MicroRNA and MET in lung cancer
Introduction
What are miRNAs?
MicroRNAs (miRNAs) are 19 to 25 nucleotide-long non coding RNA molecules that regulate the gene expression at the level of messenger RNA degradation and translation. miRNAs are mainly transcribed by RNA polymerase II as long primary transcripts characterized by hairpin structures (pri-miRNA) and processed in the nucleus by RNAse III Drosha in a 70-nucleotide-long pre-miRNA. This precursor molecule is exported by the Exportin 5 to the cytoplasm, where RNAse III Dicer generates a dsRNA of approximately 22 nucleotides, named miR/miR*. The mature single stranded microRNA product is then incorporated in the complex, known as microRNA-containing RNA-induced silencing complex (RISC), whereas the other strand is likely subjected to degradation (1).
miRNAs regulate the gene expression binding through partial complementarity for the most part to the 3’UTR of target mRNAs. The level of homology between guide and mRNA target determines which silencing mechanism will be employed: perfect matching of miRNAs to target sequences leads to transcriptional repression by cleaving and degrading the mRNA transcripts, while a limited base pairing inhibits the protein translation (Figure 1).
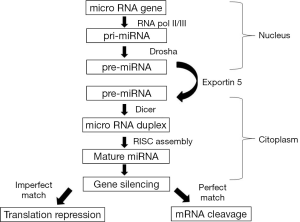
miRNA genes represent approximately 1% of the genome and it has been calculated that the human genome contains more than 1,800 miRNAs (2) with the potential of regulating around 30% of human genes (3).
By targeting multiple transcripts a single miRNA can regulate many fundamental cellular processes such as cell proliferation, apoptosis, differentiation, and migration; in the same way any gene can be regulated by multiple miRNAs.
Several databases are available to predict miRNA target genes, however the verification of these is essential, because the interaction between miRNA and the target is complex, with a poor overlap between databases (4). A reporter assay is one of the verification methods for miRNA target, because an alteration in the luciferase expression indicates whether a miRNA can bind to a target mRNA; using miRNA mimics and inhibitors and then measuring predicted targets of miRNAs is another method.
Although there is currently no gold standard for measuring miRNA expression (5) oligonucleotide microarray (microchip) and quantitative real-time reverse transcription polymerase chain reaction (PCR) (qRT-PCR) are actually two of the most common methods to evaluate known miRNAs.
miRNA and lung cancer
miRNAs are involved in a variety of biological processes including cell cycle regulation, differentiation, development, metabolism, neuronal patterning and aging. Alterations in miRNA expression are not simply an effect of tumorigenesis but it has a causative role in cancer development: they are involved in the initiation, progression and metastasis of human tumors. Several miRNAs are dysregulated in cancers and a single miRNA can have multiple targets involved in different oncogenic pathways.
Given this powerful biology, numerous miRNAs have been implicated as either tumor suppressors or oncogenes (“oncomirs”) in many different tumor types. Genomically they are frequently found to be at fragile sites in the human genome, but there are myriad additional mechanisms by which miRNAs can become dysregulated in cancer (6).
In 2002, a seminal study by Calin et al. (7) showed that miR 15 a/16-1 cluster is frequently deleted in chronic lymphocytic leukemia, implicating these miRNAs as tumor suppressor; after this discovery, several studies reported that miRNAs are aberrantly expressed in the majority of solid cancers and the diagnostic and prognostic value of miRNA expression in lung cancer has been intensely studied in recent years.
A first category of studies was conducted comparing miRNA expression of tumor samples to matched non-cancerous lung tissue: a report of Takamizawa et al. (8) showed that let-7 expression was reduced in lung cancer and this finding was confirmed later on by the group of Yanaihara et al. (9) where let-7 was part of a signature of miRNAs deregulated in tumor tissues compared to normal lung.
Recently a comprehensive meta-analysis of 20 miRNA expression studies in lung cancer, including a total of 598 tumors and 528 non-cancerous control samples, was published (10): using a robust rank aggregation method, the authors identified a statistically significant miRNA meta-signature of seven upregulated (miR-21, miR-210, miR-182, miR-31, miR-200b, miR-205 and miR-183) and eight downregulated (miR-126-3p, miR-30a, miR-30d, miR-486-5p, miR-451a, miR-126-5p, miR-143 and miR-145) miRNAs. Some of the upregulated miRNAs have a well-known role in cancer development: miR-21 is associated with worse prognosis of lung cancer patients and it was shown that directly target the tumor-suppressor PTEN, while miR-210 increases radioresistance of non-small cell lung cancer (NSCLC) cells by stabilizing HIF-1A. Oppositely, the downregulated miRNAs have a protective role: for example miR-126-3p and miR-126-5p work as regulators of angiogenesis and cell proliferation and miR-30a regulates the epithelial to mesenchymal transition by targeting the SNAI1 gene.
An alternative process to miRNA profiling of tumor tissues is measuring miRNA levels in relation to the outcome. This approach had principally been applied to address the question of whether miRNA signatures can be developed to stratify early-stage patients (those who have undergone surgical resection) into high or low risk for recurrence.
