
A STING-related prognostic score predicts high-risk patients of colorectal cancer and provides insights into immunotherapy
Introduction
Colorectal cancer (CRC) is the second deadliest cancer worldwide (1) that has multiple etiologies (2). Promoted by improved knowledge of the interaction between tumor cells and the immune system, significant roles of adaptive and innate immune factors in the development and progression of human cancers have been recognized (3,4). Immune checkpoint blockades (ICBs) have been shown applicable for several tumors, including CRC. However, ICBs could benefit only a small fraction of patients with high microsatellite instability (MSI-high) markers (5). New biomarkers and treatment strategies for CRC patients are still urgently needed.
STING (stimulator of interferon genes), one of the most intensively studied nucleic acid-sensing pattern recognition receptors (PRRs), is crucial in controlling antiviral responses (6) and detecting tumor formation (7,8). After sensing the tumor-derived DNA in the cytoplasm, the cytoplasmic nucleotide transferase cGAS in dendritic cells (DCs) catalyzes cyclic GmP-AmP (GAmP) formation to bind and activate STING, which successively stimulates the type I IFN response to initiate antitumor responses (9). Emerging studies have shown that STING was involved in tumorigenesis and treatment resistance, and could serve as a promising treatment target for CRC patients (10-15). For example, Xia et al. revealed recurrently suppressed STING signaling in CRC, and the loss of STING signaling impaired the DNA damage response (13). Yang et al. reported disrupted cGAS-STING-IFNB signaling and the prognostic value of cGAS, RNASEH2B, RNASEH2C, and SAMHD1 in CRC (14). Chon et al. also elucidated the independent prognostic value of STING, which could be harnessed as a potential therapeutic target to enhance anticancer immune responses in CRC (15). Moreover, STING has also been associated with tumor progression, metastasis, and radiotherapy resistance in CRC (10-12).
For CRC, targeted therapeutic strategies have long been limited and biomarkers mostly functioned as negative indicators (2). The consensus molecular subtype (CMS) of CRC is currently the best descriptor of the heterogeneity at the gene expression level (16). The CMS1 subtype is characterized by hypermutation, MSI, and robust immune activation with a notably enhanced infiltration of CD8+ T cells, whereas the other three subtypes are primarily immune-inert (16). The activation of the STING signaling pathway is related to better prognosis in CRC (17). By contrast, an impaired STING pathway could lead to the failure to recognize tumor-associated antigens and jeopardize T cell priming. Therefore, repairing or strengthening the STING pathway provides a niche for the antitumor immunotherapy. Early-phase clinical trials employing human STING agonists are currently underway in patients with advanced and/or metastatic solid tumors or lymphomas (NCT02675439, NCT03010176, and NCT03172936) (18).
Large public databases and machine learning algorithms have significantly fueled medical research (19). In this study, we searched PubMed using the term “STING AND colorectal cancer” without other restrictions and did not find any related high-throughput research articles. Therefore, to our knowledge, this is the first study based on a high-throughput dataset to investigate the STING pathway in the setting of CRC. We present the following article in accordance with the TRIPOD reporting checklist (available at http://dx.doi.org/10.21037/atm-20-2430).
Methods
Clinicopathologic characteristics of the TCGA cohort
We analyzed 431 CRC samples with complete information from the TCGA database, of which 48 samples had paired normal tissues. Expression profiles (FPKM) and clinical information were downloaded using the “TCGAbiolinks” R package. We only kept genes with an average expression over 1 FPKM across all samples, and low-abundance sequencing data were removed. Additionally, duplicated genes were processed by the “avereps” function of the “limma” R package to obtain the average expression value. Other information, including the CMS molecular subtype, BRAF status, KRAS status, and MSI status, were obtained from a previous resource (16). All the clinicopathologic characteristics of CRC samples are listed in Table 1.
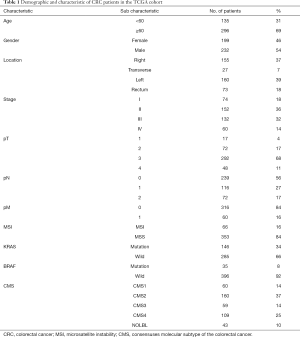
Full table
GEO dataset
The expression profile and the clinical information of the GSE87211 dataset were downloaded using the “getGEO” function of the “GEOquery” R package. We mapped each gene symbol to the corresponding probe, and the duplicated gene symbols were processed using the “avereps” function of the “limma” R package to calculate the mean value.
