H, Wu C, Wu T, et al. Inhibition of AKT Sensitizes Cancer Cells to Antineoplastic Drugs by Downregulating Flap Endonuclease 1. Mol Cancer Ther 2019;18:2407-20. [Crossref] [PubMed]
- Wang Y, Li S, Zhu L, et al. Letrozole improves the sensitivity of breast cancer cells overexpressing aromatase to cisplatin via down-regulation of FEN1. Clin Transl Oncol 2019;21:1026-33. [Crossref] [PubMed]
- He L, Yang H, Zhou S, et al. Synergistic antitumor effect of combined paclitaxel with FEN1 inhibitor in cervical cancer cells. DNA Repair (Amst) 2018;63:1-9. [Crossref] [PubMed]
- Lu X, Liu R, Wang M, et al. MicroRNA-140 impedes DNA repair by targeting FEN1 and enhances chemotherapeutic response in breast cancer. Oncogene 2020;39:234-47. [Crossref] [PubMed]
- Li C, Zhou D, Hong H, et al. TGFβ1- miR-140-5p axis mediated up-regulation of Flap Endonuclease 1 promotes epithelial-mesenchymal transition in hepatocellular carcinoma. Aging 2019;11:5593-612. [Crossref] [PubMed]
- Xie C, Wang K, Chen D. Flap endonuclease 1 silencing is associated with increasing the cisplatin sensitivity of SGC7901 gastric cancer cells. Mol Med Rep 2016;13:386-92. [Crossref] [PubMed]
- Gutierrez MC, Detre S, Johnston S, et al. Molecular changes in tamoxifen-resistant breast cancer: relationship between estrogen receptor, HER–2, and p38 mitogen-activated protein kinase. J Clin Oncol 2005;23:2469-76. [Crossref] [PubMed]


