
This article has an erratum available at: http://dx.doi.org/10.21037/atm-2021-3 the article has been update on 2021-03-23 at here.
Inhibition of IL-6/JAK/STAT3 pathway rescues denervation-induced skeletal muscle atrophy
Introduction
Skeletal muscle atrophy results from a variety of physiopathological conditions, including aging, immobilization, starvation, denervation, sepsis, diabetes, kidney failure, chronic heart failure, and cachexia (1,2). Peripheral nerve injury is becoming more common, resulting in a high incidence of denervated skeletal muscle atrophy, which lead to reduced quality of life, increased morbidity and mortality. In the last 20 years, new insights have been made into the etiology and therapy of skeletal muscle atrophy, but the treatment options for this disorder are still limited and generally unsatisfactory (3-5). Therefore, a deeper understanding of the molecular mechanisms underlying denervated skeletal muscle atrophy is critically required for developing targeted therapy and retarding the concomitant loss of skeletal muscle mass.
The protein homeostasis in skeletal muscles can be disrupted by increased protein degradation and decreased protein synthesis, leading to skeletal muscle atrophy (6,7). The ubiquitin-proteasome pathway and autophagy-lysosome pathway (mitophagy) are two main pathways involved in protein degradation (8-10). The muscle ring-finger-1 (MuRF-1) and muscle atrophy F-box (MAFbx)/atrogin-1, as two muscle-specific E3 ubiquitin ligases, are increasingly expressed during the skeletal muscle atrophy induced by different stimuli, including dexamethasone, disuse, hindlimb suspension, denervation, and cancer cachexia (4,11-16). Autophagy and mitophagy have been proven to play crucial roles in various types of skeletal muscle atrophy. The levels of Bcl2/adenovirus E1B 19 kDa protein-interacting protein 3, microtubule-associated protein 1 light chain 3 (LC3), p62, and PTEN-induced kinase 1 (PINK1) are elevated during chronic kidney disease-induced skeletal muscle atrophy (17). Mitochondrial dysfunction occurs in several types of skeletal muscle atrophy induced by disuse, aging, diabetes, sepsis, and denervation according to the significantly increased expressions of p62, microtubule-associated protein 1 light chain 3-II (LC3-II), and Beclin 1 (6,18-22). Thus, skeletal muscle atrophy is usually accompanied by the activation of proteolytic pathway, and it is important to know which molecules or events regulate the proteolytic pathways.
As an important pathogenic factor, inflammation contributes to the dysfunction of skeletal muscles (23). Inflammation induces proinflammatory cytokine production, which activates the expression of the proteolytic pathway-related genes during skeletal muscle atrophy (24). A proinflammatory cytokine interleukin-1 beta (IL-1β) increases the expression of atrogenes (atrogin 1 and MuRF1) during skeletal muscle atrophy (7). IL-6, as a pleiotropic cytokine, exerts either pro- or anti-inflammatory effects according to different local immune milieus (25,26). IL-6 is also able to regulate muscle metabolic processes, precipitating muscle hypertrophy or muscle wasting (25). For example, IL-6 levels were found to be elevated during muscle wasting in mice suffering from colon-26 carcinoma (27), while IL-6 inhibition was shown to prevent cancer-induced muscle mass loss (28). It is widely acknowledged that IL-6 binds to ubiquitously expressed type I cytokine receptor glycoprotein 130 (GP130) to activate Janus kinases (JAKs), which subsequently activate signal transducers and activators of transcription 3 (STAT3) (27). Moreover, in cultured myotubes and skeletal muscle, chronically elevated IL-6 binding to GP130 can induce increased dynamin-1-like protein (DRP-1) and mitochondrial fission 1 protein (FIS-1) expression, which are key proteins in the process of mitochondrial fission (29). Inflammation-sensitive nuclear factor kappa-B (NF-κB) and STAT3 pathways play an essential role in type 2 diabetes mellitus-induced muscle atrophy (30). Accordingly, IL-6, JAK, and STAT3 are all associated with skeletal muscle atrophy, but the detailed regulation of denervation-induced skeletal muscle atrophy by IL-6/JAK/STAT3 is still unclear.
In this study, we used microarray to show that IL-6/JAK/STAT3, an inflammatory signaling pathway, was strongly activated during denervated skeletal muscle atrophy. The in vitro and in vivo evidence demonstrated that the high expression of IL-6 intensified skeletal muscle atrophy through the activation of JAK/STAT3, while the inhibition of IL-6/JAK/STAT3 pathway attenuated skeletal muscle atrophy through depressing the protein degradation pathway. The results suggest that IL-6/JAK/STAT3 pathway may be an important mediator of denervated skeletal muscle atrophy, and thus anti-inflammation could serve as targeted therapy strategy for skeletal muscle atrophy.
We present the following article in accordance with the ARRIVE reporting checklist (available at http://dx.doi.org/10.21037/atm-20-7269).
Methods
Animals and treatments
All experiments involving animals were performed under a project license (No. S20200312-003) granted by the ethics board of Nantong University, in compliance with the national guidelines for the care and use of animals. Healthy adult male Institute of Cancer Research (ICR) mice (body weight, ~20 g) and adult male Sprague-Dawley (SD) rats (body weight, ~200 g) were provided by the Experimental Animal Center of Nantong University, Nantong, China. Animals were maintained in 22 °C temperature environment, with a 12-h light-dark cycle and free access to standard rodent chow and water.