Construction of the STING-related prognostic score (SPS)
We adopted a previous calculating strategy to construct the STING-related prognostic model and calculated the SPS for each sample (20). First, we evaluated the prognostic value of each of the 96 STING-related genes using univariate Cox regression analysis. Genes with a P-value lower than 0.05 were considered statistically significant. To construct the SPS model, we used the “scale” function in R software to calculate the Z-score of POLR1D, IRF2, and DHX9 for each sample, where the mean gene expression was 0 and the standard deviation (SD) was 1. Using the 3-gene panel, we performed multivariate Cox regression analyses, and the backward stepAIC strategy was adopted using the “MASS” R package. AIC stands for Akaike Information Criteria, and stepAIC is one of the most common methods for feature selection. The optimal model was defined as the model with the lowest stepAIC value. The SPS was then calculated as the gene expression multiplied by the corresponding coefficients of the final model:
SPS = POLR1D×0.1973+IRF2 *0.2785-DHX9×0.3104
We harnessed the “surv_cutpoint” function of the “survminer” R package to obtain a uniform cutoff value to stratify the patients. Samples with SPS ≥0.15 were assigned to the high SPS group, and samples with SPS <0.15 were assigned to the low SPS group. We used the log-rank test and Kaplan-Meier survival analysis to assess the predictive ability of the SPS.
Estimation of immune cell infiltration
Immune cell infiltration was calculated using CIBERSORT to estimate both the relative and absolute abundances of 22 immune cell types based on the gene expression profile (21). For relative abundances, the fractions of all 22 estimated immune cell subtypes are summed up to 1 for each sample. Evaluations of T helper 1, 2, and 17 cells (Th1, Th2, Th17 cells) were obtained from a previously published study (https://ars.els-cdn.com/content/image/1-s2.0-S1074761318301213-mmc2.xlsx) (22).
Gene set enrichment analysis
First, the “limma” R package was applied to identify differentially expressed genes (DEGs) and calculate the log fold change (LFC) for each DEG to obtain a ranked gene list. Next, the gene set enrichment analysis (GSEA) was performed using the “GSEA” function in the “clusterProfiler” R package (23). We used the hallmark gene sets (h.all.v7.0.symbols.gmt) from the GSEA Molecular Signatures Database (MSigDB) to interpret biological functions (24,25). The threshold was set at P=0.05. Significantly enriched gene sets were plotted using the “gseaplot2” function of the “enrichplot” R package.
Construction and evaluation of the nomogram
Nomogram is a user-friendly graphical interface that enables clinicians to estimate survival for each individual (26). In our study, the nomogram was constructed based on SPM to individualize the predicted survival probability for 1-year, 3-year, and 5-year in the TCGA CRC cohort. The calibration plot and concordance index (C-index) were harnessed to assess the performance of the nomogram.
We used the “rms” R package (https://www.rdocumentation.org/packages/rms) to generate the nomogram and calibration plots. Calibration is typically assessed by the predicted probabilities calculated by the nomogram versus the actual probabilities. A nomogram with perfect accuracy would result in a calibration plot in which x- and y-axis are separately observed and predicted probabilities with nearly all points falling along the 45-degree line. The distance between the pairs and the 45-degree line measures the absolute error of the nomogram’s prediction.
We used the “rcorr.cens” function in the “Hmisc” R package (https://cran.r-project.org/web/packages/Hmisc/) to generate the C-index. The discrimination or predictive accuracy of a model is defined as the performance to separate patients with different outcomes, which could be measured via the C-index. The C-index denotes the proportion of pairs, with the responders having a higher predicted probability of response than the nonresponders. The C-index was assessed by comparing nomogram-predicted versus observed Kaplan-Meier estimates of survival probabilities based on a bootstrap approach with 1,000 resamples.
Statistical analyses
Categorical variables were compared using the χ2 test or Fisher’s exact test. Continuous variables were compared using the t-test or Mann-Whitney U test for variables with an abnormal distribution. The paired t-test was used to identify the difference between tumors and paired normal tissues. Survival curves were analyzed using the Kaplan-Meier method and compared using the log-rank test. Principal component analysis (PCA) was performed and plotted using R software. The statistical significance threshold was set at 0.05 if not explicitly mentioned. All statistical analyses were implemented using R software (ver. 3.6.1). All data analyzed in this study are available in https://cdn.amegroups.cn/static/public/atm-20-2430-3.xlsx.
