Circulating donor-derived cell-free DNA: a true biomarker for cardiac allograft rejection?
Heart transplantation remains a lifesaving therapy for adults and children with end-stage heart disease. Since the first heart transplant was performed in 1967, advances in peri-operative care and long term immunosuppression have dramatically improved post-transplant outcomes (1). Currently, adult recipients can expect a median allograft survival of greater than 10 years and infant recipients can expect a median allograft survival that is greater than 20 years (2,3). Beginning with the first transplant, detection of acute allograft rejection was of paramount concern. In a letter to Dr. Christiaan Barnard dated December 4, 1967 that is on display in the Heart of Cape Town Museum, Dr. Norman Shumway suggested an early biomarker. He wrote, “Be certain to watch the R-wave of the EKG during the next several weeks for indices of rejection. It appears to be the earliest herald of important graft invasion.” Since 1967, many advances have improved the precision with which cardiac allograft rejection is diagnosed, allowing the clinician to balance the competing risks of rejection, infection, and drug side effects. In addition, our ability to prevent rejection with calcineurin inhibitors, proliferation signal inhibitors, anti-proliferatives, and cytolytic cell therapy has dramatically improved outcomes (1-3). Diagnostic advances have included the development of a minimally invasive technique to obtain endomyocardial biopsy samples by Dr. Philip Caves, standardization of pathologic grading of acute cellular rejection (ACR) in 1990 and 2004 (4), standardization of pathologic grading of antibody mediated rejection (AMR) in 2013 (5), and the development of a clinically validated gene-expression profiling test to modulate biopsy screening frequency (6).
In their original research article entitled “Circulating Cell-Free DNA Enables Noninvasive Diagnosis of Heart Transplant Rejection,” De Vlaminck et al. (7) describe the performance characteristics of circulating cell-free donor-derived DNA (cfdDNA) as a diagnostic test for ACR and AMR in a prospective cohort of pediatric and adult heart transplant recipients. The study leverages the dramatic decrease in cost of deep DNA sequencing technology to identify the proportion of circulating DNA that is donor derived in serially collected plasma samples. The authors demonstrate a good understanding of the limitations of the Illumina platform for deep DNA sequencing. By comparing loci known to be similar between donors and recipients, the authors were able to identify the baseline rate for incorrect sequence calls, eliminate loci with high error rates, and correct for the baseline error rate in the final analysis. Given the low fraction of donor derived DNA, errors in sequence calls at discrepant (informative) loci would be expected to result in a disproportionately high number of donor DNA calls, thus correcting for baseline error is of considerable importance in reducing the potential for false positive results. An alternative approach taken by other groups is to model the minor allele frequency for informative loci as a binomial distribution and define the cfdDNA fraction as the frequency peak from the model (8). After developing the test methodology, the authors then demonstrate that the cfdDNA fraction is elevated on post-transplant day 1, that the cfdDNA fraction rapidly drops to baseline with a half-life of 2.4 days, and that a low sustained cfdDNA baseline of 0.06%±0.11% is reached within the study cohort by 1 week post-transplantation.
Threshold selection and test performance characteristics
While most of the study methodology is strong, the authors do not report how the cutoff threshold for cfdDNA fraction was selected. Thresholds for a quantitative test can be set to maximize sensitivity, specificity or area under the receiver operating characteristic curve. It is not clear which, if any, of these outcomes was maximized with the chosen cutoff of 0.25%. In addition, a given study cohort is prone to sampling error when compared to the overall population of heart transplant recipients. This can lead to selection of a cutoff value with overly optimistic performance characteristics. To deal with this issue, data is usually either divided into training and validation sets or statistical methods, such as cross-validation, are used to account for this concern.
Developing a diagnostic test for rejection is complicated by the relatively low event rate in the current era for ACR and AMR. Reviewing our own institutional data, the rejection rate is approximately 2 rejection episodes per 100 patient years of follow-up. Our biopsy protocol calls for up to 9 biopsies in the first post-transplant year. As a result of a large denominator, only 3.2% of biopsies performed at our center in 2012 were classified as ACR 2R/3R or AMR. Using the reported cfdDNA test performance characteristics (sensitivity of 58% and specificity of 93%) and a rejection rate of 3.2% of tests, the positive predictive value (PPV) of the cfdDNA test is only 30.5% and the negative predictive value (NPV) is 97.6% (Table 1). With such a low event rate, nearly any diagnostic test would struggle to provide a perfect classification of rejection. As can be seen with the table, a higher event rate would increase the PPV significantly while hurting the NPV as well. Depending upon cost and ease of use, it is quite possible that the cfdDNA test will be used more frequently than endomyocardial biopsy. Assuming that the event rate for rejection will remain constant and the number of cfdDNA screens is doubled, the PPV will decrease even further (to 18%).
