
Bioinformatics analysis of key genes in triple negative breast cancer and validation of oncogene PLK1
Introduction
Breast cancer is the most common malignant tumor in women and is the biggest threat to women’s health worldwide. Triple-negative breast cancer (TNBC), which is negative for estrogen (ER), progesterone (PR), and human epidermal growth factor receptor 2 (HER-2) expression, has attracted widespread attention in recent years (1,2). It typically exhibits the following characteristics: early-onset, more common in premenopausal women under 50 years old, earlier local recurrence and distant metastasis, visceral metastasis (as opposed to bone metastasis), insensitivity to endocrine and targeted therapies, and poor prognosis (3). The poor prognosis of TNBC is thought to be related to its invasive biological behavior and resistance to existing therapies (4). However, the exact pathogenesis of breast cancer is still unclear. In order to provide screening markers and novel therapeutic targets for the prevention and treatment of breast cancer, understanding its pathogenesis at the molecular level is critical (5).
Relevant studies have shown that TNBC with different molecular markers is closely related to the treatment of TNBC (6,7). At present, some molecular biomarkers related to TNBC have been reported. Studies have shown that in TNBC, a higher level of vascular endothelial growth factor is associated with a shorter survival (6,8). In addition, EGFR signaling cascades play an important role in cell proliferation, angiogenesis, metastasis and proliferation, and apoptosis inhibition. EGFR expression in TNBC patients is related to poor chemotherapy response and poor survival rate. Therefore, EGFR is a biomarker of TNBC (7,9,10). The level of Ki67 in TNBC was significantly increased, and its expression was directly related to tumor size and grade in TNBC patients. A higher level of Ki67 was also associated with an increased risk of death from TNBC (11-13).
Bioinformatics is an interdisciplinary field (14-16). Using cluster analysis, pathway analysis, and other methods, we are able to screen large-scale gene chip data, mine potentially significant molecules, and theoretically discuss disease pathogenesis. With the development of high throughput technology, the application of gene chip and gene sequencing has become a necessary and efficient method to study tumor diseases (17). Presently, in the era of data sharing, there are abundant genetic testing and analysis results in major databases; yet, accurate and effective data mining is lacking.
Polo-like kinase 1 (PLK1) belongs to a family of mitotic serine/threonine kinases that is highly conserved in eukaryotes (18) and plays a key role in cell cycle regulation. PLK1 contains a conservative N-terminal kinase catalytic domain and a C-terminal POLO box domain (PBD), which are involved in substrate binding (19). PLK1 regulates almost every stage of cell division including mitosis initiation, centrosome maturation, bipolar spindle formation, sister chromatid separation, mitotic withdrawal, and cytokinesis (20,21). In addition, recent studies have shown that PLK1 plays an important role in regulating microtubule dynamics, deoxyribonucleic acid (DNA) replication, chromosome dynamics, tumor protein (p53) activity, and G2DNA damage repair (22,23). Furthermore, it is overexpressed in a variety of tumors, and its expression level is associated with a high cell proliferation rate, high metastasis potential, and poor prognosis (24,25).
In this study, differential genes were screened from the data set downloaded from the Gene Expression Omnibus (GEO) database, and PLK1 was screened through a protein-protein interaction (PPI) network. PLK1 was found to play a key role in the incidence of TNBC. The importance of PLK1 was further verified by The Cancer Genome Atlas Program (TCGA) data set. Subsequently, the mechanism of PLK1 promotion of the malignant evolution of breast cancer was verified through cell and animal level experiments. This study provides a theoretical basis for exploring potential therapeutic targets for breast cancer.
We present the following article in accordance with the ARRIVE reporting checklist (available at http://dx.doi.org/10.21037/atm-20-6873).
Methods
GEO data downloads
In this study, GEO breast cancer microarray data GSE62931 gene expression profile chip was selected as the research object (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE62931), which was Affymetrix Human Genome U1332.0 chip [HGU133_Plus_2]u (Agilent GPL15048 platform). The GSE62931 data set consisted of 53 breast cancer tumor tissue chips and 53 TNBC tumor tissue chips.
Analysis of differentially expressed genes (DEGs)
The difference in gene expression was expressed as fold change (FC). The screening criteria utilized in this study were: P<0.05 and |logFC|≥1 (screening DEGs with expression difference of more than two times).
Gene ontology (GO) knowledgebase and Kyoto Encyclopedia of Genes and Genomes (KEGG) analyses
The DEG data were imported into the DAVID online functional annotation bioinformatics microarray analysis (https://david.ncifcrf.gov/). Functional enrichment of DEGs was carried out using GO analysis to examine the biological processes (BP) in which these genes are primarily involved. False discovery rate (FDR) <0.05 was used as the inclusion criteria. Analysis mainly involved cancer related pathways using the ConsensusPathDB molecular functional interaction database (http://cpdb.molgen.mpg.de/).
Construction of PPI networks
The online data analysis software Search Tool for the Retrieval of Interacting Genes (STRING) was used to analyze the protein interacting pairs and to build the PPI networks. The lowest interaction score was selected as high credibility, and the interaction score of >0.7 was selected for study.