Availability of data and materials
The datasets generated and/or analyzed during the current study are available in the TCGA database, https://portal.gdc.cancer.gov/, and the GEO database, https://www.ncbi.nlm.nih.gov/gds/?term=GSE87211. CMS subtyping calls, BRAF status, KRAS status and MSI status were all obtained from doi: 10.7303/syn2623706. Evaluations of Th1, Th2, Th17 cells were obtained from a previously published research (https://ars.els-cdn.com/content/image/1-s2.0-S1074761318301213-mmc2.xlsx).
Results
Investigation of STING-related genes in CRC
We investigated 431 CRC tumors and 48 paired normal tissues from the TCGA database. The overall expression levels of STING in tumor cells were higher than paired normal tissues (P=0.049; Figure 1A). Since recurrently suppressed STING had been suggested in CRC (13), we considered that the increased levels of STING might be due to the heterogeneity of CRC. Further research in the context of CRC subtypes showed that the expression of STING was exclusively increased in the CMS1 subtype (P=0.036, paired t-test) and was suppressed in the CMS2, CMS3, and CMS4 subtypes (Figure 1B; Table S1).
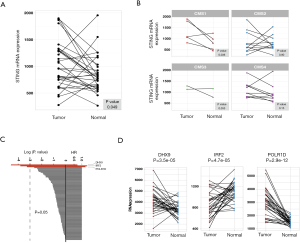
To explore the prognostic value of STING, we selected 96 STING-related genes of five pathways from MSigDB (24,25). These pathways are generally related to innate immune responses to viral and bacterial infections, detection of cytosolic nucleic acids, activation of type I IFN responses, apoptotic signaling through the type II major histocompatibility (MHC II) complex, and initiation of the adaptive immune response (Table S2). We performed univariate Cox regression analysis for each gene, and POLR1D, DHX9 and IRF2 showed significant prognostic value (Figure 1C, Table S3). DHX9 (HR =0.72, 95% CI, 0.56–0.92, P=0.01) showed a positive association, whereas POLR1D (HR =1.23, 95% CI, 1.01–1.49, P=0.038) and IRF2 (HR =1.34, 95% CI, 1.04–1.73, P=0.022) showed negative associations with the prognosis in CRC samples. Moreover, DHX9 and POLR1D were significantly overexpressed, compared to the decreased IRF2 in tumor tissues (Figure 1D).
To assess whether POLR1D, IRF2, and DHX9 were associated with conventional features (age, sex, stage, pT, pN, pM and location), the expression levels of POLR1D, IRF2, and DHX9 in all 431 CRC samples were de-dimensioned to the x- and y-axes in the PCA analysis. As a result, none of these conventional features showed significant associations with POLR1D, IRF2, and DHX9 (Figure S1).
Construction of the SPS
Using the three prognostic indicators, we built a multivariate Cox regression model to predict outcomes of CRC. DHX9 and IRF2 remained significant in the model (DHX9, HR =0.73, P=0.0263 and IRF2, HR =1.32, P=0.0347), and POLR1D reached borderline significance (HR =1.22, P=0.0561; Table S3). We calculated SPS and divided CRC patients into a high SPS group and a low SPS group according to the optimal cutoff value (cutoff value =0.15, Methods). The low SPS group containing 256 samples showed a better outcome, while the high SPS group including 175 samples showed worse prognosis (P=2e-4, log-rank test; Figure 2A), with a 2.67-fold higher risk than the low SPS group (95% CI, 1.53–4.67, P=0.0006).
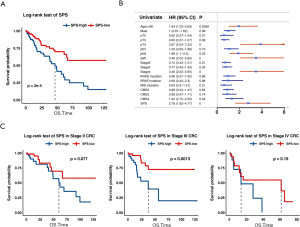
In the univariate Cox regression analyses, only age, pT, pN, pM, stage, and SPS showed significant prognostic value (Figure 2B, more data available online: https://cdn.amegroups.cn/static/public/atm-20-2430-1.xlsx). We classified CRC patients into stage I-IV and performed subset analyses. For patients with stage III CRC, the high SPS group showed significantly worse prognosis than the low SPS group (P=0.0013; Figure 2C). The low SPS group of stage II and IV CRC tended to have better outcomes, though the p value did not reach statistical significance (P=0.077 in stage II and P=0.19 in stage IV; Figure 2C). By comparison, SPS was unsatisfactory in distinguishing patients at risk in stage I (P=0.6; Figure S2), which may be due to the relatively small volume of samples. Thus, the SPS showed prognostic potential, especially for stage III CRC patients.