Landi et al. (11) reported a five-miRNA signature (miR-25, miR-34c-5p, miR-191, let-7e and miR-34a) that was first of all able to differentiate squamous from adenocarcinoma and then to correlate with poor overall survival among squamous patients; also Yu et al. (12) validated a 5-miRNA signature, including let-7a, miR-221, miR-137, miR-372 and miR-182, that was an independent predictor of cancer relapse and survival after surgery.
Recently several authors (13,14) have studied miRNAs measurement also from patient serum or plasma: MiR-21, in combination with miR-210 and miR-486-5p, was shown to be expressed significantly higher in the plasma of patients with malignant solitary pulmonary nodules compared to those with benign nodules (15) and MiR-155 with miR-197 and miR-182 was able to distinguish between NSCLC patients and control samples by real time PCR of plasma (16). This technique has the advantage to repeatedly measure miRNAs during the course of treatment or in post-treatment surveillance and to overcome the problem of small amounts of tissue in lung cancer patients but it needs to be validated.
MET and miRNA in lung cancer
In the last two decades, it has been consistently proven that one of the players of the intricate scenario leading to tumorigenesis is MET, the tyrosine kinase receptor for hepatocyte growth factor (HGF). MET can be activated either by binding to its ligand HGF, overexpression/amplification, mutation or decreased degradation. Deregulated activation of the MET-driven “invasive-growth” program confers unrestricted proliferation and metastatic properties to cancer cells (17) and recently it was demonstrated that the expression of an activated MET is required also for the maintenance and sustaining of metastatic colonies in the lung (18). MET ability to control all these ways has an explanation in its signal transducing properties. The receptor, in fact, is known to concomitantly activate both the Ras-MAPK and the phosphatidylinositide 30-kinase-AKT pathways controlling together growth, resistance to apoptosis and cytoskeletal rearrangement.
MET has been predicted and shown to be the target gene of multiple miRNAs (Tables 1,2), which play a crucial role in controlling its activity in a stimulatory or inhibitory sense, creating an intricate system that we have started to know only from few years and that opens new therapeutic possibilities, too. In this review the studies on miRNAs that control MET, which have been mainly published in later years, will be reported.
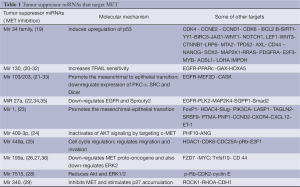
Full table

Full table
MiR-34 family controls MET acting as an effector in the p53 tumor suppressor network
MiR34 and MET
MiR-34 family consists of three members: miR-34a, miR-34b and miR-34c and MET is negatively regulated by all of them. MiR-34a is encoded in the second exon of a gene located on chromosome 1p36.22, whereas miR-34b and miR-34c share a common host gene located on chromosome 11q23.1. All members of miR-34 family, targeting more than 77 target mRNAs, were shown to suppress tumor growth and metastasis by inhibiting the processes that stimulate the cancer development, including cell cycle, EMT, metastasis, stemness and by promoting the processes that inhibit carcinogenesis, such as apoptosis (38). MiR-34 family is frequently decreased in expression in solid tumors including NSCLC and this is a prognostically negative factor. CpG hypermethylation of miR-34a has been reported in multiple types of cancer including NSCLC and could decrease the miR-34a expression level (39); also miR-34b/c was found to undergo specific hypermethylation-associated silencing and this has a role in the metastasis formation in NSCLC (40).
Gallardo et al. (41) examined the role of miR-34 in 70 patients who underwent surgical resection for NSCLC: miR-34a and miR-34b were significantly repressed versus paired normal tissue and low levels of miR-34a in tumor samples correlated with a high rate of relapse; the authors also reported that the miR34b expression was lower in NSCLC tissue compared to that in pericarcinous tissue and confirmed that lower miR34b expression was correlated with higher lymph node metastasis. In 2008, Migliore et al. (42) showed that miR 34 b-c (and miR 199) contribute to control MET activity: they first confirmed that these miRNAs bind to the 3’UTR of MET and then transfected human carcinoma cells with chemically synthesized 34b-c miRNA precursors showing a significant reduction of MET protein even in cells displaying overexpression and MET gene amplification; furthermore the miRNA34 transfected cells were unable to migrate and to scatter (to break intercellular junctions) in response to HGF. Conversely, the inhibition of these endogenous miRNAs, by use of antagomiRs, resulted in increased expression of MET.
p53 and MET
The tumor-suppressor protein p53 is a master regulator of the stress response and provides a key barrier to cellular transformation and tumorigenesis; the p53 activation results in cell cycle arrest, apoptosis, or senescence, depending on the cellular context and the type of stimulus (43). p53 is mutated in more than 50% of NSCLC (44) and the loss of p53 signaling leads to uncontrolled cellular division and apoptotic avoidance.