PLK1 prognostic analysis
In this study, patients with PLK1 gene were analyzed using the Gene Expression Profiling Interactive Analysis (GEPIA) data analysis tool (http://gepia.cancer-pku.cn/index.html). GEPIA is a database site for online analytics. The GEPIA website contains extensive sequencing data, including results from 8,587 tumor tissues and 9,736 normal tissues.
Clinical tissue collection
Tumor tissues and adjacent paracancerous control tissues were collected from the Xinhua Hospital Affiliated to Medical College of Shanghai Jiao Tong University from January 2009 to May 2020. Twenty female patients, with an average age of 56.2 years and complete clinicopathological data were selected. None of the patients received preoperative radiotherapy, chemotherapy, or endocrine therapy. According to the immunohistochemistry results, the classification was based on the Nielsen standard (26). All patients signed the informed consent. This study was approved by the ethics committee of Xinhua Hospital Affiliated to Medical College of Shanghai Jiao Tong University (No. XHEC-D-2020-179) and in line with Declaration of Helsinki (as revised in 2013).
Cell culture
Breast cancer cell lines were purchased from American Type Culture Collection (ATCC, Manassas, VA, USA). All cells were incubated at a constant temperature of 37 °C and 5% carbon dioxide (CO2) under humid conditions. Cell growth was observed daily, and the culture medium was replaced as required. Cell lines were transfected with PLK1 small interfering ribonucleic acid (siRNA) (20 nM) and overexpressed plasmid. Lipofectamine 2000 (Life Technologies, Rockville, MA, USA) was used, and negative control siRNA (si-NC) and si-PLK1 were transferred into the cells for 6 h. The liquid was subsequently changed according to the manufacturer’s instructions. The transfected cells were used 48 h later.
Quantitative real-time polymerase chain reaction (QRT-PCR)
Total RNA was obtained using a RNA extraction kit and the concentration was determined. Reverse transcription kits were used to reverse transcribe RNA into complementary DNA (cDNA). SYBR (5 µL), upstream and downstream primers (0.3 µL each), diethyl pyrocarbonate (DEPC) water (3.4 µL), and cDNA (1 µL) were mixed and sampled. Machine test: 95 °C for 5 min, 39 cycles (95 °C for 10 s, 58 °C for 30 s, 72 °C for 20 s). β-actin, forward: CTGGAACGGTGAAGGTGACA, reverse: AAGGGACTTCCTGTAACAATGCA. Results were analyzed using the BIO-RAD CFX Manager. The β-actin was used as an internal reference for the analysis of target gene expression. Experimental results using 2−ΔΔCt algorithm to calculate. The experiment was repeated three times.
Proliferation experiment
The cells were inoculated into a 96-well plate, and were then transfected. Following continuous culture for 72 h, cell proliferation in each group was detected using the cell counting kit 8 (CCK-8) (Beyotime, Shanghai, China) by microplate reader (MultiskanEX, Lab systems, Helsinki, Finland) according to the manufacturer’s instructions (27).
Transwell migration assay experiment
The transwell migration assay test was carried out for 24-well plates. 5×104 cells/mL of treated cell suspensions in the logarithmic growth phase were added to the transwell chamber (Corning, USA). The transwell chamber was pre-mixed with Matrigel (Thermo Fisher Scientific, Waltham, MA, USA). Fetal bovine serum (FBS) medium (at a concentration of 500 µL) was added to the 24-well transwell chamber. After 24 h of culture, the transwell chamber was removed. A wet swab was rubbed off and no cells were inserted above the transwell chamber, thereby avoiding interference from cells that do not penetrate. The cells were then fixed with 4% paraformaldehyde (Sigma-Aldrich, St. Louis, MO, USA) for 10 min. Crystal violet staining solution was used for 15 min. It was then rinsed with phosphate buffered saline (PBS) and dried. Cells passing through the membrane were examined using an inverted microscope. Cells were counted and an average was calculated. Finally, a picture was taken (28).
Scratch test
The cells were spread in a 6-well plate with 3×105 cells/well. After the cell confluence reached 80%, a 10 µL suction head was used to draw a “horizontal line” in the orifice plate. The serum-free cell culture medium was then replaced. The scratch healing was observed and recorded at regular intervals, and the data were analyzed.
Clone formation experiment
A single-cell suspension was prepared by digestion with 0.25% trypsin. The cells were suspended in Roswell Park Memorial Institute (RPMI) 1640 culture medium containing 10% FBS. A 1 mL cell suspension (cell density: 200/mL) was placed in a 6 cm culture dish, and the cells were incubated for 2–3 weeks. The culture was ceased when visible clones appeared in the petri dish. The supernatant was discarded and washed with phosphate buffer solution (PBS) three times. The cells were fixed with 4% paraformaldehyde (5 mL), and the fixed solution was discarded after 15 min. An appropriate amount of GIEMSA staining solution was added for 10–30 min. The dye was then washed slowly with running water and allowed to air dry. The petri dish was turned upside down and overlay with a sheet of transparent grid film. The number of clones larger than eight cells was counted under the microscope and the clone formation rate was subsequently calculated.