SPS is an independent risk factor for CRC
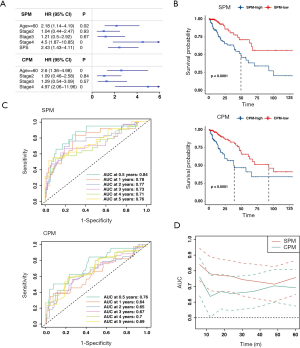
To explore whether SPS is an independent risk factor, we conducted a multivariate Cox regression analysis. After adjusting for age, sex, stage, pT, pN, pM, MSI state, KRAS state, BRAF state, and CMS molecular subtype, SPS remained an independent prognostic factor (HR =2.9, P=0.00013). The optimal multivariate prognostic model (SPM) was selected by the backward stepAIC (Methods), containing age, stage, and SPS (Figure 3A, more data available online: https://cdn.amegroups.cn/static/public/atm-20-2430-2.xlsx). We additionally compared the SPM to an extra clinicopathological prognostic model (CPM) that didn’t include SPS (Figure 3A). The difference between the high SPM group and low SPM group, and the difference between the high CPM group and the low CPM group were all statistically significant (log-rank test, P<0.001; Figure 3B). However, SPM achieved a higher concordance index (CI =0.74 for SPM; CI =0.681 for CPM). Time-dependent receiver operating characteristic curve (ROC) analyses and time-dependent AUC curves also identified the superior accuracy of the SPM in predicting the prognosis of CRC (Figure 3C,D).
Validation of SPS
To avoid overfitting, we randomly sampled three genes from the remaining 93 STING-related genes to generate 1000 extra multivariate Cox regression models. The prognostic significance of these random models was then compared with the SPS. We found that the DHX9-IRF2-POLR1D-based SPS prominently outperformed random classifiers (Figure 4A). Additionally, we tested the performance of SPS in an external cohort (GSE87211) that consisted of 363 CRC patients. After normalizing the expression of POLR1D, IRF2, and DHX9, samples were divided into a high SPS group and a low SPS group (Methods). The low SPS group showed significantly longer survival than the high SPS group (P=0.006, log-rank test; Figure 4B). Moreover, the risk of death was 3.02-fold higher in the high-risk group than in the low-risk group (HR =3.02, 95% CI, 1.35–9.04, P= 0.01).

Features of high-risk CRC and implications in immunotherapy
To obtain a better understanding of the biological background of the high SPS group, we compared the two groups in terms of genomic aberrations, transcriptional features, and the immune landscape.
Genomic aberrations
Genomic aberrations that can increase cytoplasmic DNA augment the ability of the host’s immune system to detect the tumor. We found that a higher SPS was associated with lower cancer/testis antigen (CTA) levels (P=0.006, r=−0.134; Spearman’s correlation test; Figure S3) (27,28). We also found that the homologous recombination defect (HRD), which is considered the most effective target of defects in DNA repair (29), was negatively related to the SPS (P=0.04, r=−0.1, Spearman’s correlation test; Figure S3).
CMS subtypes
We found that the SPS risk classification was not independent of the CMS molecular subtypes. The low SPS group had a higher frequency of patients with the CMS2 subtype, while the high SPS group had a higher frequency of patients with the CMS4 subtype (P=0.014, Fisher’s exact test; Figure 5A). Additionally, the subtype NOLBL, the mixed CRC subtype, primarily overlapped with the four subtypes. The CMS1 subtype showed fewer associations with the CMS2 and CMS4 subtypes (Figure S4).