To create peripheral nerve injury, the animals were anesthetized with an intraperitoneal injection of mixed narcotics (100 mg/kg ketamine plus 10 mg/kg xylazine), and then the right sciatic nerve was transected to produce a 10-mm long nerve defect. The mice were randomized into groups to receive different treatments for 14 days. The animals were grouped as follows (n=6/group): group Den + Rux, mice administered with vehicle plus ruxolitinib (75 mg/kg/d, a JAK1/2 inhibitor); group Den + C188-9, mice administered with vehicle plus C188-9 (a STAT3 inhibitor, 12.5 mg/kg/d); and group Den, mice administered with vehicle alone. In a separate set of experiments, animals were grouped as follows (n=4/group): group Den + STAT3i, tibialis anterior muscle of mice injected with STAT3 short hairpin RNA (shRNA) lentivirus; and group Den, tibialis anterior muscle of mice injected with viral vectors. A control (Ctrl) group consisting of the mice that were subjected to sham operation before treatment with viral vectors for 14 days was also included.
After different treatments, all mice were killed by cervical decapitation, and the tibialis anterior muscle was harvested to be rapidly frozen in liquid nitrogen and stored at –80 °C for subsequent experiments.
Local infusion
An osmotic pump was implanted for delivering recombinant murine IL-6 (rIL-6) to maintain the sustained high level of IL-6 in animal skeletal muscles as described in a previous publication (31). In brief, we subcutaneously implanted (s.c.) osmotic minipumps (Alzet Model 2002; Alza Corp., Mountain View, CA, USA) in normal mice in order to infuse rIL-6 (Beyotime, Nantong, China) to their tibialis anterior muscle through a catheter (i.d. 0.006 in, Alza) at a flow rate of 4.5 pg/h for each muscle. For catheter implantation, the incisions were made in the skin overlying the tibialis anterior muscle, and on the back, and following this, two further small cuts were made into the fascia of the muscle; a catheter was then tunneled under the fascia and secured. The proximal end of the catheter was mated with the pump, which was placed under the skin via the back incision, and then both incisions were closed. The mice infused with vehicle (saline) plus tocilizumab (TCZ, an anti-IL-6 receptor antibody, 5 mg/kg,) constituted the Den + TCZ group. The vehicle (saline)-infused mice served as the control (n=6/group). After 14 days, the mouse tibialis anterior muscle was harvested under anesthesia, weighed, snap-frozen in liquid nitrogen, and stored at –80 °C.
Microarray analysis and ingenuity pathway analysis (IPA)
For microarray analysis, SD rats were randomly divided into 11 experimental groups and 1 control group (n=3). The origin and management of animals were the same as those in a previously described study (2). For the experimental groups, the sciatic nerve of rats was transected to leave a 10-mm long defect, and the rats were then euthanized under anesthesia at 0.25, 0.5, 3, 6, 12, 24 h, and at 3, 7, 14, 21, 28 d post-surgery. For control group, the rats were subjected to sham surgery and then sacrificed in the same way. Following rat sacrifice, the tibialis anterior muscle of rats was rapidly dissected and homogenized to extract RNA using a RNasey Mini Kit (Qiagen, Valencia, CA, USA) according to the kit guidelines. For microarray analysis of RNA samples, standard protocols were followed (2). After total RNA was transcribed into complement RNA (cRNA), the cRNA labeled with Cy3 was assembled into the Agilent SurePrint G3 Rat GE 8×60 k microarray (Agilent, Santa Clara, CA, USA). The GeneSpring software (version 13.1) was used for normalization and transformation of raw data. The messenger RNA (mRNA) expression in the experimental groups (corresponding to different time points post-surgery) was compared to that in the control group (0 h post-surgery). The differentially expressed genes were considered to be those having an mRNA expression with a fold difference of >2 and a P value of <0.05. The differentially expressed inflammation-related genes were subjected to gene interaction analysis using IPA software (Ingenuity, Redwood, CA, USA).
Cell culture and treatments
To induce the differentiation of C2C12 myoblast cells into myotubes, the cells were cultured in the presence of 2% horse serum [American Type Culture Collection (ATCC), Manassas, VA, USA] for 7 days with a medium change completed every 2 days (6). Then, the resulting C2C12 myotubes were treated for 12 h in the presence or absence of IL-6, or treated for 12 h with IL-6 plus JAK1/2 inhibitor, or with IL-6 plus STAT3 inhibitor, before morphometric examination and biochemical assessments.
Quantitative real-time polymerase chain reaction (qRT-PCR)
A RNeasy kit (Qiagen, Valencia, CA, USA) was used to extract total RNA from rat tibialis anterior muscles. The cDNA was synthesized and qRT-PCR was conducted using iTaq Fast SYBR Green Supermix (BioRad, Hercules, CA, USA) as per the manufacturer’s instructions under the following conditions: 42 °C for 20 min, 40 cycles at 95 °C for 5 min, 94 °C for 20 s, 72 °C for 42 s (4). The expression level was calculated using the 2–ΔΔCt method as previously reported (32). The primers used were the following: rat IL-6R (NM_017020) 5'-GGCAACCTTAGTGCTCAT-3', 5'-CTGTCTGCTCCAGCTTGTTA-3'; rat JAK2 (NM_031514) 5'-GAGCTACTGAAGAACAACGG-3', 5'-TGAAAGAGGGACGTTGGTTGA-3'; rat STAT3 (NM_012747) 5'-GTCTGAATTAAGGGCAGTGAG-3', 5'-CAGGGAAGGGAGAGCAATGA-3'; rat ACTB (NM_031144.2) 5'-CCACCATGTACCCAGGCATT-3', 5'-CGGACTCATCGTACTCCTGC-3'.