Xenograft model
Nude mice were subcutaneously injected with breast cancer cells (5×105/0.1 mL) under the armpit skin to establish an animal model of tumor-bearing mice. Tumor volume in mice was measured during the experiment. The mice were euthanized with CO2 at the end of the experiment. The tumor mass was dissected and weighed, and the tumor inhibition rate was calculated as follows: tumor inhibition rate (%) = (average tumor weight of the control group − average tumor weight of the administration group)/ average tumor weight of the control group ×100%.
Immunohistochemistry
Rabbit anti-human PLK1 and Ki-67 monoclonal antibodies were purchased from Abcam, USA. The ready-to-use MaxVisionTM detection kit and amino benzidine color development kit were purchased from Fuzhou Maixin Biological and Biotechnology Co., LTD. The specimens were treated with 10% neutral formalin fixation, embedded in paraffin, cut into 4 µm thick continuous sections, and then subjected to immunohistochemical MaxVision two-step staining (the specific operational steps were performed according to the kit instructions). PBS replaced primary antibody as the negative control, and the gastric cancer tissues with known PLK1 positive were used as the positive control.
Terminal deoxynucleotidyl transferase dUTP nick end labelling (TUNEL) staining
The tissue slices were initially incubated with deparaffin and protease K. After 2 min, they were washed with PBS three times. Next, they were incubated with terminal deoxynucleotidyl transferase (TdT) enzyme at 37 °C in the dark for 60 min, and subsequently washed in PBS three times. The slides were then incubated in horseradish peroxidase-labeled streptavidin followed by detection with stable chromagen 3,3'-diaminobenzidine (DAB). Finally, the slices were observed using a confocal laser scanning microscope (LSM880, Germany).
Statistical analysis
Statistical results are expressed as mean ± standard deviation. All data were analyzed using SPSS 19.0 software (SPSS Inc., Chicago, IL, USA). The statistical method used was the independent sample t-test. One-way analysis of variance (ANOVA) was used for comparison among groups. P<0.05 was considered statistically significant.
Results
Screening of DEGs between TNBC and non-TNBC
The results showed that 824 genes with DEGs were screened by analyzing GSE62931 chip data, among which 405 genes were up-regulated and 419 genes were down-regulated (Figure 1 and https://cdn.amegroups.cn/static/public/10.21037atm-20-6873-1.xlsx).
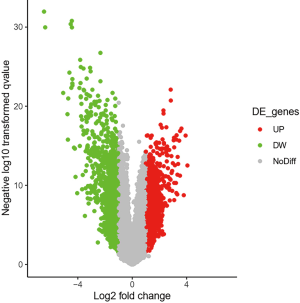
GO and KEGG enrichment analysis of down-regulated genes
Functional enrichment analysis of selected DEGs was performed using DAVID. The GO function annotation results of down-regulated expressed genes showed that these DEGs mainly participated in the following BP: proteolysis, multicellular development, protein glycosylation, neuropeptide signaling pathway, negative regulation of protein kinase activity, digestion, negative regulation of the Wingless-related integration site (Wnt) signaling pathway, and response to peptide hormone (Figure 2A). The primary cell composition (CC) involved included the extracellular space, the extracellular region, the integral component of plasma membrane, the extracellular matrix and the proteinaceous extracellular matrix (Figure S1). The key molecular functions (MF) involved were calcium ion binding, carbohydrate binding, heparin binding, serine-type endopeptidase inhibitor activity, heme binding, microtubule motor activity, metallocarboxypeptidase activity, polypeptide N-acetyl galactosaminyl transferase activity, and dopamine receptor binding (Figure S2). Analysis mainly involved cancer related pathways using the ConsensusPathDB molecular functional interaction database (http://cpdb.molgen.mpg.de/). KEGG pathway analysis showed that multiple DEGs were enriched into signaling by receptor tyrosine kinases, biological oxidations, estrogen signaling receptor (ESR)-mediated signaling, innate immune system, platelet activation, signaling and aggregation, and post-translational protein modification (Figure 2B).