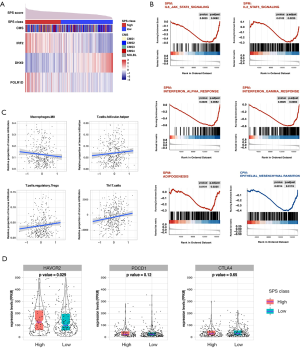
Altered pathways
CRC samples were sorted according to SPM risk level and evenly divided into a high SPM group (215 samples) and a low SPM group (216 samples). We compared the difference between the transcriptional landscapes of these groups. As a result, 26 of 50 hallmark gene sets showed significant enrichment in both, of which 15 gene sets were exclusively enriched in the high SPM group and 11 gene sets were enriched in the low SPM group (Table S4). As a control, we also analyzed the difference between the high CPM group (214 samples) and the low CPM group (217 samples), derived from the model only based on age and stage. The high CPM group enriched eleven gene sets, and the low CPM group enriched six gene sets (Table S4). Significantly, five high-risk related hallmark gene sets overlapped between the high SPM group and the high CPM group, including pathways associated with P53, hypoxia, and KRAS signaling upregulation, the inflammatory response, and TNFa signaling via NFKB (Figure S5, Table S4). The IL6-JAK-STAT3, IL2-STAT5, IFNa, IFNr, and adipogenesis pathways were found to be exclusively enriched in the high SPM group (Figure 5B, Table S4). By contrast, the epithelial-mesenchymal transition pathway was only detected in the high CPM group (Figure 5B, Table S4).
Immune landscape
We harnessed Cibersort to assess the absolute and relative infiltration of 22 types of immune cells in the TIME of CRC (Figure S6). The infiltration of M0 macrophages was negatively associated with the SPS, while the infiltration of follicular helper T cells and regulatory T cells were positively associated with the SPS (M0: r=−0.152, P=0.0015; follicular helper T cells: r=0.115, P=0.017; Tregs, r=0.131, P=0.0064; Pearson’s correlation test; Figure 5C). Th17 cells, which are also differentiated from CD4+ T cells (30), showed a significantly positive correlation with the SPS (r=0.132, P=0.007, Pearson’s correlation test; Figure 5C). Additionally, we observed that TIM-3 was significantly upregulated in the high SPS group, while CTLA-4 and PDCD1 were not significantly different between the two groups (Figure 5D).
Construction and evaluation of the nomogram
To extend the usage of SPS in the clinic, we constructed a nomogram predicting OS for CRC patients (Figure 6A). The bias between predictions and actual observations was referred to as calibration (26), and the calibration curves of survival achieved satisfactory performance for CRC of 1, 3 and 5 years (Figure 6B,C,D). Moreover, the CI of the nomogram was 0.736. We further separately constructed two nomograms: one with removed SPS and the other based on TNM, which were less accurate than the SPS-based nomogram (SPS: 0.382, age: 0.409, stage: 0.317, age + stage: 0.713, TNM: 0.712).
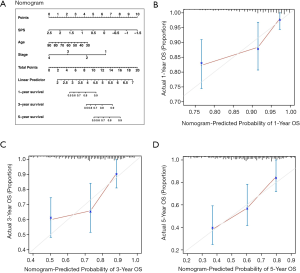
To use the nomogram: First, map the information of an individual patient to the corresponding variable axis. Second, draw a line upward to identify the corresponding value on the “Points” axis, which presents as the predicted probabilities scaled from 0 to 10. Third, manually calculate the sum of the three values and localize the final point of an individual on the “Total Points” axis. Likelihoods of 1-, 3- or 5-year survival can be obtained on the corresponding survival axis.
Discussion
In our study, the expression of STING was upregulated exclusively in the CMS1 subtype and impaired in the other three subtypes. The impairment might explain the lack of immune infiltration in the CMS2, CMS3, and CMS4 subtypes. Additionally, POLR1D, IRF2, and DHX9 showed significant prognostic value among the 96 STING-related genes. DHX9 has multiple functions in regulating transcription, translation, RNA processing and transport, DNA replication and the maintenance of genomic stability (31-37). A recent study indicated that DHX9 inhibited epithelial-mesenchymal transition (EMT) by regulating STAT3 in human lung adenocarcinoma cells (37). Accordingly, we observed overexpressed DHX9 in tumors, and higher DHX9 was related to better outcomes in CRC. By comparison, higher levels of POLR1D and IRF2 were associated with adverse outcomes in CRC. This was in line with that POLR1D plays a role in the synthesis of ribosomal RNA precursors and small RNAs. Also, a recent study reported a positive correlation between the expression level of POLR1D and tumor size and predicted poor outcomes in CRC patients. The study also demonstrated that POLR1D affected cell proliferation, migration, and apoptosis in vitro, and influenced tumor growth in vivo (38). IRF2 is a member of the interferon regulatory transcription factor (IRF) family that competitively inhibits the IRF1-mediated transcriptional activation of IFN-α/β (39). It was reported that IRF2 participated in the KRAS-IRF2-CXCL3-CXCR2 axis, and CRC with higher IRF2 expression exhibited a better response to anti-PD-1 therapy (40). Though these single predictors are significant, we considered that a combined gene panel could provide more comprehensive information to guide the clinical practice. It turned out that the SPS derived from a multivariate model was proven to be an independent risk factor and outperformed current clinicopathological features in predicting CRC outcomes.