Western blot analysis
Western blot analysis was performed to determine the protein expression as previously described (33). The total proteins were extracted with radioimmunoprecipitation assay (RIPA) lysis buffer containing protease and phosphatase inhibitor cocktail (Beyotime), and quantified using BCA Kit (Beyotime), followed by separation on SDS-PAGE and transferring to polyvinylidene difluoride membranes (Millipore, Billerica, MA, USA), which were then reacted with different primary antibodies at 4 °C overnight. The primary antibodies included mouse anti-myosine heavy chain (MHC) antibody (R&D Systems, Minneapolis, MN, USA), rabbit anti-MuRF1, rabbit anti-MAFbx (Fbx32), rabbit anti-LC3B, rabbit anti-PINK1, rabbit anti-BNIP3, rabbit anti-ATG7, rabbit anti-Beclin 1, rabbit anti-beta tubulin (Abcam, Cambridge, UK), mouse anti-pJak1 (Tyr1034/1035)/Jak2 (Tyr1007/1008), rabbit anti-pSTAT3 (Tyr705), and rabbit anti-Stat3 (Cell Signaling Technology, Danvers, MA, USA). Then, the membranes were incubated with horse radish peroxidase (HRP)-conjugated secondary immunoglobin G (IgG) antibodies for 2 h. The target proteins were detected using enhanced chemiluminescence (Thermo Fisher Scientific, USA), and the band intensity was quantified and normalized against loading control.
Determination of muscle fiber cross-sectional area (CSA)
The muscle fiber CSA was measured using laminin staining as previously described (4,6). Briefly, all mouse tibialis anterior muscles were fixed, flash-frozen, and cryosectioned into 10-µm thick sections. The middle third of each muscle was sliced. The sections were incubated in the presence of anti-laminin antibody (Abcam) at 4 °C for 12 h, and then incubated with fluorescent secondary antibody (Alexa Fluor, Invitrogen, USA) at room temperature for 1 h. Five slices were selected from each muscle sample. The slides were observed and imaged under fluorescence microscopy (ZEISS, Oberkochen, Germany) and the CSA was determined using ImageJ software [National Institutes of Health (NIH), Bethesda, MD, USA] through blinded analysis of five randomly captured images under each detection condition.
Determination of myotube diameter
The myotube diameter was determined by MHC staining as previously described (6). Briefly, C2C12 myotubes were fixed, permeabilized, blocked, and incubated with mouse anti-MHC antibody (R&D Systems) at 4 °C overnight. Subsequently, C2C12 myotubes were rinsed and incubated with dye-conjugated goat anti-mouse antibody (Alexa Fluor, Invitrogen) at room temperature for 1 h, and were visualized under fluorescence microscopy (ZEISS). The diameter of the myotubes was measured with ImageJ software (NIH), and calculated as the mean value from three measurements (34).
Enzyme-linked immunosorbent assay (ELISA)
ELISA was conducted for measuring the IL-6 content as previously described (4). In brief, ELISA plates (Beyotime) were incubated in 100 µL lysates of the mouse tibialis anterior muscles or in 100 µL serum at 37 °C for 2 h. The plates were then washed and incubated in the presence of biotinylated polyclonal anti-IL-6 antibody at 37 °C for 1 h. The plates were further washed and incubated in the presence of HRP-conjugated streptavidin at 37 °C in dark for 20 min. Finally, the absorbance was read at 450 nm using a spectrophotometer, and the enzyme activity was calculated to determine the IL-6 content.
Transmission electron microscopy (TEM) analysis
TEM was used to observe the mitochondrial changes in muscle samples as previously described (19). The samples were fixed with a solution of 3% glutaraldehyde in phosphate-buffered saline (PBS). Following orientation and reduction to an optimal size of 1–2 mm3, samples were post-fixed in 1% osmium tetroxide (OsO4) in cacodylate buffer, dehydrated in graded ethanol, and embedded in ARALDITE® (Huntsman Corp., TX, USA). The sections were stained with uranyl acetate and lead citrate, and visualized under TEM (Hitachi, Tokyo, Japan).
Statistical analysis
The data are expressed as means ± SD and were analyzed by one-way ANOVA plus Tukey’s post hoc pairwise comparisons through GraphPad Prism software (version 8.0.1, San Diego, CA, USA). A P value <0.05 was considered as statistically significant.
Results
Activation of IL-6/JAK/STAT3 signaling pathway in denervated skeletal muscle atrophy
Inflammation is known to be related to skeletal muscle atrophy in critically ill patients (7,23,35). Our previous report has also indicated that inflammation plays a role in skeletal muscle atrophy induced by denervation (2). In this study, the heatmap from microarray data showed a differential expression profile of inflammation-related genes in denervated rat tibialis anterior muscles. The expression of inflammation-related genes can be divided into two main patterns. One pattern was featured by time-dependently increased expression of genes, including JAK2, JAK3, STAT3, NF-κB2, and IL17RA, especially at 24 h post denervation. Another pattern displayed an opposite trend, specifically, a time-dependent decrease in the expression of the CAV1, CaSR, ALOX15, and MAPKPK genes, especially at 24 h post denervation (Figure 1A). These data suggested that inflammation was markedly induced. To further analyze the interaction between these inflammation-related genes, IPA revealed that IL-6R, JAK2, and STAT3 were all activated and there was a connection between them (Figure 1B). The mRNA levels of IL-6R, JAK2, or STAT3 during denervation-induced muscle atrophy were also determined by qRT-PCR, and the results were consistent with microarray data (Figure 1C,D,E), which provided further evidence for IL-6/JAK/STAT3 signaling pathway being activated during denervation-induced muscle atrophy.

As a well-known inflammatory signaling pathway, IL-6/JAK/STAT3 exerts a crucial function during cachexia-induced skeletal muscle atrophy (27). To determine the activation of this signaling pathway during denervated skeletal muscle atrophy, we determined the IL-6 content in serum or tibialis anterior muscles of mice through ELISA, and found that the IL-6 content was time-dependently increased at different times after denervation (Figure 2A). We further determined the expression of phosphorylated JAK1/2 (pJAK1/2) and pSTAT3 through western blot analysis, and found that the expression of pJAK1/2 and pSTAT3 was time-dependently increased in mouse tibialis anterior muscles after denervation (Figure 2B,C,D). Taken together, these results suggested that IL-6/JAK/STAT3 signaling pathway might be substantively activated during denervation-induced muscle atrophy.