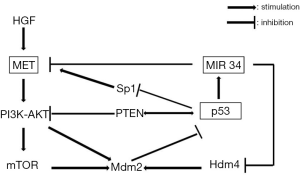
MET is a critical player in p53-mediated control of motility and invasion and antagonizes p53 through Mdm2 upregulation via PI3K-AKT and mTOR (45) (Figure 2); similarly the wild type of p53 negatively regulates MET expression by two mechanisms: suppression at the transcriptional level with inhibition of SP1 binding to MET promoter and transactivation of miR-34 (46). In fact the two genomic loci encoding the three miR-34 family members have each one a p53 binding site in their promoter and their expression is induced by oncogenic stress or DNA damage (47). They directly act repressing HDM4, a potent negative regulator of p53, which positively regulates Mdm2 to promote p53 degradation through polyubiquitination. So, p53 and miR-34 form a positive feedback loop to strengthen the downstream effects of p53 and these miRNAs seem to act in parallel with the other p53 effectors, creating a sort of signal strengthening (48).
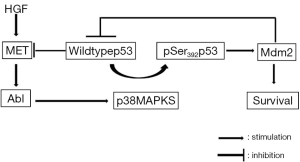
Furlan et al. (49) identified for the first time in HCC cells the c-Abl as a signaling node interconnecting MET and p53 core pathways and showed that its inhibition impairs MET-dependent tumorigenesis. MET ensures cell survival through a new pathway in which c-Abl leads to wt p53 phosphorylation on Ser392 by p38-MAPK, leading to Mdm2 upregulation and finally stimulating survival and tumorigenesis (Figure 3).
Wang et al. (50) confirmed these results in lung cancer cells, showing that there is an association between lower miR34b and overexpression of Phospho-MET, Phospho p53 (on S392) and Mdm2.
The authors noticed also that there was no increased expression levels of miR-34b corresponding to the overexpression of p53S392 observed in the samples with NSCLC, probably because the phosphorylation on S392 decrease the transcriptional activities of p53 in lung cancer tissues and its capacity to activate miR-34 family.
Okada et al. (51) discovered another mechanism that human lung adenocarcinoma acts escaping miR-34 control: the cancer cells are able to generate an elevated level of a short HDM4 isoform that lacks miR-34-binding sites evading thereby miR-34 regulation to disable the p53-miR-34 positive feedback.
This is one of the mechanisms known as p53 mutations, deregulation of miR-34 through either genomic deletion or promoter hypermethylation to disrupt the p53-miR-34 positive feedback and ultimately contribute to tumor development.
MiR 222/221 and MiR 130a
MiR-222/221 cluster is among the most dysregulated miRNA implicated in cancer.
The expression of miR-222/221 is highly upregulated in a variety of solid tumors and their oncogenic role is known in NSCLC and many others cancers. In 2008, Garofalo et al. (52) reported that NSCLC cells overexpressing miR-221&222 are TRAIL-resistant and show an increase in migration and invasion capabilities. The TNF-related apoptosis inducing ligand (TRAIL) is a cytokine of the TNFα family that induces apoptosis by binding to death receptors (DR) 4 and 5 on the cell surface, which then leads to a cascade of death-inducing signaling complex (DISC) formation, caspase activation and ultimately the execution of the apoptotic program.
The same authors founded that MET, through the upregulation of miR 221&222 expression, confer resistance to TRAIL-induced-cell death and enhance tumorigenicity of lung and cancer cells (53). Indeed, upon HGF stimulation, MET increased phosphorylation of ERK1/2 and JNK; phosphorylated JNKs activate the oncoprotein c-Jun, which forms the activator protein-1(AP-1) transcription factor as a homodimer; they show that c-Jun has one binding site in the miR 221/222 promoter region and that miR 221&222, after c-Jun stimulation, represses PTEN and TIMP3 expression (Figure 4).
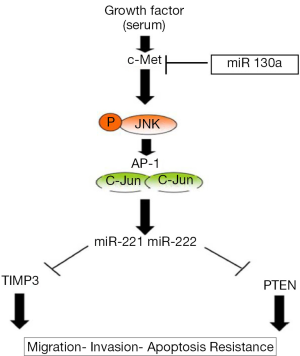
PTEN is an oncosuppressor that acts directly antagonizing the activity of PI3 kinase (PI3K) (54). Its inactivation results in constitutive activation of the PI3K/AKT pathway and in a subsequent increase in protein synthesis, cell cycle progression, migration and survival; TIMP3 (55) is a member of the tissue inhibitors of metalloproteinases (MMPs) that inhibits the activity of MMPs by binding with a 1:1 stoichiometry to the active site. Previous studies (56,57) have shown that the overexpression of TIMP3 in vascular smooth muscle cells and melanoma cell lines inhibits invasion and promotes apoptotic cell death.