The high SPS group and the low SPS group were different in several biological features. We observed lower levels of CTA and HRD in the high SPS group. CTA could elicit immune responses in tumors, thus being a target for immunotherapy (27,28). HRD impairs normal DNA damage repair and results in genomic loss of heterozygosity (LOH) defined as loss or duplication of chromosomal regions (29). Genome instability that leads to carcinogenesis and tumor progression currently represents a biomarker of ICBs. Therefore, the high SPS group may be less benefited from ICBs. Moreover, the high SPS group was more immunologically tolerant, as we observed decreased M0 macrophages and increased TIM-3. By comparison, CTLA-4 and PDCD1 that primarily inhibit overactivated T cell responses showed no difference between the two groups. It’s known that TIM-3 encodes the Th1-specific cell surface protein to regulate macrophage activation, leading to immunological tolerance (41). Besides, as the high SPS group had a higher frequency of the CMS4 subtype, it may reflect features of the CMS4 subtype to some extent, for example, increased TGF-β activation (16). In terms of pathway analysis, five hallmark gene sets were enriched in both SPM-high and CPM-high CRC, which have all been suggested to contribute to unfavorable outcomes in CRC (42-45). The IL6-JAK-STAT3 and the IL2-STAT5 signaling pathway were enriched exclusively in the high SPM group, and a higher SPS was associated with enhanced Th17 and Treg cells. This could be explained by facts that Tregs develop and express STAT5 under the influence of IL-2 and TGF-β and that IL-6 could induce the development of Th17 cells (46). In terms of the immunotherapy for CRC, ICBs currently are used limited among patients with dMMR/MSI-H (5), which might widely represent the CMS1 subtype. For the CMS2, CMS3, and CM4 subtypes, the Th17 axis may be a more promising target for the high SPS group, and immune interventions including STING agonists and immune interventions targeting the Th17 axis may reverse the impaired innate immune response phase (47).
To better facilitate individualized medical assessments, we also provided a nomogram which we believe could be a useful tool for clinicians in the future. Finally, there are several limitations of this study, such as the use of retrospective datasets from the TCGA and GEO databases. Thus, the results should be further strengthened by more prospective studies.
Conclusions
In conclusion, for the first time, we identified and validated a STING-related independent risk factor and built a prognostic score, named SPS, to improve the prediction accuracy of prognosis in CRC. The relevant nomogram could be a promising tool for individual assessment in the future. In addition, the SPS provides insights into the immunotherapy in CRC. While ICBs may be beneficial to CRC patients of the CMS1 subtype, we suggest that STING agonists and immunotherapies targeting the Th17 axis could benefit the CMS2, CMS3, and CMS4 subtypes in the high SPS CRC group.
Acknowledgments
Funding: Clinical Research and Cultivation Project of Shanghai ShenKang Hospital Development Center (Grant No. SHDC12017X01).
