
RNA sequencing (RNA-seq) analysis of gene expression provides new insights into hindlimb unloading-induced skeletal muscle atrophy
Introduction
As one of the important organs in the body, skeletal muscle bears many important physiological functions. Antigravity muscle plays important roles in supporting body weight, supporting posture and balance, and movement, and the disappearance of gravity can cause disuse atrophy of this type of muscle. For example, spaceflight, especially medium- and long-term space flight, will cause significant structural and functional changes in astronauts’ skeletal muscle, manifesting as a reduction in muscle mass, a reduction in muscle fiber cross-sectional area, the conversion of muscle fiber type, a reduction in muscle protein contents, and decreased muscle contractility (1-3). Insufficient effective measures to address these effects will seriously affect astronauts’ physical health and working ability in flight and their adaptability after returning to Earth. In addition to weightlessness, muscle atrophy may occur under various physiological and pathological conditions, including denervation, starvation, aging, and cancer. Therefore, it is necessary to determine the mechanisms underlying skeletal muscle atrophy and effectively protect against atrophy, urgent challenges to be addressed in space medicine and rehabilitation medicine.
Under normal physiological conditions, the synthesis and degradation of protein in skeletal muscle are in dynamic equilibrium. When the balance is disrupted, and protein degradation becomes dominant, muscle atrophy occurs (4,5). Activation of the ubiquitin-proteasome hydrolysis system in skeletal muscle is the key cause of weightlessness-induced muscular atrophy (6,7). The expression of 2 muscle-specific E3 ubiquitin ligases, muscle atrophy F-box (MAFbx) and muscle RING finger 1 (MuRF1), significantly increase in various disuse-atrophy models (8,9). The expression of MuRF1 and MAFbx is regulated by a complex signaling network that includes insulin-like growth factor I (IGF1)/phosphatidylinositol 3-kinase (PI3K), p(PI3K)/protein kinase B (AKT)/forkhead box O (FOXO) and tumor necrosis factor-alpha (TNF-α)/nuclear factor kappa-B (NF-κB) (10,11). In recent years, many studies have used transcriptomics to analyze differentially expressed genes (DEGs) in the process of muscle atrophy, providing a substantial amount of gene expression data for the study of muscle atrophy (12-15). Our recent study systematically analyzed the DEGs of the tibialis anterior muscle at different time points after denervation. It classified muscle atrophy into 4 phases: the oxidative stress phase, the inflammation phase, the atrophic phase, and the atrophic fibrosis phase (16). In subsequent studies, we used various drugs to intervene and found they attenuated muscle atrophy processes through a mechanism closely related to the inhibition of oxidative stress and inflammatory responses in the early phase of muscular atrophy (17-23). The main issue with transcriptome analysis for weightlessness-induced muscle atrophy is that the analyses’ time points which have been selected need to be tuned further. For example, in most literatures, only one-time point (2 weeks) or multiple time points starting three days after weightlessness have been selected (24-27). Only Stevenson et al. used an Affymetrix gene microarray to detect DEGs in rat soleus muscle 1, 4, 7, and 14 days after weightlessness. They revealed gene expression changes related to different biological processes, including ubiquitination, glycolysis, and oxidative stress during muscle atrophy (28). To obtain more valuable information on gene expression changes in weightlessness-induced muscle atrophy, especially in the early phase of muscular atrophy, it is necessary to conduct new multi-time point transcriptome analyses.
Currently, in models of weightlessness-induced muscle atrophy, the tail suspension model with rats can simulate a series of physiological changes in hindlimb muscle during space flight and is one of the most extensively used animal models in studies of muscular dystrophy (29,30). Studies have shown that the soleus muscle exhibits significant atrophy after suspension for one week, and the muscle wet weight ratio decreases by approximately 50% after two weeks (31,32). In this study, RNA sequencing (RNA-seq) analysis was used to analyze DEGs in rat soleus muscle at 12, 24, 36 hours, three days, and seven days after tail suspension. Cluster analysis and bioinformatics methods were applied in the systematic analysis of DEGs to systematically and comprehensively study the molecular mechanisms underlying weightlessness-induced muscular atrophy. This study supplies potential targets for the prevention and treatment of weightlessness-induced muscle atrophy in the clinic.
We present the following article in accordance with the ARRIVE reporting checklist (available at http://dx.doi.org/10.21037/atm-20-7400).