In 2012, Acunzo et al. (58) showed that miR-130, an oncosuppressor expressed at low level in lung cancer cell lines, was able to increase TRAIL sensitivity in NSCLC TRAIL resistant cell lines; miR 130a, decreasing MET expression, is able to reduce the binding of c-Jun to the miRNA-221 and miRNA-222 promoter region inducing TRAIL sensitivity in NSCLC cells.
miRNAs targeting MET implicated in EGFR resistance
The epidermal growth factor receptor, EGFR (ErbB1/Her-1), is a member of the ErbB family of tyrosine kinase growth factor receptors that is frequently found to be activated by mutation or amplification in epithelial malignancies. In lung cancer it is mutated in approximately 10% of Caucasian patients and 30% of Asian patients. The EGFR tyrosine kinase inhibitors (TKIs) are effective clinical therapies for advanced NSCLC patients with EGFR activating mutations; however, despite initial dramatic benefits from EGFR TKIs, all of these patients ultimately develop resistance (referred to as acquired resistance) to these agents. In EGFR mutant lung cancers more than 50% of resistance cases are due to the occurrence of a secondary T790M mutation in EGFR. Amplification of the MET receptor has also been shown to maintain ERBB3/PI3K/AKT signaling in presence of gefitinib and cause resistance to EGFR targeted therapies in approximately 20% of NSCLC patients (59). In fact, clinical trials using combined EGFR and MET inhibitors in NSCLC patients with acquired resistance to gefitinib/erlotinib are currently underway. Garofalo et al. (60) studied the miRNAs implicated in EGFR resistence: to identify EGFR- and MET-regulated-miRNAs, they first silenced EGFR and MET in Calu-1 cells (a NSCLC cell line) using shRNA lentiviral particles and found 8 miRNAs that are regulated by both EGFR and MET (miRNA 221-222-30b/c-21-29a/c-100). Then they focused on miR-30b-c and miR 221-222 showing the highest downregulation after both MET and EGFR silencing: they demonstrated that in sensitive cells the gefitinib treatment triggers programmed cell death through downregulation of miR 30b-c and miR 221-222 and the consequent upregulation of APAF-1 (apoptotic peptidase activating factor 1) and BIM (BCL2-like 11); MET overexpression is able to induce resistance to gefitinib through the upregulation of miR 30b-c and miR 221-222 making uneffective the EGFR inhibition alone. They also showed that this resistance could be overcome using MET inhibitors or anti-miRNA 221-222 and anti-30c which strongly increase gefitinib-sensitivity in xenograft mouse models in vivo.
Another oncoMir regulated by both EGFR and MET is miR-21; data from transgenic tumor models show that it drives tumorigenesis by repressing negative regulators of the Ras/MEK/ERK, Ras/PI3K/Akt and Ras/RalGDS/JNK pathways (61); when it is overexpressed it represses PTEN inducing gefitinib resistance (62), while its blockade is able to reverse the EMT phenotype associated with EGFR-TKI resistance (63).
Recently Shen et al. (64) showed that the upregulation of miR-21 and the downregulation of PTEN in tumor tissues from human NSCLC patients negatively correlate with shorter disease free survival; furthermore, by analyzing miR-21/PTEN expression levels and clinical response to TKI treatment in 46 NSCLC patients, they found that high miR-21 expression levels and low PTEN protein levels were associated with poor clinical response to TKIs and shorter overall survival.
When miR-21 was knockdown, the TKI sensitivity was restored both in vitro and in vivo (mouse models), by inactivation of ERK and AKT pathways and PTEN upregulation. MiR 103 and miR 203 work as tumor suppressors promoting the mesenchymal to epithelial transition (65). They are upregulated after MET silencing, inducing apoptosis in gefitinib-resistant NSCLC. They reduce mesenchymal markers and increase epithelial cell junction proteins, by downregulating the expression of PKC-ε, SRC and Dicer that exert pro-survival effects and contribute to gefitinib resistance by activating AKT and ERK signaling pathways (66,67).
MiR 27a is an oncosuppressor that is tipically downregulated in NSCLC patients (68); it belongs to the miR-23a-27a-24-2 cluster, whose expression is induced through ELK1 after HGF binding to MET.
Recently, Acunzo et al. (69) have showed that MiR-27a negatively regulates MET and EGFR in NSCLC with direct and indirect mechanisms: it directly targets MET and EGFR 3UTRs downregulating their expression; miR-27a also downregulates Sprouty2, that normally increases MET and EGFR levels by attenuating their degradation through ubiquitination.
The lack of miR 27 regulatory action on Met-EGFR axis can then lead to an uncontrolled proliferation.
Mir130a, as we have seen in the previous section, is an oncosuppressor that increases TRAIL sensitivity directly targeting MET and it is also implicated in the EGFR resistance.
Zhou et al. (70) demonstrated that miR-130a expression was negatively correlated with that of MET and that is overexpressed in gefitinib-sensitive NSCLC cell lines, but is low in gefitinib-resistant NSCLC cell lines.
This group also showed that overexpression of miR-130a increased cell apoptosis and inhibited proliferation of NSCLC cells treated with gefitinib, whereas lowering the expression of miR-130a decreased cell apoptosis and promoted cell proliferation after treatment with gefitinib in both gefitinib-sensitive and -resistant NSCLC cell lines; they also demonstrated that miR-130 overcomes gefitinib resistance downregulating MET protein levels.