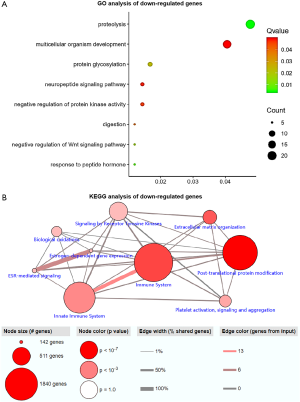
GO and KEGG enrichment analysis of up-regulated genes
The GO function annotation results of up-regulated genes showed that these DEGs are mainly involved in the following BP: chromosome segregation, nuclear chromosome segregation, sister chromatid segregation, cell cycle, cell cycle process, mitotic cell cycle, nuclear division, mitotic cell cycle process, mitotic sister chromatid segregation, cell division, mitotic nuclear division, and chromosome organization (Figure 3A). The primary CC involved included: cytoplasm, cytoskeleton, chromosome, chromosomal region, kinetochore, spindle midzone, and kinesin complex (Figure S3). The key MF involved were double-stranded DNA binding, structural molecule activity, structural constituent of cytoskeleton, chemokine activity, microtubule binding, chemokine receptor binding, CXCR chemokine receptor binding, receptor for advanced glycation end products (RAGE) receptor binding, G protein-coupled receptor (GPCR) binding, protein binding, sequence-specific DNA binding, tubulin binding, and axon guidance receptor activity (Figure S4). Analysis mainly involved cancer related pathways using the ConsensusPathDB molecular functional interaction database (http://cpdb.molgen.mpg.de/). KEGG pathway analysis revealed that the enrichment of the channel was primarily: PLK1 signaling events, epidermal growth factor receptor 1 (EGFR1) signaling, signaling by GPCR, signaling by Rho guanosine triphosphatases (GTPases), RNA polymerase II transcription, and interleukin (IL)-17 signaling pathway (Figure 3B).
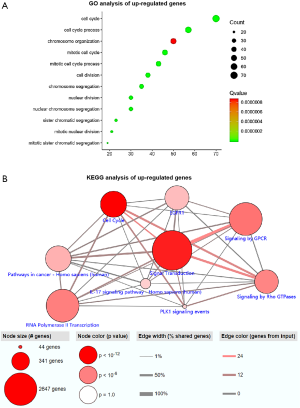
PPI network construction and screening and identification of Hub genes
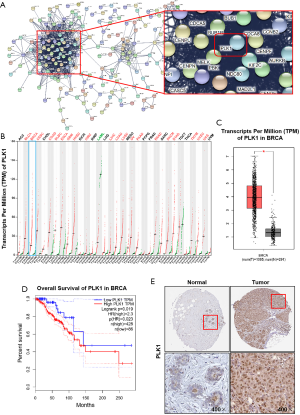
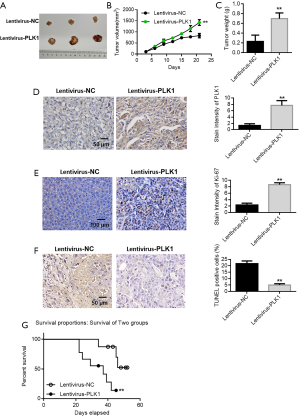
Screened DEGs were input into the STRING website to build the PPI network (Figure 4A). The results showed that the most closely connected gene in the network was PLK1, which was the Hub gene with a high degree of connectivity. The expression of PLK1 was further analyzed and verified, and results showed that PLK1 was highly expressed in multiple tumors (Figure 4B). The expression of PLK1 in breast cancer was also investigated, and results showed that PLK1 was significantly up-regulated in the tumor group compared with the normal group (Figure 4C). Survival analysis of breast cancer patients with clinical and prognostic information on TCGA was performed using The Human Protein Atlas (online). Patients were divided into high expression group and low expression group according to the gene expression level. The results indicated that the expression level of PLK1 was negatively correlated with the overall survival of patients (Figure 4D). Further immunohistochemistry analysis showed that PLK1 expression increased in breast cancer tumor tissues (Figure 4E). Therefore, we speculate that the expression level of PLK1 may play an important role in the prognosis of breast cancer patients, and a high expression level of PLK1 may be a risk factor for poor prognosis.
PLK1 is highly expressed in breast cancer
In order to verify the accuracy of the analysis results, we collected the tumor tissues and adjacent paracancerous control tissues of 20 breast cancer patients to detect the expression level of PLK1. QRT-PCR analysis showed that the expression level of PLK1 in breast cancer tissues was increased compared with the paracancerous control tissue (Figure 5A). The PLK1 expression levels in normal human breast cells (MCF10A) and breast cancer cells (MDA-MB-436, MDA-MB-231, T-47D, and BT-474) were also detected (Figure 5B). The results showed that the expression of PLK1 varied as follows: MDA-MB-436 > MDA-MB-231 > T-47D > BT-474 > MCF10A. In the subsequent experiments, MDA-MB-436 and MDA-MB-231 were used to verify the function of PLK1.
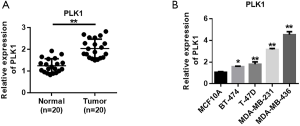
Overexpression of PLK1 promotes the malignant evolution of MDA-MB-436 and MDA-MB-231 in breast cancer
To verify the role of PLK1, we used siRNA and plasmid to knock down and overexpress PLK1, respectively. The efficiency of PLK1 expression in MDA-MB-436 cells showed that siRNA could effectively reduce the expression of PLK1, and overexpressed plasmid could successfully up-regulate the expression of PLK1 (Figure 6A). The efficiency of PLK1 expression in MDA-MB-231 cells was consistent with the trend in MDA-MB-436 cells (Figure 6B). The CCK-8 experiment was subsequently used to verify the effect of PLK1 on the proliferation of MDA-MB-436 and MDA-MB-231 cells. The results showed that the overexpression of PLK1 promoted the proliferation of breast cancer cells compared with the control group. Moreover, PLK1 knockdown inhibited the proliferation of breast cancer cells (Figure 6C,D). Next, transwell migration and scratch experiments were used to verify the effect of PLK1 on the invasion and migration of breast cancer cells. The results showed that the overexpression of PLK1 promoted the invasion and migration of breast cancer cells compared with the control group, while PLK1 knockdown inhibited the invasion and migration of breast cancer cells (Figure 6E,F,G,H). The cell clone formation experiment also showed that overexpression of PLK1 promoted the clone formation of MDA-MB-436 and MDA-MB-231 cells, while PLK1 knockdown inhibited clonal formation of MDA-MB-436 and MDA-MB-231 cells (Figure 6I,J).