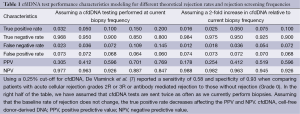
Full table
Due to the low event rate, a diagnostic strategy which first involves a screening test, followed by a confirmatory test is most appropriate. The CARGO II study provided clinicians with a gene-expression profile based score cutoff that has a NPV >99% (9). By setting the diagnostic cut-off such that NPV was optimized, the risk that clinical rejection will be missed is significantly lowered. By screening in this way the majority of the biopsies performed will still be negative for rejection, however the overall number of biopsies can be reduced approximately 3-fold without any change in clinical outcomes (6). While the IMAGE study demonstrated that shifting from a protocol biopsy schedule to a “for-cause” biopsy practice pattern in conjunction with gene-expression profiling yielded similar clinical outcomes, it failed to test the hypothesis that standard clinical assessments and a “for-cause” biopsy protocol triggered by clinician assessment, ECG, and echocardiography may have yielded the same results (10).
Physiologic relevance or “The Sniff Test”
cfdDNA fraction is appealing as a biomarker as it may prove to be proportional to the degree of myocardial injury sustained during the rejection injury and repair response. As a quantitative test which can be trended non-invasively over time, it may also serve as a measure of therapeutic response. The paper by De Vlaminck et al. (7) presents data on several individual patients with rejection. While the cfdDNA fraction trended down in each case, it did not return to baseline. The authors elegantly demonstrate that cfdDNA fraction is high at the time of transplant, presumably due to ischemia reperfusion injury, but that it rapidly comes down to the baseline by the end of the first week. Since the cfdDNA fraction remains elevated after treatment for rejection, this suggests that allograft injury/rejection is ongoing and/or that treatment is sub-optimal. Perhaps it is overly optimistic to assume that targeted anti-rejection treatment is able to acutely halt the rejection response in most cases when, in fact, myocardial injury may actually be an ongoing process following the acute rejection event. Studies which address patients with “recurrent rejection” may actually be identifying patients with persistent rejection and a waxing and waning clinical response to therapy (11,12). One could hypothesize that the rate at which cfdDNA fraction returns to baseline will correlate with the development of allograft failure or dysfunction. If that is borne out in future studies, then cfdDNA may serve as a viable proxy for treatment response, with persistently elevated levels of cfdDNA indicating a need for intensification of anti-rejection therapy.
While biomarkers can be useful without developing a nuanced understanding of their role in pathophysiology, it would be helpful to understand the cellular source of the cfdDNA. Cardiomyocytes do not divide rapidly and have a low proportion of DNA relative to their total cellular content. The endothelium is the primary target of the rejection response and has a relatively high DNA content (13). It is reasonable to hypothesize that a substantial portion of the cfdDNA derives from the allograft endothelium when it is damaged, and subsequently repaired, during the rejection process. Endothelial repair occurs both from local proliferation of endothelial cells and recruitment of recipient endothelial progenitor cells from the circulation. This results in formation of a chimeric vascular endothelium over time within the allograft (14). As a result of this phenomenon, cfdDNA may become less sensitive in the late post-transplant period after a greater number of recipient endothelial progenitor cells are incorporated within the allograft endothelium. In addition, measurement of the total circulating cell-free DNA content must be incorporated as a quality control. Since the cfdDNA is reported as a percentage of total cell-free DNA, it is possible that increased amounts of total cell-free DNA could dilute the donor specific content and falsely skew the results. Such a scenario was recently reported in a transplant patient with sepsis (8).