Zhou et al. (71) assessed the anti-tumour effect of miR-34a in a gefitinib- resistant lung cancer mouse model and showed that the combination of miR-34a and gefitinib caused dramatic regression of tumours compared with either gefitinib or miR-34a monotherapy. In the mouses treated with the combination therapy they found that the expression of MET and p-ERK were decreased, suggesting that miR-34 replacement therapies might sensitize resistant tumors to EGFR-TKIs by suppressing MET expression and its activation of oncogenic signaling pathways.
Therapeutic applications of miRNAs targeting MET
The increasing evidence of the important role of miRNAs in cancer development and the capacity of a single miRNA to control many known oncogenes have strengthen the interest in using miRNAs in cancer treatment.
miRNA therapeutic approaches can be divided into two different categories: miRNA inhibition therapy when the target miRNA is overexpressed and miRNA replacement therapy when the miRNA is repressed.
miRNA antagonists are generated to inhibit endogenous miRNAs that show a gain-of-function in diseased tissues. This technique involves the introduction of a highly chemically-modified miRNA passenger strand (anti-miR) that binds with high affinity to the active miRNA strand. Since binding is frequently irreversible, the new miRNA duplex is unable to be processed by RISC and/or degraded.
By reintroducing a tumor suppressor miRNA, miRNA replacement therapy seeks to restore a loss-of-function in cancer and to reactivate cellular pathways driving a therapeutic response.
Although successful delivery is an obstacle to effective miRNA-based therapeutics, new findings from recent trials and the rapid advances in systemic drug delivery systems led to a rapid progress in this field (72).
In 2010, Wiggins et al. (73) described the development of a therapeutic formulation using chemically synthesized miR-34a and a lipid-based delivery vehicle. They first demonstrated that intratumoral delivery of formulated miR-34a block lung tumor growth in mice inducing a specific inhibitory effect in tumor cells with an accumulation of miR-34a and concurrent repression of its direct target genes (MET, CDK4 and Bcl-2); then they evidenced that, similarly to the intratumoral injection of miR-34a, also the intravenous delivery of formulated miR-34a specifically blocked the tumor growth and did not induce an elevation of liver and kidney enzymes and an immune response. They also noticed that miR-34a inhibited tumor cells that showed normal levels of endogenous miR-34a, indicating that the therapeutic application of tumor suppressor miRNAs is not limited to replacement.
Also Xue et al. (74) recently investigated the effects of Mir-34a delivery on lung cancer development in a genetically engineered mouse model (called “KP”) of lung cancer based on loss of p53 and Kras activation.
Tumor bearing KP mice were treated with intravenous Mir-34a in a lipid/polimer nanoparticle at a dose of 1.5 mg/kg twice each week for 4 weeks and 10 weeks after the first injection were scanned with microCT. They noticed that the therapeutic delivery of Mir-34a delayed tumor progression compared with control animals and tumors showed reduced levels of ki67 without increasing toxicity. They also demonstrated that the combination of Mir-34 and siRNA targeting Kras is possible and improved the therapeutic response compared to miR-34a or siKras alones. Furthermore, they analized the combination of chemotherapy with nanoparticles: they treated tumor bearing Kp mice with either cisplatin, nanoparticles carrying both Mir-34a mimics and siKras or a combination of the two. The KP mice treated with the combination therapy survived significantly longer than the others, suggesting that restoring p53 functions with Mir-34a could be important to obtain an improved response to chemotherapy.
Xu et al. (75) used synthesized locked nucleic acid (LNA) antimiR-21 (Oncomir regulated by both c-MET and EGFR) showing the increased sensitivity of lung cancer cells to Cisplatin in vitro and in vivo and confirming that the combining chemotherapy with anti-Mir (or suppressor MiR) might be a potential strategy for the treatment of human NSCLC.
A combinatorial therapeutic approach was also studied by Kasinski et al. (76) who used for the first time a combination of two tumor suppressive miRNAs: the well-known miR 34a and the let7 (that targets Kras, cMYC, HMGA2 and the LIN 28 isoforms). They treated the aggressive “KP” lung cancer mouse models with systemic nanodelivery of both miRNAs showing that it is feasible and that they act reducing tumor proliferation and invasiveness in a synergistic manner: the average tumor volumes from the dual treated animals were significantly smaller than the average volumes from each of the single miRNA treated group. They show also that the combinatorial approach was able to downregulate more target genes than the use of each miRNA alone, suggesting that the combinatorial treatment, by simultaneously targeting multiple key factors in the cancer growth, could overcome the development of secondary resistance.
In summary, miRNA-based therapy has a great potential to be a more powerful tool in tumor treatment by the simultaneous modulation of multiple genes involved in distinct tumor-related signaling networks. Actually the first clinical trial with microRNA replacement therapy in cancer is ongoing: this is a phase I study (77) with miR34 (MRX34) in patients with liver cancer or solid cancer with liver involvement. It is delivered using a liposomal formulation and the first safety data from 26 patients show that it has a manageable safety profile.
Acknowledgements
Disclosure: The author declares no conflict of interest.