Footnote
Reporting Checklist: The authors have completed the TRIPOD reporting checklist. Available at http://dx.doi.org/10.21037/atm-20-2430
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/atm-20-2430). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. The relevant data provided by TCGA and GEO database are publicly available and open-ended, and do not require the approval of the local ethics committee.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Bray F, Ferlay J, Soerjomataram I, et al. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin 2018;68:394-424. [Crossref] [PubMed]
- Dekker E, Tanis PJ, Vleugels JLA, et al. Colorectal cancer. Lancet 2019;394:1467-80. [Crossref] [PubMed]
- Corrales L, McWhirter SM, Dubensky TW Jr, et al. The host STING pathway at the interface of cancer and immunity. J Clin Invest 2016;126:2404-11. [Crossref] [PubMed]
- Demaria O, Cornen S, Daëron M, et al. Harnessing innate immunity in cancer therapy. Nature 2019;574:45-56. [Crossref] [PubMed]
- Ganesh K, Stadler ZK, Cercek A, et al. Immunotherapy in colorectal cancer: rationale, challenges and potential. Nat Rev Gastroenterol Hepatol 2019;16:361-75. [Crossref] [PubMed]
- Burdette DL, Monroe KM, Katia ST, et al. STING is a direct innate immune sensor of cyclic di-GMP. Nature 2011;478:515. [Crossref] [PubMed]
- Woo SR, Fuertes M, Corrales L, et al. STING-Dependent Cytosolic DNA Sensing Mediates Innate Immune Recognition of Immunogenic Tumors. Immunity 2014;41:830-42. [Crossref] [PubMed]
- Fuertes MB, Kacha AK, Justin K, et al. Host type I IFN signals are required for antitumor CD8+ T cell responses through CD8{alpha}+ dendritic cells. J Exp Med 2011;208:2005-16. [Crossref] [PubMed]
- Bode C, Fox M, Tewary P, et al. Human plasmacytoid dentritic cells elicit a Type I Interferon response by sensing DNA via the cGAS-STING signaling pathway. Eur J Immunol 2016;46:1615-21. [Crossref] [PubMed]
- Marill J, Mohamed Anesary N, Paris S. DNA damage enhancement by radiotherapy-activated hafnium oxide nanoparticles improves cGAS-STING pathway activation in human colorectal cancer cells. Radiother Oncol 2019;141:262-6. [Crossref] [PubMed]
- Zhu Q, Man SM, Gurung P, et al. Cutting edge: STING mediates protection against colorectal tumorigenesis by governing the magnitude of intestinal inflammation. J Immunol 2014;193:4779-82. [Crossref] [PubMed]
- Jiang X, Liu G, Hu Z, et al. cGAMP inhibits tumor growth in colorectal cancer metastasis through the STING/STAT3 axis in a zebrafish xenograft model. Fish Shellfish Immunol 2019;95:220-6. [Crossref] [PubMed]
- Xia T, Konno H, Ahn J, et al. Deregulation of STING Signaling in Colorectal Carcinoma Constrains DNA Damage Responses and Correlates With Tumorigenesis. Cell Rep 2016;14:282-97. [Crossref] [PubMed]
- Yang CA, Huang HY, Chang YS, et al. DNA-Sensing and Nuclease Gene Expressions as Markers for Colorectal Cancer Progression. Oncology 2017;92:115-24. [Crossref] [PubMed]
- Chon HJ, Kim H, Noh JH, et al. STING signaling is a potential immunotherapeutic target in colorectal cancer. J Cancer 2019;10:4932-8. [Crossref] [PubMed]
- Guinney J, Dienstmann R, Wang X, et al. The consensus molecular subtypes of colorectal cancer. Nat Med 2015;21:1350-6. [Crossref] [PubMed]
- Naito Y, Saito K, Shiiba K, et al. CD8+ T cells infiltrated within cancer cell nests as a prognostic factor in human colorectal cancer. Cancer Res 1998;58:3491-4. [PubMed]
- Ramanjulu JM, Pesiridis GS, Yang J, et al. Design of amidobenzimidazole STING receptor agonists with systemic activity. Nature 2018;564:439-43. [Crossref] [PubMed]
- Brenner C. Applications of Bioinformatics in Cancer. Cancers (Basel) 2019;11:1630. [Crossref] [PubMed]
- Shen S, Wang G, Zhang R, et al. Development and validation of an immune gene-set based Prognostic signature in ovarian cancer. EBioMedicine 2019;40:318-26. [Crossref] [PubMed]
- Newman AM, Liu CL, Green MR, et al. Robust enumeration of cell subsets from tissue expression profiles. Nature Methods 2015;12:453-7. [Crossref] [PubMed]
- Thorsson V, Gibbs DL, Brown SD, et al. The Immune Landscape of Cancer Immunity 2018;48:812-30.e14.