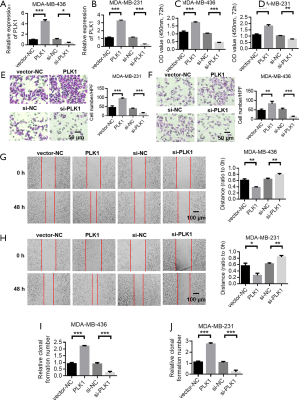
The tumor bearing experiment in nude mice showed that PLK1 promoted the proliferation of breast cancer cells.
We constructed a nude mouse xenograft model to further investigate the cancer-promoting effect of PLK1. The results indicated that PLK1 overexpression promoted the proliferation of breast cancer (Figures 7A,B). The tumor weight detection results in nude mice also showed that overexpression of PLK1 resulted in an increased tumor weight (Figure 7C). PLK1 immunohistochemical detection results indicated that PLK1 was significantly up-regulated in the lentivirus group (Figure 7D). Ki-67 detection results of cell proliferation showed that overexpression of PLK1 promoted the proliferation of tumor cells (Figure 7E). TUNEL staining detected apoptosis of tumor tissues, while experimental results showed that overexpression of PLK1 inhibited the apoptosis of breast cancer tumor cells (Figure 7F). Subsequently, we validated the cancer-promoting effect of PLK1 in mice that survived tumor injection through caudal vein. Results indicated that overexpression of PLK1 reduced the survival time of mice (Figure 7G).
Discussion
Breast cancer is a common malignancy with highly heterogeneous biological characteristics (29). Patients differ greatly in pathological manifestations, molecular biology, and treatment regimens (30). TNBC accounts for approximately 10–17% of all breast cancers (31). It is aggressive, has an earlier onset, earlier local recurrence and distant metastasis, and visceral metastases of the brain, liver, lung and other organs are more prevalent. It has a peak recurrence of between 1 and 3 years after treatment, and a high mortality rate within 5 years (32).
Tumor research is the most applied field of expression profile gene chip in disease research (15,33). The gene expression profile chip provides a powerful tool for analyzing gene differences between tumor and normal tissues and searching for novel tumor markers (34). It is critical to the screening of genetic markers for tumor classification and typing, and potential targets for drug therapy (16,28,35,36). In this study, the gene expression profile chip GSE62931 in the GEO database was used to screen out a total of 824 genes with significant differences in expression in non-TNBC and breast cancer tissues. In this study, GO and KEGG enrichment analysis was also performed on the screened DEGs. The results showed that these genes were primarily involved in the BP of cell and mitotic division; tumor growth depends in part on an increase in mitotic events. At the same time, these genes were also principally involved in the composition of the extracellular matrix, actin cytoskeleton and other cells, in addition to the binding of proteins and actin, as well as other MF. These are the basic biological conditions that regulate cell proliferation, migration, and invasion. However, the abnormality is the basic characteristic that leads to tumor formation. Through PPI analysis, it was found that the key Hub gene, namely the PLK1 gene, was closely related to tumorigenesis.
King et al. found that PLK1 was correlated with ER-α expression and TNBC (37). Maire et al. found that PLK1 expression level was higher in TNBC compared with adeno-cavity Type A, adeno-cavity Type B, and HER-2 overexpression type in breast cancer, and suggested that PLK1 was a potential therapeutic target for TNBC (38). The results of this study showed that PLK1 has a higher expression rate in the ER negative breast cancer molecular subtype compared with the ER positive breast cancer molecular subtype. In addition, there was a significant negative correlation with ER expression and a notable positive correlation with Ki-67 expression. The results suggested that PLK1 overexpression could play a key role in the carcinogenesis of ER negative breast cancer with high proliferative activity. We found that the overall survival of breast cancer patients with high PLK1 expression was lower than breast cancer patients with low PLK1 expression (39).