References
- Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell 2004;116:281-97. [PubMed]
- Available online: http://www.mirbase.org/
- Lewis BP, Burge CB, Bartel DP. Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microRNA targets. Cell 2005;120:15-20. [PubMed]
- Bernardo BC, Charchar FJ, Lin RC, et al. A microRNA guide for clinicians and basic scientists: background and experimental techniques. Heart Lung Circ 2012;21:131-42. [PubMed]
- Nelson PT, Wang WX, Wilfred BR, et al. Technical variables in high-throughput miRNA expression profiling: much work remains to be done. Biochim Biophys Acta 2008;1779:758-65.
- Di Leva G, Garofalo M, Croce CM. MicroRNAs in cancer. Annu Rev Pathol 2014;9:287-314. [PubMed]
- Calin GA, Dumitru CD, Shimizu M, et al. Frequent deletions and down-regulation of micro- RNA genes miR15 and miR16 at 13q14 in chronic lymphocytic leukemia. Proc Natl Acad Sci U S A 2002;99:15524-9. [PubMed]
- Takamizawa J, Konishi H, Yanagisawa K, et al. Reduced expression of the let-7 microRNAs in human lung cancers in association with shortened postoperative survival. Cancer Res 2004;64:3753-6. [PubMed]
- Yanaihara N, Caplen N, Bowman E, et al. Unique microRNA molecular profiles in lung cancer diagnosis and prognosis. Cancer Cell 2006;9:189-98. [PubMed]
- Võsa U, Vooder T, Kolde R, et al. Meta-analysis of microRNA expression in lung cancer. Int J Cancer 2013;132:2884-93. [PubMed]
- Landi MT, Zhao Y, Rotunno M, et al. MicroRNA expression differentiates histology and predicts survival of lung cancer. Clin Cancer Res 2010;16:430-41. [PubMed]
- Yu SL, Chen HY, Chang GC, et al. MicroRNA signature predicts survival and relapse in lung cancer. Cancer Cell 2008;13:48-57. [PubMed]
- Chen X, Ba Y, Ma L, et al. Characterization of microRNAs in serum: a novel class of biomarkersfor diagnosis of cancer and other diseases. Cell Res 2008;18:997-1006. [PubMed]
- Hu Z, Chen X, Zhao Y, et al. Serum microRNA signatures identified in a genome-wide serum microRNA expression profiling predict survival of non-small-cell lung cancer. J Clin Oncol 2010;28:1721-6. [PubMed]
- Shen J, Liu Z, Todd NW, et al. Diagnosis of lung cancer in individuals with solitary pulmonary nodules by plasma microRNA biomarkers. BMC Cancer 2011;11:374. [PubMed]
- Zheng D, Haddadin S, Wang Y, et al. Plasma microRNAs as novel biomarkers for early detection of lung cancer. Int J Clin Exp Pathol 2011;4:575-86. [PubMed]
- Comoglio PM, Trusolino L. Invasive growth: from development to metastasis. J Clin Invest 2002;109:857-62. [PubMed]
- Corso S, Migliore C, Ghiso E, et al. Silencing the MET oncogene leads to regression of experimental tumors and metastases. Oncogene 2008;27:684-93. [PubMed]
- Bader AG. miR-34 - a microRNA replacement therapy is headed to the clinic. Front Genet 2012;3:120. [PubMed]
- Chen Y, Gorski DH. Regulation of angiogenesis through a microRNA (miR-130a) that down-regulates antiangiogenic homeobox genes GAX and HOXA5. Blood 2008;111:1217-26. [PubMed]
- Zhou X, Xu G, Yin C, et al. Down-regulation of miR-203 induced by Helicobacter pylori infection promotes the proliferation and invasion of gastric cancer by targeting CASK. Oncotarget 2014;5:11631-40. [PubMed]
- Bao Y, Chen Z, Guo Y, et al. Tumor suppressor microRNA-27a in colorectal carcinogenesis and progression by targeting SGPP1 and Smad2. PLoS One 2014;9:e105991. [PubMed]
- Han C, Yu Z, Duan Z, et al. Role of microRNA-1 in human cancer and its therapeutic potentials. Biomed Res Int 2014;2014:428371.
- Wan L, Zhu L, Xu J, et al. MicroRNA-409-3p functions as a tumor suppressor in human lung adenocarcinoma by targeting c-Met. Cell Physiol Biochem 2014;34:1273-90. [PubMed]
- Luo W, Huang B, Li Z, et al. MicroRNA-449a is downregulated in non-small cell lung cancer and inhibits migration and invasion by targeting c-Met. PLoS One 2013;8:e64759. [PubMed]
- Kim S, Lee UJ, Kim MN, et al. MicroRNA miR-199a* regulates the MET proto-oncogene and the downstream extracellular signal-regulated kinase 2 (ERK2). J Biol Chem 2008;283:18158-66. [PubMed]
- Zhou J, Liu R, Wang Y, et al. miR-199a-5p regulates the expression of metastasis-associated genes in B16F10 melanoma cells. Int J Clin Exp Pathol 2014;7:7182-90. [PubMed]
- Lee JM, Yoo JK, Yoo H, et al. The novel miR-7515 decreases the proliferation and migration of human lung cancer cells by targeting c-Met. Mol Cancer Res 2013;11:43-53. [PubMed]
- Fernandez S, Risolino M, Mandia N, et al. miR-340 inhibits tumor cell proliferation and induces apoptosis by targeting multiple negative regulators of p27 in non-small cell lung cancer. Oncogene 2014. [Epub ahead of print]. [PubMed]
- Spizzo R, Nicoloso MS, Croce CM, et al. SnapShot: MicroRNAs in Cancer. Cell 2009;137:586-586.e1.