- Yu G, Wang LG, Han Y, et al. clusterProfiler: an R package for comparing biological themes among gene clusters. OMICS 2012;16:284-7. [Crossref] [PubMed]
- Liberzon A, Birger C, Thorvaldsdóttir H, et al. The Molecular Signatures Database Hallmark Gene Set Collection. Cell Syst 2015;1:417-25. [Crossref] [PubMed]
- Subramanian A, Tamayo P, Mootha VK, et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci U S A 2005;102:15545-50. [Crossref] [PubMed]
- Iasonos A, Schrag D, Raj GV, et al. How To Build and Interpret a Nomogram for Cancer Prognosis. J Clin Oncol 2008;26:1364-70. [Crossref] [PubMed]
- Salmaninejad A, Zamani MR, Pourvahedi M, et al. Cancer/Testis Antigens: Expression, Regulation, Tumor Invasion, and Use in Immunotherapy of Cancers. Immunol Invest 2016;45:619-40. [Crossref] [PubMed]
- Gordeeva O. Cancer-testis antigens: Unique cancer stem cell biomarkers and targets for cancer therapy. Semin Cancer Biol 2018;53:75-89. [Crossref] [PubMed]
- Motegi A, Masutani M, Yoshioka KI, et al. Aberrations in DNA repair pathways in cancer and therapeutic significances. Semin Cancer Biol 2019;58:29-46. [Crossref] [PubMed]
- Zhu J, Yamane H, Paul WE. Differentiation of effector CD4 T cell populations Annu Rev Immunol 2010;28:445-89. [Crossref] [PubMed]
- Lee T, Paquet M, Larsson O, et al. Tumor cell survival dependence on the DHX9 DExH-box helicase. Oncogene 2016;35:5093-105. [Crossref] [PubMed]
- Cao S, Sun R, Wang W, et al. RNA helicase DHX9 may be a therapeutic target in lung cancer and inhibited by enoxacin. Am J Transl Res 2017;9:674-82. [PubMed]
- He L, Chen Y, Wu Y, et al. Nucleic acid sensing pattern recognition receptors in the development of colorectal cancer and colitis. Cell Mol Life Sci 2017;74:2395-411. [Crossref] [PubMed]
- Hong H, An O, Chan THM, et al. Bidirectional regulation of adenosine-to-inosine (A-to-I) RNA editing by DEAH box helicase 9 (DHX9) in cancer. Nucleic Acids Res 2018;46:7953-69. [Crossref] [PubMed]
- Ding X, Jia X, Wang C, et al. A DHX9-lncRNA-MDM2 interaction regulates cell invasion and angiogenesis of cervical cancer. Cell Death Differ 2019;26:1750-65. [Crossref] [PubMed]
- Wang YL, Liu JY, Yang JE, et al. Lnc-UCID Promotes G1/S Transition and Hepatoma Growth by Preventing DHX9-Mediated CDK6 Down-regulation. Hepatology 2019;70:259-75. [Crossref] [PubMed]
- Yan X, Chang J, Sun R, et al. DHX9 inhibits epithelial-mesenchymal transition in human lung adenocarcinoma cells by regulating STAT3. Am J Transl Res 2019;11:4881-94. [PubMed]
- Wang M, Niu W, Hu R, et al. POLR1D promotes colorectal cancer progression and predicts poor prognosis of patients. Mol Carcinog 2019;58:735-48. [Crossref] [PubMed]
- Taniguchi T, Ogasawara K, Takaoka A, et al. IRF family of transcription factors as regulators of host defense. Annu Rev Immunol 2001;19:623-55. [Crossref] [PubMed]
- Liao W, Overman MJ, Boutin AT, et al. KRAS-IRF2 Axis Drives Immune Suppression and Immune Therapy Resistance in Colorectal Cancer. Cancer Cell 2019;35:559-72.e7. [Crossref] [PubMed]
- Wolf Y, Anderson AC, Kuchroo VK. TIM3 comes of age as an inhibitory receptor. Nat Rev Immunol 2020;20:173-85. [Crossref] [PubMed]
- Baritaki S, de Bree E, Chatzaki E, et al. Chronic Stress, Inflammation, and Colon Cancer: A CRH System-Driven Molecular Crosstalk. J Clin Med 2019;8:1669. [Crossref] [PubMed]
- Patel M, Horgan PG, McMillan DC, et al. NF-κB pathways in the development and progression of colorectal cancer. Transl Res 2018;197:43-56. [Crossref] [PubMed]
- Nagaraju GP, Bramhachari PV, Raghu G, et al. Hypoxia inducible factor-1α: Its role in colorectal carcinogenesis and metastasis. Cancer Lett 2015;366:11-8. [Crossref] [PubMed]
- Huang D, Sun W, Zhou Y, et al. Mutations of key driver genes in colorectal cancer progression and metastasis. Cancer Metastasis Rev 2018;37:173-87. [Crossref] [PubMed]
- Knochelmann HM, Dwyer CJ, Bailey SR, et al. When worlds collide: Th17 and Treg cells in cancer and autoimmunity. Cell Mol Immunol 2018;15:458-69. [Crossref] [PubMed]
- Vitiello GA, Miller G. Targeting the interleukin-17 immune axis for cancer immunotherapy. J Exp Med 2020;217:e20190456. [Crossref] [PubMed]