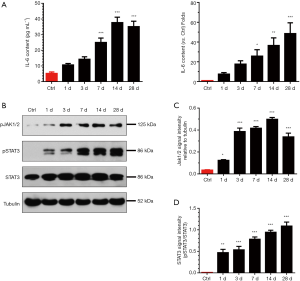
Importance of IL-6 in denervated skeletal muscle atrophy
To explore the role of IL-6 in denervated skeletal muscle atrophy, osmotic pumps were implanted in normal mice to deliver rIL-6, thus maintaining high levels of IL-6 in tibialis anterior muscles. This method does not significantly increase the level of circulating IL-6. We noted that delivering rIL-6 into mice tibialis anterior muscle led to tibialis anterior muscle atrophy, as evidenced by the smaller muscle wet weight and the smaller mean CSA of the tibialis anterior muscles in mice administered with rIL-6 compared with those in mice administered with vehicle (saline) (Figure 3A,B,C,D). These data suggested that the impact of IL-6 on skeletal muscles was similar to that of denervation on skeletal muscles. Meanwhile, the IL-6 content was high in the denervated tibialis anterior muscles and in the rIL-6-treated tibialis anterior muscles (Figure 3E). To determine whether IL-6 blocking could alleviate denervation-induced skeletal muscle atrophy, we noted that tocilizumab, an anti-IL-6 receptor antibody, could reduce the tibialis anterior muscle atrophy induced by denervation (Figure 3A,B,C,D), indicating the critical importance of IL-6 in denervation-induced skeletal muscle atrophy.
Interestingly, the expression of pJAK1/2 or pSTAT3 in denervated and rIL-6-administered tibialis anterior muscles was significantly increased compared to that in vehicle (saline)-administrated tibialis anterior muscles. The expression of the ubiquitin proteasome system (MAFbx and MuRF1) and autophagy lysosomal system (PINK1, BNIP3) also displayed a similar change (Figure 3F,G). Inversely, the MHC expression in denervated and rIL-6-administered tibialis anterior muscles was decreased compared to that in vehicle (saline)-administered tibialis anterior muscles. Importantly, all of the above expression changes could be reversed by application of tocilizumab in denervated tibialis anterior muscles (Figure 3F,G). These observations suggested that IL-6 was important in denervated skeletal muscle atrophy and critical for the activation/phosphorylation of JAK/STAT3.
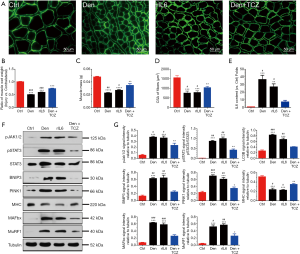
Effects of JAK/STAT3 inhibition on IL-6 induced C2C12 myotube atrophy
In determining the in vitro effects of JAK/STAT3 inhibition on IL-6 induced skeletal muscle atrophy, we observed that exposure of C2C12 myotubes to rIL-6 (100 ng/mL) for 24 h could elicit a significant decrease in the diameter of C2C12 myotubes compared to exposure to vehicle (saline) alone for 24 h (Figure 4A). The wasting of C2C12 myotubes elicited by rIL-6 was accompanied by the increased expression of pJAK1/2, pSTAT3, MAFbx, MuRF1, and the decreased expression of MHC (Figure 4B,C). When C2C12 myotubes were treated with 400 nM ruxolitinib (a JAK1/2 inhibitor) or 10 µM C188-9 (a STAT3 inhibitor) in the presence of 100 ng/mL rIL-6 for 24 h, the signaling of JAK/STAT3 was blocked, the expression of MAFbx and MuRF1 decreased, the expression of MHC increased, and the atrophy of C2C12 myotubes was inhibited (Figure 4A,B,C). These results suggested that pharmacological depression of JAK/STAT3 could block IL-6-induced C2C12 myotube atrophy in vitro.
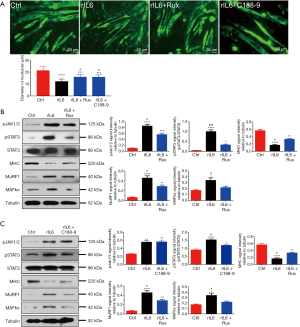
Effects of JAK/STAT3 inhibition on denervated skeletal muscle atrophy
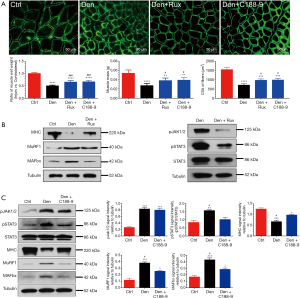
We also examined the effects of JAK/STAT3 inhibition on denervated skeletal muscle atrophy in vivo. In the mice with sciatic nerve injury, the ratio of wet weight and CSA of denervated tibialis anterior muscles were significantly decreased compared to those of control tibialis anterior muscles from sham operation mice. After administration with ruxolitinib or C188-9 for 14 days, results showed that JAK/STAT3 inhibition treatment significantly reversed the above-mentioned decrease in mice with sciatic nerve injury when compared to the control group (Figure 5A); meanwhile, JAK/STAT3 inhibition with Rux and C188-9 did not significantly change the mass of the contralateral muscles. Concomitantly, the expression of MAFbx and MuRF1 was inhibited while MHC content was elevated in denervated tibialis anterior muscles in the JAK/STAT3 inhibition treatment group compared to that in the control group. Moreover, ruxolitinib and C188-9 dramatically inhibited JAK/STAT3 signaling, as shown by the decreased expression of pJAK1/2 or pSTAT3 in denervated muscles (Figure 5B,C). In addition to using STAT3 inhibitors, STAT3 shRNA lentivirus (STAT3i) were used to knockdown STAT3 in denervated muscles. Similar to STAT3 inhibitors, STAT3i could also significantly improve the decrease of the wet weight and CSA of the denervated muscles, decrease the expression of pSTAT3, MAFbx, and MuRF1, and increase the content of MHC in muscles after denervation (Figure 6A,B,C), which suggested that STAT3 activation was necessary for denervated skeletal muscle atrophy in vivo. These data demonstrated that JAK/STAT3 inhibition could alleviate denervated skeletal muscle atrophy in vivo.