- Li J, Donath S, Li Y, et al. miR-30 regulates mitochondrial fission through targeting p53 and the dynamin-related protein-1 pathway. PLoS Genet 2010;6:e1000795. [PubMed]
- Kim C, Lee H, Cho YM, et al. TNFα-induced miR-130 resulted in adipocyte dysfunction during obesity-related inflammation. FEBS Lett 2013;587:3853-8. [PubMed]
- Li M, Liu Z, Zhang Z, et al. miR-103 promotes 3T3-L1 cell adipogenesis through AKT/mTOR signal pathway with its target being MEF2D. Biol Chem 2015;396:235-44. [PubMed]
- Tian Y, Fu S, Qiu GB, et al. MicroRNA-27a promotes proliferation and suppresses apoptosis by targeting PLK2 in laryngeal carcinoma. BMC Cancer 2014;14:678. [PubMed]
- Pan W, Wang H, Jianwei R, et al. MicroRNA-27a promotes proliferation, migration and invasion by targeting MAP2K4 in human osteosarcoma cells. Cell Physiol Biochem 2014;33:402-12. [PubMed]
- Song J, Gao L, Yang G, et al. MiR-199a regulates cell proliferation and survival by targeting FZD7. PLoS One 2014;9:e110074. [PubMed]
- Wu T, Zhou H, Hong Y, et al. miR-30 family members negatively regulate osteoblast differentiation. J Biol Chem 2012;287:7503-11. [PubMed]
- Rokavec M, Li H, Jiang L, et al. The p53/miR-34 axis in development and disease. J Mol Cell Biol 2014;6:214-30. [PubMed]
- Lodygin D, Tarasov V, Epanchintsev A, et al. Inactivation of miR-34a by aberrant CpG methylation in multiple types of cancer. Cell Cycle 2008;7:2591-600. [PubMed]
- Lujambio A, Calin GA, Villanueva A, et al. A microRNA DNA methylation signature for human cancer metastasis. Proc Natl Acad Sci U S A 2008;105:13556-61. [PubMed]
- Gallardo E, Navarro A, Viñolas N, et al. miR-34a as a prognostic marker of relapse in surgically resected non-small-cell lung cancer. Carcinogenesis 2009;30:1903-9. [PubMed]
- Migliore C, Petrelli A, Ghiso E, et al. MicroRNAs impair MET-mediated invasive growth. Cancer Res 2008;68:10128-36. [PubMed]
- Zilfou JT, Lowe SW. Tumor suppressive functions of p53. Cold Spring Harb Perspect Biol 2009;1:a001883. [PubMed]
- Herbst RS, Heymach JV, Lippman SM. Lung cancer. N Engl J Med 2008;359:1367-80. [PubMed]
- Moumen A, Patané S, Porras A, et al. Met acts on Mdm2 via mTOR to signal cell survival during development. Development 2007;134:1443-51. [PubMed]
- Hwang CI, Matoso A, Corney DC, et al. Wild-type p53 controls cell motility and invasion by dual regulation of MET expression. Proc Natl Acad Sci U S A 2011;108:14240-5. [PubMed]
- He L, He X, Lim LP, et al. A microRNA component of the p53 tumour suppressor network. Nature 2007;447:1130-4. [PubMed]
- Migliore C, Giordano S. MiRNAs as new master players. Cell Cycle 2009;8:2185-6. [PubMed]
- Furlan A, Stagni V, Hussain A, et al. Abl interconnects oncogenic Met and p53 core pathways in cancer cells. Cell Death Differ 2011;18:1608-16. [PubMed]
- Wang LG, Ni Y, Su BH, et al. MicroRNA-34b functions as a tumor suppressor and acts as a nodal point in the feedback loop with Met. Int J Oncol 2013;42:957-62. [PubMed]
- Okada N, Lin CP, Ribeiro MC, et al. A positive feedback between p53 and miR-34 miRNAs mediates tumor suppression. Genes Dev 2014;28:438-50. [PubMed]
- Garofalo M, Quintavalle C, Di Leva G, et al. MicroRNA signatures of TRAIL resistance in human non-small cell lung cancer. Oncogene 2008;27:3845-55. [PubMed]
- Garofalo M, Di Leva G, Romano G, et al. miR-221&222 regulate TRAIL resistance and enhance tumorigenicity through PTEN and TIMP3 downregulation. Cancer Cell 2009;16:498-509. [PubMed]
- Leevers SJ, Vanhaesebroeck B, Waterfield MD. Signalling through phosphoinositide 3-kinases: the lipids take centre stage. Curr Opin Cell Biol 1999;11:219-25. [PubMed]
- Bode W, Reinemer P, Huber R, et al. The X-ray crystal structure of the catalytic domain of human neutrophil collagenase inhibited by a substrate analogue reveals the essentials for catalysis and specificity. EMBO J 1994;13:1263-9. [PubMed]