The results of this study showed that the positive expression rate of PLK1 in breast cancer was increased compared with that of the paracancerous control group. Meanwhile, the overexpression of PLK1 was found to promote the proliferation, invasion, and migration of breast cancer. In vitro animal experiments further confirmed that overexpression of PLK1 promoted tumor proliferation and inhibited tumor cell apoptosis. This suggests that PLK1 overexpression may be closely related to the incidence and development of breast cancer. The expression of PLK1 was observed to be related to the invasion and metastasis of breast cancer, suggesting that PLK1 may be an effective indicator and potential target for evaluating the prognosis of TNBC. Previous studies found that PLK1-targeted siRNA could effectively inhibit the proliferation of various malignant tumors, block tumor cells at the G2/M stage, and induce tumor cell apoptosis and sensitivity to radiotherapy and chemotherapy (40-42). Therefore, the development of PLK1 inhibitors as therapeutic targets may provide novel ideas regarding an individualized treatment for basal-cell type breast cancer (43-46).
There have been some reports on the cancer-promoting mechanism of PLK1. Studies on liver cancer and pancreatic cancer have found that the high expression of PLK1 mainly occurs in the early stage of tumor (47). In addition, PLK1 may be expressed as an inhibition of p53 activity by the G2 and S-phase-expressed 1 protein (GTSE1) and Topors, a ubiquitinized SUMOE3 ligase. PLK1 helps DNA-damaged cells escape the checkpoint block (48). Therefore, overexpression of PLK1 can directly promote cell proliferation, and on the other hand, can increase chromosomal instability and eventually lead to the occurrence of tumors by assisting the arrest of DNA-damaged cells to escape from the monitoring sites. Some downstream products and proteins of PLK1 action include FoxM1, Myc, Mdm2 and β-catenin (49-52). At the same time, PLK1 can also regulate some tumor suppressor factors such as P53, pRb, Brca2 and PTEN (53-55).
Conclusions
PLK1 gene screened by GSE62931 gene chip is abnormally expressed in a variety of cancers, and has been clearly shown to play a key role in the occurrence and development of certain tumors. The PLK1 gene is located at the key node position in the expression spectrum network composed of breast cancer DEGs. Thus, it can be used as the key Hub gene in the process of breast cancer occurrence, providing guidance for the further research on the molecular mechanism of breast cancer occurrence and development and the screening of molecular markers.
Acknowledgments
Funding: None.
Footnote
Reporting Checklist: The authors have completed the ARRIVE reporting checklist. Available at http://dx.doi.org/10.21037/atm-20-6873
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/atm-20-6873). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. All patients signed the informed consent. This study was approved by the ethics committee of Xinhua Hospital Affiliated to Medical College of Shanghai Jiao Tong University (No. XHEC-D-2020-179) and in line with Declaration of Helsinki (as revised in 2013).
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Schmid P, Cortes J, Pusztai L, et al. Pembrolizumab for early triple-negative breast cancer. N Engl J Med 2020;382:810-21. [Crossref] [PubMed]
- Wang M, Hu Y, Hou L, et al. A clinical study on the use of Huaier granules in post-surgical treatment of triple-negative breast cancer. Gland Surg 2019;8:758-65. [Crossref] [PubMed]
- Qin JJ, Yan L, Zhang J, et al. STAT3 as a potential therapeutic target in triple negative breast cancer: a systematic review. J Exp Clin Cancer Res 2019;38:195. [Crossref] [PubMed]
- Quereda V, Bayle S, Vena F, et al. Therapeutic targeting of CDK12/CDK13 in triple-negative breast cancer. Cancer Cell 2019;36:545-58.e7. [Crossref] [PubMed]
- Zhong W, Hou H, Liu T, et al. Cartilage Oligomeric Matrix Protein promotes epithelial-mesenchymal transition by interacting with Transgelin in Colorectal Cancer. Theranostics 2020;10:8790-806. [Crossref] [PubMed]
- Linderholm B, Hellborg H, Johansson U, et al. Significantly higher levels of vascular endothelial growth factor (VEGF) and shorter survival times for patients with primary operable triple-negative breast cancer. Ann Oncol 2009;20:1639-46. [Crossref] [PubMed]
- Naito Y, Urasaki T. Precision medicine in breast cancer. Chin Clin Oncol 2018;7:29. [Crossref] [PubMed]