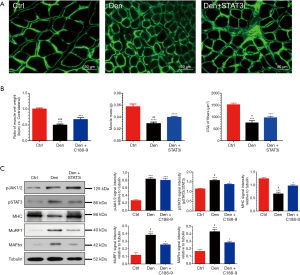
Effects of STAT3 knockdown on mitophagy in denervated skeletal muscles
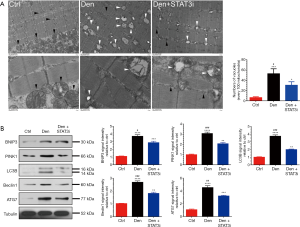
Mitophagy is associated with the degree of muscle atrophy (36,37). In exploring the effects of STAT3 knockdown on mitophagy in denervated skeletal muscle atrophy, we noted that autophages and autophagic vesicles could be easily observed in denervated muscles under TEM (Figure 7A). Meanwhile, the expression of the mitophagy markers, including BNIP3, PINK1, LC3B, Beclin 1, and ATG7, were increased in denervated muscles (Figure 7B). On the other hand, STAT3 knockdown significantly inhibited mitophagy, as indicated by the decrease in the number of autophages and autophagic vesicles, and the significantly reduced expression of PINK1, BNIP3, LC3B, ATG7, and Beclin 1 (Figure 7A,B). These results suggested that STAT3 knockdown could block mitophagy in denervated skeletal muscles.
Discussion
Dysregulation in the muscle microenvironment is liable to induce skeletal muscle atrophy (4,38), and inflammation, as an important factor in changing the muscle microenvironment (39), should thus also play a critical role in skeletal muscle atrophy (24). An abundance of research has attested to the associations of skeletal muscle atrophy with inflammation. For instance, IL-1β was found to increase atrogene expression to induce skeletal muscle atrophy in sepsis (7), an elevated level of tumor necrosis factor alpha (TNF-α) was demonstrated to inhibit the regenerative potential of satellite cells in dystrophic muscle by silencing Notch 1 (25), and the IL-6/JAK/STAT3 pathway has been shown to be aberrantly hyperactivated in many types of cancer (40,41), leading to skeletal muscle atrophy in cancer cachexia (27,28). Madaro et al. found that IL-6 was significantly induced in fibro-adipogenic progenitors of muscles after denervation and figured prominently in muscle atrophy (42).
Despite the above findings, whether or not the inflammatory IL-6/JAK/STAT3 pathway is markedly activated in denervated skeletal muscle atrophy has not been thoroughly examined. Our previous study indicated the inflammatory response to be widely activated in tibialis anterior muscles of rats undergoing sciatic nerve transection (2). This finding is consistent with the previously reported results regarding the involvement of inflammation in sepsis or cancer-induced skeletal muscle atrophy (7,23,35). In this study, we attempted to further analyze the distinct expression profiles of inflammation-related genes during denervated muscle atrophy. The expression profiles could be divided into two main patterns: a primarily time-dependent increase of expression for proinflammatory factors, and a primarily time-dependent decrease of expression for anti-inflammatory factors. These two opposing trends suggested that inflammation was linked to denervated skeletal muscle atrophy. IPA of these differentially expressed genes indicated that IL-6R, JAK2, and STAT3 might be activated and interconnected. It is known that the binding of IL-6 to its receptor, IL-6R, can activate three signaling pathways: SHP2/PI3K/AKT2, SHP2/MEK/ERK, and JAK/STAT3 (43-45). The microarray data in this study showed that IL-6R, JAK2, and STAT3 were all upregulated during denervated skeletal muscle atrophy, prompting speculation that the binding of IL-6 to IL-6R could activate JAK/STAT3 signaling pathway to act on denervation-induced skeletal muscle atrophy. We found that the expression of pJAK1/2, pSTAT3, and particularly IL-6 was increased in denervated muscles, implying that IL-6/JAK/STAT3 signaling pathway was significantly activated during denervation-induced muscle atrophy. This finding is consistent with other studies (27,28,46,47). For instance IL-6 was demonstrated to induce cachexia, resulting in muscle wasting, while IL-6 inhibition could retard cachexia-induced skeletal muscle wasting (28). Another study showed that STAT3 was a primary mediator of skeletal muscle wasting in cancer cachexia, while STAT3 inhibition retarded cancer-induced skeletal muscle wasting (27,48).
We found that IL-6 administration led to skeletal muscle atrophy, indicating that the influence of IL-6 on skeletal muscles was similar to the that of denervation on skeletal muscles. In contrast, inhibition of IL-6 signaling prevented denervation-induced muscle atrophy, suggesting that IL-6 was both sufficient and necessary for denervated skeletal muscle atrophy. The impact of IL-6 on denervated muscle atrophy was consistent with the impact of IL-6 on cachexia as described previously (28). Evidence from clinical trials, however, indicated that inhibition of IL-6 signaling could not completely retard cancer-induced skeletal muscle atrophy, which might be due to the many other factors involved in cancer-induced skeletal muscle atrophy (49).