Prior probabilities and the Bayesian approach to biomarker studies
Clinicians are taught throughout their medical training to take a Bayesian approach to diagnostic and treatment decisions by constantly modifying the prior probability of disease based on new information from multiple sources. Our analysis methods for biomarker studies rarely take an integrative Bayesian approach but instead rely upon comparison of the novel biomarker to a “gold standard” diagnostic test. In reality, the rejection event rates reported in clinical studies represent an average event rate for the study population. Depending upon time since transplant, levels of immunosuppression, presence of donor specific antibodies, presence of heart failure symptoms, appearance of the echocardiogram, and assessment of patient compliance, the pre-test probability for rejection will vary substantially for an individual patient (10,15). A more nuanced understanding about when diagnostic tests can meaningfully influence the probability of rejection is what is most important. Maximizing or changing diagnostic test cutoff thresholds based on the pre-test probability would be tremendously helpful to the heart transplant clinician. The adult cardiology community has performed this type of analysis effectively for treatment of coronary artery disease. Using tools readily available to the primary care clinician, 10-year risk of coronary artery disease is determined (16). This baseline risk then informs primary prevention treatment strategies. When patients present with an acute coronary syndrome, the prior probability of coronary artery disease helps determine which diagnostic tests are utilized to shift those probabilities and guide treatment decisions (17).
While pathological grading of endomyocardial biopsies has remained the gold standard in assessing biomarkers of rejection in heart transplant recipients, the biopsy serves as one more diagnostic test which modifies the prior probability of rejection. In a patient with a low likelihood of rejection, a biopsy which barely qualifies as an ACR 2R may not clinch the diagnosis of rejection. Conversely, patients can present with hemodynamic compromise and a compelling clinical rejection syndrome despite low rejection grades on biopsy. For example, when rejection with hemodynamic compromise was examined in a pediatric heart transplant cohort, one-third of biopsies were graded as ACR 0R or 1R (15). In addition, despite improved ability to diagnose AMR, biopsy negative rejection continues to be a recognized phenomenon in heart transplant recipients with LV dysfunction (18). Multiple studies have demonstrated that the intraobserver variability between pathologists for grading endomyocardial biopsies is high in blinded comparisons (19). However, in the real world pathologists do not function as a core lab, divorced from clinical information or the prior probability of rejection. When a pathologist is deciding whether to call a marginal biopsy a 1R or a 2R, the indication for the biopsy, the center’s clinical protocols, and other clinical information, have the potential to bias the final biopsy grade.
Comparing new diagnostic tests to an imperfect gold standard introduces uncertainty into the estimates of test performance. Mengel et al. (20) have pointed out that molecular analysis of endomyocardial biopsy specimens and correlation of findings with clinically relevant rejection outcomes allows for refinement of the gold standard and increases the reliability of novel diagnostic test performance estimates. The heart transplant community has to take a creative and pragmatic approach to solve this problem. When designing comprehensive rejection biomarker studies, the outcome variable should be strictly defined but not limited to pathological endomyocardial biopsy grade. A carefully defined Bayesian algorithm or composite rejection score for classifying patients as rejectors and non-rejectors should outperform pathological grading of endomyocardial biopsies specimens alone as a study outcome variable. Only then will studies be able to discover and validate biomarkers that are most predictive of clinically meaningful rejection and poor patient outcomes.
Conclusions
Cell-free donor-specific DNA is an intriguing non-invasive biomarker for ACR and AMR in heart transplant patients. The study by De Vlaminck et al. is an important early step in understanding the performance characteristics when compared to pathologic diagnosis of ACR and AMR by endomyocardial biopsy (7). It will be vital to validate that the performance characteristics hold up in a larger cohort of heart transplant recipients and to demonstrate that clinical use of cfdDNA improves the care and outcomes for heart transplant recipients. Further study is also necessary to determine if cfdDNA fraction may provide independent clinical prognostic information and if cfdDNA can be used to monitor therapeutic response to anti-rejection therapy. It is time for our community to take a more integrative Bayesian approach to the definition of rejection outcome measures in biomarker studies and to study how new diagnostic tests can complement routine clinical care, rather than to replace it.
Acknowledgements
Funding: Dr. Daly has been supported by NIH grants L40 HL110356, T32 HL07572, and K12 HD052896-06, the Alexia Clinton Research Fund, and the Boston Children’s Hospital Cardiac Transplant and Education Fund.
Disclosure: The author declares no conflict of interest.