- Zhao Z, Li Y, Shukla R, et al. Development of a biocompatible copolymer nanocomplex to deliver VEGF siRNA for triple negative breast cancer. Theranostics 2019;9:4508-24. [Crossref] [PubMed]
- Nakai K, Hung MC, Yamaguchi H. A perspective on anti-EGFR therapies targeting triple-negative breast cancer. Am J Cancer Res 2016;6:1609-23. [PubMed]
- Nakajima H, Ishikawa Y, Furuya M, et al. Protein expression, gene amplification, and mutational analysis of EGFR in triple-negative breast cancer. Breast Cancer 2014;21:66-74. [Crossref] [PubMed]
- Kashiwagi S, Yashiro M, Takashima T, et al. Advantages of adjuvant chemotherapy for patients with triple-negative breast cancer at Stage II: usefulness of prognostic markers E-cadherin and Ki67. Breast Cancer Res 2011;13:R122. [Crossref] [PubMed]
- Li H, Han X, Liu Y, et al. Ki67 as a predictor of poor prognosis in patients with triple-negative breast cancer. Oncol Lett 2015;9:149-52. [Crossref] [PubMed]
- Guestini F, Ono K, Miyashita M, et al. Impact of Topoisomerase IIα, PTEN, ABCC1/MRP1, and KI67 on triple-negative breast cancer patients treated with neoadjuvant chemotherapy. Breast Cancer Res Treat 2019;173:275-88. [Crossref] [PubMed]
- Zhai Q, Li H, Sun L, et al. Identification of differentially expressed genes between triple and non-triple-negative breast cancer using bioinformatics analysis. Breast Cancer 2019;26:784-91. [Crossref] [PubMed]
- Xi X, Chu Y, Liu N, et al. Joint bioinformatics analysis of underlying potential functions of hsa-let-7b-5p and core genes in human glioma. J Transl Med 2019;17:129. [Crossref] [PubMed]
- Zhong W, Yang W, Qin Y, et al. 6-Gingerol stabilized the p-VEGFR2/VE-cadherin/beta-catenin/actin complex promotes microvessel normalization and suppresses tumor progression. J Exp Clin Cancer Res 2019;38:285. [Crossref] [PubMed]
- Chen WX, Yang LG, Xu LY, et al. Bioinformatics analysis revealing prognostic significance of RRM2 gene in breast cancer. Biosci Rep 2019;39:BSR20182062. [Crossref] [PubMed]
- Reda M, Ngamcherdtrakul W, Gu S, et al. PLK1 and EGFR targeted nanoparticle as a radiation sensitizer for non-small cell lung cancer. Cancer Lett 2019;467:9-18. [Crossref] [PubMed]
- Ueda A, Oikawa K, Fujita K, et al. Therapeutic potential of PLK1 inhibition in triple-negative breast cancer. Lab Invest 2019;99:1275-86. [Crossref] [PubMed]
- Barr FA, Silljé HH, Nigg EA. Polo-like kinases and the orchestration of cell division. Nat Rev Mol Cell Biol 2004;5:429-40. [Crossref] [PubMed]
- Llamazares S, Moreira A, Tavares A, et al. polo encodes a protein kinase homolog required for mitosis in Drosophila. Genes Dev 1991;5:2153-65. [Crossref] [PubMed]
- Liu XS, Song B, Liu X. The substrates of Plk1, beyond the functions in mitosis. Protein Cell 2010;1:999-1010. [Crossref] [PubMed]
- Song B, Liu XS, Davis K, et al. Plk1 phosphorylation of Orc2 promotes DNA replication under conditions of stress. Mol Cell Biol 2011;31:4844-56. [Crossref] [PubMed]
- Strebhardt K. Multifaceted polo-like kinases: drug targets and antitargets for cancer therapy. Nat Rev Drug Discov 2010;9:643-60. [Crossref] [PubMed]
- Cholewa BD, Liu X, Ahmad N. The role of polo-like kinase 1 in carcinogenesis: cause or consequence? Cancer Res 2013;73:6848-55. [Crossref] [PubMed]
- Nielsen TO, Hsu FD, Jensen K, et al. Immunohistochemical and clinical characterization of the basal-like subtype of invasive breast carcinoma. Clin Cancer Res 2004;10:5367-74. [Crossref] [PubMed]
- Wang H, Zhong W, Zhao J, et al. Oleanolic Acid Inhibits Epithelial-Mesenchymal Transition of Hepatocellular Carcinoma by Promoting iNOS Dimerization. Mol Cancer Ther 2019;18:62-74. [Crossref] [PubMed]
- Xiao T, Zhong W, Zhao J, et al. Polyphyllin I suppresses the formation of vasculogenic mimicry via Twist1/VE-cadherin pathway. Cell Death Dis 2018;9:906. [Crossref] [PubMed]
- McCann KE, Hurvitz SA, McAndrew N. Advances in Targeted Therapies for Triple-Negative Breast Cancer. Drugs 2019;79:1217-30. [Crossref] [PubMed]
- Lee KL, Kuo YC, Ho YS, et al. Triple-Negative Breast Cancer: Current Understanding and Future Therapeutic Breakthrough Targeting Cancer Stemness. Cancers (Basel) 2019;11:1334. [Crossref] [PubMed]
- Hwang SY, Park S, Kwon Y. Recent therapeutic trends and promising targets in triple negative breast cancer. Pharmacol Ther 2019;199:30-57. [Crossref] [PubMed]
- Mehanna J, Haddad FG, Eid R, et al. Triple-negative breast cancer: Current perspective on the evolving therapeutic landscape. Int J Womens Health 2019;11:431-7. [Crossref] [PubMed]