In this study, we showed that pJAK1/2 and pSTAT3 had a higher expression in the denervated and IL-6 injected skeletal muscles. Similar to pJAK1/2 and pSTAT3, the ubiquitin proteasome system and autophagy lysosomal system were also found to exhibit significantly elevated expression. Moreover, the expression changes of all these cytokines were reversed by IL-6 signaling blocking in denervated skeletal muscles, which provided further evidence that IL-6 may be critical to JAK/STAT3 activation.
IL-6 activates JAKs, which subsequently activate STAT3 to promote the transcription of several genes in cancer cachexia (27,40). In this study, the in vitro and in vivo experiments investigated the role of the JAK downstream of IL-6 in denervated skeletal muscle atrophy. The elevated expression of pJAK1/2 and pSTAT3 was observed in IL-6-induced C2C12 myotube atrophy, while ruxolitinib inhibition of JAK1/2 prevented the activation of JAK/STAT3, thus alleviating myotube atrophy. JAK1/2 inhibition could block IL-6-induced myotube atrophy in vitro. Similarly, JAK1/2 inhibition could alleviate denervation-induced skeletal muscle atrophy. These findings are consistent with previous findings in which JAK inhibition prevented STAT3 phosphorylation during controlled mechanical ventilation, thus alleviating diaphragm atrophy and blunting the activation of proteolytic pathways (50).
JAK/STAT pathway triggers muscle protein loss in cancer cachexia (51), while STAT3 accelerates the activation of the ubiquitin proteasome system and contributes to skeletal muscle atrophy in type 2 diabetes mellitus (30). Moreover, STAT3 can impair the protein synthesis in skeletal muscles of mice suffering from chronic kidney disease or cancer cachexia, while STAT3 inhibition can attenuate chronic kidney disease–induced skeletal muscle atrophy (30,52). Here, we found that STAT3 blocking significantly decreased the expression of pSTAT3 and markedly alleviated C2C12 myotube atrophy, but had no effects on JAK activation. A similar result was also observed in an in vivo model, whose creation was prompted by the discovery that blocking STAT3 relieved denervation-induced skeletal muscle atrophy, and was accompanied by the deactivation of STAT3, the decreased expression of MAFbx and MuRF1, and the increased expression of MHC. These results are also in line with those of another previous study which reported that blocking STAT3 could suppress the expression of caspase-3, MAFbx, and myostatin, thus alleviating skeletal muscle wasting in cancer cachexia (51).
Inhibition of STAT3 signaling has been known to decrease TNF-α-induced autophagy (53). Similarly, this study showed that STAT3 knockdown significantly inhibited mitophagy after denervation, indicating that the activation of STAT3 was necessary for denervation-induced skeletal muscle atrophy; conversely, the local inhibition of STAT3 in skeletal muscles could reduce mitophagy in denervated skeletal muscles.
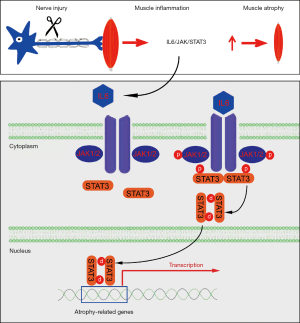
To summarize, this study revealed a previously unappreciated involvement of the IL-6/JAK/STAT3 signaling pathway in denervated skeletal muscle atrophy. Inflammatory cytokine IL-6 was triggered in denervated skeletal muscles, leading to the activation of JAK and the ensuing activation of STAT3. Phosphorylation of STAT3 resulted in nuclear localization and atrogene expression (Figure 8). The inhibition of IL-6 through tocilizumab (an anti-IL-6 receptor antibody), inhibition of JAK through ruxolitinib (a JAK1/2 inhibitor), or inhibition of STAT3 through C188-9 (a STAT3 inhibitor) or STAT3 shRNA lentivirus, could alleviate skeletal muscle atrophy through suppression of proteolysis. Our findings support IL-6/JAK/STAT3 as a potential therapeutic target for skeletal muscle atrophy.
Acknowledgments
We thank Professor Jie Liu for his help in revising the manuscript.
Funding: This work was supported by the National Natural Science Foundation of China (No. 82072160, 81871554, 81671230, and 81901933), the Priority Academic Program Development of Jiangsu Higher Education Institutions (PAPD), the Major Natural Science Research Projects in Universities of Jiangsu Province (No. 20KJA310012), and the Natural Science Foundation of Jiangsu Province (No. BK20201209).