- Ahonen M, Baker AH, Kähäri VM. Adenovirus-mediated gene delivery of tissue inhibitor of metalloproteinases-3 inhibits invasion and induces apoptosis in melanoma cells. Cancer Res 1998;58:2310-5. [PubMed]
- Baker AH, Zaltsman AB, George SJ, et al. Divergent effects of tissue inhibitor of metalloproteinase-1, -2, or -3 overexpression on rat vascular smooth muscle cell invasion, proliferation, and death in vitro. TIMP-3 promotes apoptosis. J Clin Invest 1998;101:1478-87. [PubMed]
- Acunzo M, Visone R, Romano G, et al. miR-130a targets MET and induces TRAIL-sensitivity in NSCLC by downregulating miR-221 and 222. Oncogene 2012;31:634-42. [PubMed]
- Lin Y, Wang X, Jin H. EGFR-TKI resistance in NSCLC patients: mechanisms and strategies. Am J Cancer Res 2014;4:411-35. [PubMed]
- Garofalo M, Romano G, Di Leva G, et al. EGFR and MET receptor tyrosine kinase-altered microRNA expression induces tumorigenesis and gefitinib resistance in lung cancers. Nat Med 2011;18:74-82. [PubMed]
- Hatley ME, Patrick DM, Garcia MR, et al. Modulation of K-Ras-dependent lung tumorigenesis by MicroRNA-21. Cancer Cell 2010;18:282-93. [PubMed]
- Zhang JG, Wang JJ, Zhao F, et al. MicroRNA-21 (miR-21) represses tumor suppressor PTEN and promotes growth and invasion in non-small cell lung cancer (NSCLC). Clin Chim Acta 2010;411:846-52. [PubMed]
- Han M, Liu M, Wang Y, et al. Antagonism of miR-21 reverses epithelial-mesenchymal transition and cancer stem cell phenotype through AKT/ERK1/2 inactivation by targeting PTEN. PLoS One 2012;7:e39520. [PubMed]
- Shen H, Zhu F, Liu J, et al. Alteration in Mir-21/PTEN expression modulates gefitinib resistance in non-small cell lung cancer. PLoS One 2014;9:e103305. [PubMed]
- Viticchiè G, Lena AM, Latina A, et al. MiR-203 controls proliferation, migration and invasive potential of prostate cancer cell lines. Cell Cycle 2011;10:1121-31. [PubMed]
- Weisheit S, Schäfer C, Mertens C, et al. PKCε acts as negative allosteric modulator of EGF receptor signalling. Cell Signal 2011;23:436-48. [PubMed]
- Qin B, Ariyama H, Baba E, et al. Activated Src and Ras induce gefitinib resistance by activation of signaling pathways downstream of epidermal growth factor receptor in human gallbladder adenocarcinoma cells. Cancer Chemother Pharmacol 2006;58:577-84. [PubMed]
- Heegaard NH, Schetter AJ, Welsh JA, et al. Circulating micro-RNA expression profiles in early stage nonsmall cell lung cancer. Int J Cancer 2012;130:1378-86. [PubMed]
- Acunzo M, Romano G, Palmieri D, et al. Cross-talk between MET and EGFR in non-small cell lung cancer involves miR-27a and Sprouty2. Proc Natl Acad Sci U S A 2013;110:8573-8. [PubMed]
- Zhou YM, Liu J, Sun W. MiR-130a overcomes gefitinib resistance by targeting met in non-small cell lung cancer cell lines. Asian Pac J Cancer Prev 2014;15:1391-6. [PubMed]
- Zhou JY, Chen X, Zhao J, et al. MicroRNA-34a overcomes HGF-mediated gefitinib resistance in EGFR mutant lung cancer cells partly by targeting MET. Cancer Lett 2014;351:265-71. [PubMed]
- Cho WC. MicroRNAs as therapeutic targets and their potential applications in cancer therapy. Expert Opin Ther Targets 2012;16:747-59. [PubMed]
- Wiggins JF, Ruffino L, Kelnar K, et al. Development of a lung cancer therapeutic based on the tumor suppressor microRNA-34. Cancer Res 2010;70:5923-30. [PubMed]
- Xue W, Dahlman JE, Tammela T, et al. Small RNA combination therapy for lung cancer. Proc Natl Acad Sci U S A 2014;111:E3553-61. [PubMed]
- Xu L, Huang Y, Chen D, et al. Downregulation of miR-21 increases cisplatin sensitivity of non-small-cell lung cancer. Cancer Genet 2014;207:214-20. [PubMed]
- Kasinski AL, Kelnar K, Stahlhut C, et al. A combinatorial microRNA therapeutics approach to suppressing non-small cell lung cancer. Oncogene 2014. [Epub ahead of print]. [PubMed]
- Available online: https://clinicaltrials.gov/ct2/show/NCT01829971