- Chen WX, Cheng L, Xu LY, et al. Bioinformatics analysis of prognostic value of TRIM13 gene in breast cancer. Biosci Rep 2019;39:BSR20190285. [Crossref] [PubMed]
- Casciano JC, Perry C, Cohen-Nowak AJ, et al. MYC regulates fatty acid metabolism through a multigenic program in claudin-low triple negative breast cancer. Br J Cancer 2020;122:868-84. [Crossref] [PubMed]
- Deng JL, Xu YH, Wang G, et al. Identification of potential crucial genes and key pathways in breast cancer using bioinformatic analysis. Front Genet 2019;10:695. [Crossref] [PubMed]
- Zhong W, Sun B, Gao W, et al. Salvianolic acid A targeting the transgelin-actin complex to enhance vasoconstriction. EBioMedicine 2018;37:246-58. [Crossref] [PubMed]
- King SI, Purdie CA, Bray SE, et al. Immunohistochemical detection of Polo-like kinase-1 (PLK1) in primary breast cancer is associated with TP53mutation and poor clinical outcome. Breast Cancer Res 2012;14:R40. [Crossref] [PubMed]
- Maire V, Némati F, Richardson M, et al. Polo-like kinase 1: a potential therapeutic option in combination with conventional chemotherapy for the management of patients with triple-negative breast cancer. Cancer Res 2013;73:813-23. [Crossref] [PubMed]
- Wolf G, Hildenbrand R, Schwar C, et al. Polo-like kinase: a novel marker of proliferation: correlation with estrogen-receptor expression in human breast cancer. Pathol Res Pract 2000;196:753-9. [Crossref] [PubMed]
- Wang ZX, Xue D, Liu ZL, et al. Overexpression of polo-like kinase 1 and its clinical significance in human non-small cell lung cancer. Int J Biochem Cell Biol 2012;44:200-10. [Crossref] [PubMed]
- Harris PS, Venkataraman S, Alimova I, et al. Polo-like kinase 1 (PLK1) inhibition suppresses cell growth and enhances radiation sensitivity in medulloblastoma cells. BMC Cancer 2012;12:80. [Crossref] [PubMed]
- Hu K, Law JH, Fotovati A, et al. Small interfering RNA library screen identified polo-like kinase-1 (PLK1) as a potential therapeutic target for breast cancer that uniquely eliminates tumor-initiating cells. Breast Cancer Res 2012;14:R22. [Crossref] [PubMed]
- Yan L, Zhang Y, Li K, et al. miR-593-5p inhibit cell proliferation by targeting PLK1 in non small cell lung cancer cells. Pathol Res Pract 2020;216:152786. [Crossref] [PubMed]
- Giordano A, Liu Y, Armeson K, et al. Polo-like kinase 1 (Plk1) inhibition synergizes with taxanes in triple negative breast cancer. PloS One 2019;14:e0224420. [Crossref] [PubMed]
- Xi X, Liu N, Wang Q, et al. ACT001, a novel PAI-1 inhibitor, exerts synergistic effects in combination with cisplatin by inhibiting PI3K/AKT pathway in glioma. Cell Death Dis 2019;10:757. [Crossref] [PubMed]
- Zhong W, Sun B, Ruan H, et al. Deglycosylated azithromycin targets transgelin to enhance intestinal smooth muscle function. iScience 2020;23:101464. [Crossref] [PubMed]
- Petrelli A, Perra A, Schernhuber K, et al. Sequential analysis of multistage hepatocarcinogenesis reveals that miR-100 and PLK1 dysregulation is an early event maintained along tumor progression. Oncogene 2012;31:4517-26. [Crossref] [PubMed]
- Liu XS, Li H, Song B, et al. Polo-like kinase 1 phosphorylation of G2 and S-phase-expressed 1 protein is essential for p53 inactivation during G2 checkpoint recovery. EMBO Rep 2010;11:626-32. [Crossref] [PubMed]
- Dibb M, Han N, Choudhury J, et al. The FOXM1-PLK1 axis is commonly upregulated in oesophageal adenocarcinoma. Br J Cancer 2012;107:1766-75. [Crossref] [PubMed]
- Fu Z, Malureanu L, Huang J, et al. Plk1-dependent phosphorylation of FoxM1 regulates a transcriptional programme required for mitotic progression. Nat Cell Biol 2008;10:1076-82. [Crossref] [PubMed]
- Ren Y, Bi C, Zhao X, et al. PLK1 stabilizes a MYC-dependent kinase network in aggressive B cell lymphomas. J Clin Invest 2018;128:5517-30. [Crossref] [PubMed]
- Dias SS, Hogan C, Ochocka AM, et al. Polo-like kinase-1 phosphorylates MDM2 at Ser260 and stimulates MDM2-mediated p53 turnover. FEBS Lett 2009;583:3543-8. [Crossref] [PubMed]
- Ando K, Ozaki T, Yamamoto H, et al. Polo-like kinase 1 (Plk1) inhibits p53 function by physical interaction and phosphorylation. J Biol Chem 2004;279:25549-61. [Crossref] [PubMed]
- Li Z, Li J, Bi P, et al. Plk1 phosphorylation of PTEN causes a tumor-promoting metabolic state. Mol Cell Biol 2014;34:3642-61. [Crossref] [PubMed]
- Lee M, Daniels MJ, Venkitaraman AR. Phosphorylation of BRCA2 by the Polo-like kinase Plk1 is regulated by DNA damage and mitotic progression. Oncogene 2004;23:865-72. [Crossref] [PubMed]
(English Language Editor: A. Kassem)