Footnote
Reporting Checklist: The authors have completed the ARRIVE reporting checklist. Available at http://dx.doi.org/10.21037/atm-20-7269
Data Sharing Statement: Available at http://dx.doi.org/10.21037/atm-20-7269
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/atm-20-7269). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. Experiments were performed under a project license (No. S20200312-003) granted by the ethics board of Nantong University, in compliance with the national guidelines for the care and use of animals.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Tong T, Kim M, Park T. α-Cedrene, a newly identified ligand of MOR23, increases skeletal muscle mass and strength. Mol Nutr Food Res 2018. [Epub ahead of print]. [Crossref] [PubMed]
- Shen Y, Zhang R, Xu L, et al. Microarray analysis of gene expression provides new insights into denervation-induced skeletal muscle atrophy. Front Physiol 2019;10:1298. [Crossref] [PubMed]
- Dutt V, Gupta S, Dabur R, et al. Skeletal muscle atrophy: Potential therapeutic agents and their mechanisms of action. Pharmacol Res 2015;99:86-100. [Crossref] [PubMed]
- Wu C, Tang L, Ni X, et al. Salidroside attenuates denervation-induced skeletal muscle atrophy through negative regulation of pro-inflammatory cytokine. Front Physiol 2019;10:665. [Crossref] [PubMed]
- Li J, Chan MC, Yu Y, et al. miR-29b contributes to multiple types of muscle atrophy. Nat Commun 2017;8:15201. [Crossref] [PubMed]
- Qiu J, Zhu J, Zhang R, et al. miR-125b-5p targeting TRAF6 relieves skeletal muscle atrophy induced by fasting or denervation. Ann Transl Med 2019;7:456. [Crossref] [PubMed]
- Huang N, Kny M, Riediger F, et al. Deletion of Nlrp3 protects from inflammation-induced skeletal muscle atrophy. Intensive Care Med Exp 2017;5:3. [Crossref] [PubMed]
- Qiu J, Fang Q, Xu T, et al. Mechanistic role of reactive oxygen species and therapeutic potential of antioxidants in denervation- or fasting-induced skeletal muscle atrophy. Front Physiol 2018;9:215. [Crossref] [PubMed]
- Sun H, Gong Y, Qiu J, et al. TRAF6 inhibition rescues dexamethasone-induced muscle atrophy. Int J Mol Sci 2014;15:11126-41. [Crossref] [PubMed]
- Huang Z, Zhu J, Ma W, et al. Strategies and potential therapeutic agents to counter skeletal muscle atrophy. Biotarget 2018;2:8. [Crossref]
- Kim H, Jang M, Park R, et al. Conessine treatment reduces dexamethasone-induced muscle atrophy by regulating murf1 and atrogin-1 expression. J Microbiol Biotechnol 2018;28:520-6. [Crossref] [PubMed]
- Morales MG, Abrigo J, Acuna MJ, et al. Angiotensin-(1-7) attenuates disuse skeletal muscle atrophy in mice via its receptor, Mas. Dis Model Mech 2016;9:441-9. [Crossref] [PubMed]
- Marzuca-Nassr GN, Murata GM, Martins AR, et al. Balanced diet-fed fat-1 transgenic mice exhibit lower hindlimb suspension-induced soleus muscle atrophy. Nutrients 2017;9:1100. [Crossref] [PubMed]
- Cerquone Perpetuini A, Re Cecconi AD, Chiappa M, et al. Group I Paks support muscle regeneration and counteract cancer-associated muscle atrophy. J Cachexia Sarcopenia Muscle 2018;9:727-46. [Crossref] [PubMed]
- Chacon-Cabrera A, Mateu-Jimenez M, Langohr K, et al. Role of PARP activity in lung cancer-induced cachexia: effects on muscle oxidative stress, proteolysis, anabolic markers, and phenotype. J Cell Physiol 2017;232:3744-61. [Crossref] [PubMed]
- Kaur N, Gupta P, Saini V, et al. Cinnamaldehyde regulates H2O2-induced skeletal muscle atrophy by ameliorating the proteolytic and antioxidant defense systems. J Cell Physiol 2019;234:6194-208. [Crossref] [PubMed]
- Zhang YY, Gu LJ, Huang J, et al. CKD autophagy activation and skeletal muscle atrophy-a preliminary study of mitophagy and inflammation. Eur J Clin Nutr 2019;73:950-60. [Crossref] [PubMed]
- Yeo D, Kang C, Gomez-Cabrera MC, et al. Intensified mitophagy in skeletal muscle with aging is downregulated by PGC-1alpha overexpression in vivo. Free Radic Biol Med 2019;130:361-8. [Crossref] [PubMed]
- Wang D, Sun H, Song G, et al. Resveratrol improves muscle atrophy by modulating mitochondrial quality control in STZ-induced diabetic mice. Mol Nutr Food Res 2018;62:e1700941. [Crossref] [PubMed]
- Kang C, Ji LL. PGC-1alpha overexpression via local transfection attenuates mitophagy pathway in muscle disuse atrophy. Free Radic Biol Med 2016;93:32-40. [Crossref] [PubMed]
- Paul PK, Gupta SK, Bhatnagar S, et al. Targeted ablation of TRAF6 inhibits skeletal muscle wasting in mice. J Cell Biol 2010;191:1395-411. [Crossref] [PubMed]
- Stana F, Vujovic M, Mayaki D, et al. Differential regulation of the autophagy and proteasome pathways in skeletal muscles in sepsis. Crit Care Med 2017;45:e971-9. [Crossref] [PubMed]
- Zhu X, Kny M, Schmidt F, et al. Secreted frizzled-related protein 2 and inflammation-induced skeletal muscle atrophy. Crit Care Med 2017;45:e169-83. [Crossref] [PubMed]
- Ramírez C, Russo TL, Delfino G, et al. Effect of tibiotarsal joint inflammation on gene expression and cross-sectional area in rat soleus muscle. Braz J Phys Ther 2013;17:244-54. [Crossref] [PubMed]
- Londhe P, Guttridge DC. Inflammation induced loss of skeletal muscle. Bone 2015;80:131-42. [Crossref] [PubMed]