References
- Miniati DN, Robbins RC. Heart transplantation: a thirty-year perspective. Annu Rev Med 2002;53:189-205. [PubMed]
- Dipchand AI, Edwards LB, Kucheryavaya AY, et al. The registry of the International Society for Heart and Lung Transplantation: seventeenth official pediatric heart transplantation report--2014; focus theme: retransplantation. J Heart Lung Transplant 2014;33:985-95. [PubMed]
- Lund LH, Edwards LB, Kucheryavaya AY, et al. The registry of the International Society for Heart and Lung Transplantation: thirty-first official adult heart transplant report--2014; focus theme: retransplantation. J Heart Lung Transplant 2014;33:996-1008. [PubMed]
- Stewart S, Winters GL, Fishbein MC, et al. Revision of the 1990 working formulation for the standardization of nomenclature in the diagnosis of heart rejection. J Heart Lung Transplant 2005;24:1710-20. [PubMed]
- Berry GJ, Burke MM, Andersen C, et al. The 2013 International Society for Heart and Lung Transplantation Working Formulation for the standardization of nomenclature in the pathologic diagnosis of antibody-mediated rejection in heart transplantation. J Heart Lung Transplant 2013;32:1147-62. [PubMed]
- Pham MX, Teuteberg JJ, Kfoury AG, et al. Gene-expression profiling for rejection surveillance after cardiac transplantation. N Engl J Med 2010;362:1890-900. [PubMed]
- De Vlaminck I, Valantine HA, Snyder TM, et al. Circulating cell-free DNA enables noninvasive diagnosis of heart transplant rejection. Sci Transl Med 2014;6:241ra77.
- Hidestrand M, Tomita-Mitchell A, Hidestrand PM, et al. Highly sensitive noninvasive cardiac transplant rejection monitoring using targeted quantification of donor-specific cell-free deoxyribonucleic acid. J Am Coll Cardiol 2014;63:1224-6. [PubMed]
- Deng MC, Eisen HJ, Mehra MR, et al. Noninvasive discrimination of rejection in cardiac allograft recipients using gene expression profiling. Am J Transplant 2006;6:150-60. [PubMed]
- Mehra MR, Parameshwar J. Gene expression profiling and cardiac allograft rejection monitoring: is IMAGE just a mirage? J Heart Lung Transplant 2010;29:599-602. [PubMed]
- Woodside KJ, Lick SD. Alemtuzumab (Campath 1H) as successful salvage therapy for recurrent steroid-resistant heart transplant rejection. J Heart Lung Transplant 2007;26:750-2. [PubMed]
- Chin C, Naftel DC, Singh TP, et al. Risk factors for recurrent rejection in pediatric heart transplantation: a multicenter experience. J Heart Lung Transplant 2004;23:178-85. [PubMed]
- Bruneau S, Woda CB, Daly KP, et al. Key Features of the Intragraft Microenvironment that Determine Long-Term Survival Following Transplantation. Front Immunol 2012;3:54. [PubMed]
- Simper D, Wang S, Deb A, et al. Endothelial progenitor cells are decreased in blood of cardiac allograft patients with vasculopathy and endothelial cells of noncardiac origin are enriched in transplant atherosclerosis. Circulation 2003;108:143-9. [PubMed]
- Everitt MD, Pahl E, Schechtman KB, et al. Rejection with hemodynamic compromise in the current era of pediatric heart transplantation: a multi-institutional study. J Heart Lung Transplant 2011;30:282-8. [PubMed]
- Goff DC Jr, Lloyd-Jones DM, Bennett G, et al. 2013 ACC/AHA guideline on the assessment of cardiovascular risk: a report of the American College of Cardiology/American Heart Association Task Force on Practice Guidelines. Circulation 2014;129:S49-73. [PubMed]
- Amsterdam EA, Wenger NK, Brindis RG, et al. 2014 AHA/ACC Guideline for the Management of Patients With Non-ST-Elevation Acute Coronary Syndromes: A Report of the American College of Cardiology/American Heart Association Task Force on Practice Guidelines. Circulation 2014;130:e344-426. [PubMed]
- Tang Z, Kobashigawa J, Rafiei M, et al. The natural history of biopsy-negative rejection after heart transplantation. J Transplant 2013;2013:236720.
- Angelini A, Andersen CB, Bartoloni G, et al. A web-based pilot study of inter-pathologist reproducibility using the ISHLT 2004 working formulation for biopsy diagnosis of cardiac allograft rejection: the European experience. J Heart Lung Transplant 2011;30:1214-20. [PubMed]
- Mengel M, Sis B, Kim D, et al. The molecular phenotype of heart transplant biopsies: relationship to histopathological and clinical variables. Am J Transplant 2010;10:2105-15. [PubMed]