- Ma W, Xu T, Wang Y, et al. The role of inflammatory factors in skeletal muscle injury. Biotarget 2018;2:7. [Crossref]
- Bonetto A, Aydogdu T, Jin X, et al. JAK/STAT3 pathway inhibition blocks skeletal muscle wasting downstream of IL-6 and in experimental cancer cachexia. Am J Physiol Endocrinol Metab 2012;303:E410-21. [Crossref] [PubMed]
- Zimmers TA, Fishel ML, Bonetto A. STAT3 in the systemic inflammation of cancer cachexia. Semin Cell Dev Biol 2016;54:28-41. [Crossref] [PubMed]
- Fix DK, VanderVeen BN, Counts BR, et al. Regulation of skeletal muscle DRP-1 and FIS-1 protein expression by IL-6 signaling. Oxid Med Cell Longev 2019;2019:8908457. [Crossref] [PubMed]
- Perry BD, Caldow MK, Brennan-Speranza TC, et al. Muscle atrophy in patients with type 2 diabetes mellitus: roles of inflammatory pathways, physical activity and exercise. Exerc Immunol Rev 2016;22:94-109. [PubMed]
- Haddad F, Zaldivar F, Cooper DM, et al. IL-6-induced skeletal muscle atrophy. J Appl Physiol (1985) 2005;98:911-7. [Crossref] [PubMed]
- Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods 2001;25:402-8. [Crossref] [PubMed]
- Huang Z, Fang Q, Ma W, et al. Skeletal muscle atrophy was alleviated by salidroside through suppressing oxidative stress and inflammation during denervation. Front Pharmacol 2019;10:997. [Crossref] [PubMed]
- Abrigo J, Rivera JC, Simon F, et al. Transforming growth factor type beta (TGF-beta) requires reactive oxygen species to induce skeletal muscle atrophy. Cell Signal 2016;28:366-76. [Crossref] [PubMed]
- Cella PS, Marinello PC, Borges FH, et al. Creatine supplementation in Walker-256 tumor-bearing rats prevents skeletal muscle atrophy by attenuating systemic inflammation and protein degradation signaling. Eur J Nutr 2020;59:661-9. [Crossref] [PubMed]
- Theilen NT, Kunkel GH, Tyagi SC. The role of exercise and TFAM in preventing skeletal muscle atrophy. J Cell Physiol 2017;232:2348-58. [Crossref] [PubMed]
- Wang J, Wang F, Zhang P, et al. PGC-1alpha over-expression suppresses the skeletal muscle atrophy and myofiber-type composition during hindlimb unloading. Biosci Biotechnol Biochem 2017;81:500-13. [Crossref] [PubMed]
- Talbert EE, Guttridge DC. Impaired regeneration: a role for the muscle microenvironment in cancer cachexia. Semin Cell Dev Biol 2016;54:82-91. [Crossref] [PubMed]
- Sfanos KS, Yegnasubramanian S, Nelson WG, et al. The inflammatory microenvironment and microbiome in prostate cancer development. Nat Rev Urol 2018;15:11-24. [Crossref] [PubMed]
- Johnson DE, O'Keefe RA, Grandis JR. Targeting the IL-6/JAK/STAT3 signalling axis in cancer. Nat Rev Clin Oncol 2018;15:234-48. [Crossref] [PubMed]
- Yeung YT, Aziz F, Guerrero-Castilla A, et al. Signaling pathways in inflammation and anti-inflammatory therapies. Curr Pharm Des 2018;24:1449-84. [Crossref] [PubMed]
- Madaro L, Passafaro M, Sala D, et al. Denervation-activated STAT3-IL-6 signalling in fibro-adipogenic progenitors promotes myofibres atrophy and fibrosis. Nat Cell Biol 2018;20:917-27. [Crossref] [PubMed]
- Baran P, Hansen S, Waetzig GH, et al. The balance of interleukin (IL)-6, IL-6.soluble IL-6 receptor (sIL-6R), and IL-6.sIL-6R.sgp130 complexes allows simultaneous classic and trans-signaling. J Biol Chem 2018;293:6762-75. [Crossref] [PubMed]
- Roy B, Curtis ME, Fears LS, et al. Molecular mechanisms of obesity-induced osteoporosis and muscle atrophy. Front Physiol 2016;7:439. [Crossref] [PubMed]
- Takaesu G. Two types of TRAF6-dependent TAK1 activation in the IL-1 signaling pathway. Biotarget 2018;2:2. [Crossref]
- Ma JF, Sanchez BJ, Hall DT, et al. STAT3 promotes IFNgamma/TNFalpha-induced muscle wasting in an NF-kappaB-dependent and IL-6-independent manner. EMBO Mol Med 2017;9:622-37. [Crossref] [PubMed]
- Narsale AA, Carson JA. Role of interleukin-6 in cachexia: therapeutic implications. Curr Opin Support Palliat Care 2014;8:321-7. [Crossref] [PubMed]
- Guadagnin E, Mazala D, Chen YW. STAT3 in skeletal muscle function and disorders. Int J Mol Sci 2018;19:2265. [Crossref] [PubMed]
- Bayliss TJ, Smith JT, Schuster M, et al. A humanized anti-IL-6 antibody (ALD518) in non-small cell lung cancer. Expert Opin Biol Ther 2011;11:1663-8. [Crossref] [PubMed]
- Smith IJ, Godinez GL, Singh BK, et al. Inhibition of Janus kinase signaling during controlled mechanical ventilation prevents ventilation-induced diaphragm dysfunction. FASEB J 2014;28:2790-803. [Crossref] [PubMed]
- Silva KA, Dong J, Dong Y, et al. Inhibition of Stat3 activation suppresses caspase-3 and the ubiquitin-proteasome system, leading to preservation of muscle mass in cancer cachexia. J Biol Chem 2015;290:11177-87. [Crossref] [PubMed]
- Ranganathan P, Jayakumar C, Ramesh G. Proximal tubule-specific overexpression of netrin-1 suppresses acute kidney injury-induced interstitial fibrosis and glomerulosclerosis through suppression of IL-6/STAT3 signaling. Am J Physiol Renal Physiol 2013;304:F1054-65. [Crossref] [PubMed]
- Wang L, Wang Y, Du M, et al. Inhibition of Stat3 signaling pathway decreases TNF-alpha-induced autophagy in cementoblasts. Cell Tissue Res 2018;374:567-75. [Crossref] [PubMed]
(English Language Editor: J. Gray)