Methods
Animal experiment
Adult male Sprague-Dawley (SD) rats (weighing ~200 g) were provided by the Experimental Animal Center (Nantong, China). Rats were fed food and water ad libitum and were housed at 23±2 °C with a 12-hour light/dark cycle. The experiments involving animals were conducted following the animal care guidelines of Nantong University and ethically approved by the Jiangsu Administration Committee for Experimental Animals (No. 20180305-004). The animals were randomly divided into five experimental groups and a control group (n=3 in each group).
Tail suspension hind limb unloading
The tail suspension is the most used animal model for non-weight bearing skeletal muscle. Individual rats were acclimated to single-cage housing for three days before tail suspension experiments. The tail suspension was performed according to a previously published protocol (33). Briefly, the tail was wrapped with nonabrasive adhesive tape, and then, the animal was suspended by attaching a swivel hook to the metal bars on the top of the cage. The rats were free to rotate in all directions within the cage and had access to food and water. Control mice were separated into individual cages with the same conditions as the unloading groups but without tail suspension (referred to as 0 hours). At the end of the indicated time point (12, 24, 36 hours, three days, and seven days), CO2 asphyxiation euthanized experimental and control rats and cervical dislocation. The soleus muscle of the hindlimbs was harvested and quickly immersed in liquid nitrogen for later experiments.
RNA’s extraction and RNA-seq analysis
According to the kit guidelines, each soleus muscle sample was homogenized, and RNA was extracted with a mirVana™ miRNA ISOlation Kit (Ambion-1561, Thermo Scientific, Delaware, ME, United States). The total RNA quantity was measured using a NanoDrop (ND-2000 Thermo Scientific, Delaware, ME, United States). Libraries were constructed using a TruSeq Stranded mRNA LTSample Prep Kit (Illumina, San Diego, CA, USA) according to the manufacturer’s instructions. In brief, 50 µL of total RNA (4 µg) was purified and isolated. Using interrupted mRNA as a template, first-strand cDNA and double-stranded cDNA were synthesized and then purified. A poly(A) tail was added to double-stranded cDNA and then ligated with an RNA adaptor, followed by purification and PCR amplification. The constructed libraries were evaluated using an Agilent 2100 Bioanalyzer (Agilent Technologies, Santa Clara, CA, USA). The libraries were then sequenced on the Illumina sequencing platform (Illumina HiSeq X Ten), and 125 bp/150 bp paired-end reads were generated. Raw data in FASTQ format were processed using Trimmomatic for quality control. The reads holding low-quality reads and poly-N sequences were removed to obtain clean reads. Then, the clean reads were mapped to a reference genome using hisat. The FPKM value for each gene was calculated using cuff-links, and the read counts for each gene were obtained using htseq-count. DEGs were found using the DESeq package in R. P value <0.05, and fold-change >1.5 were set as the thresholds for significantly differential expression.
Time-specific pattern identification
RNA-seq data mining was performed using Pearson correlation heatmaps and principal component analysis (PCA) to identify time-specific patterns as described previously (16). In brief, Pearson correlation coefficients were calculated using the core function in the ‘stats’ package in R (http://www.r-project.org), and the results were generated using the ‘pheatmap’ package in R. PCA was performed using the ‘prcomp’ function in the ‘stats’ package in R (http://www.r-project.org); the first 2 PCs were selected to view the expression patterns of samples.
Gene function analysis
The Kyoto Encyclopedia of Genes and Genomes (KEGG) database and Gene Ontology (GO) category database were applied for functional annotation of DEGs. Enrichment analysis of KEGG and GO categories was performed using DAVID online tools (https://david.ncifcrf.gov). Upregulated and downregulated genes were separated for analysis. KEGG or GO categories with a P value less than 0.05 were considered significantly enriched. The average expression profile was calculated as described previously (34).
Regulatory network analysis
Ingenuity pathway analysis (IPA) (http://ingenuity.com/index.html, Ingenuity Systems, Redwood City, CA) online software was used to identify canonical pathways and gene networks. The Ingenuity Pathway Knowledge Base was used for a deep analysis of molecular networks and revealed interactions among DEGs (FC >2, P<0.01) (34,35). The Build Connect tool in IPA was used to create connections between molecules, with direct (solid line) or indirect (dot line) relationships shown.
Statistical Analysis
The values of gene expression are calculated from the average of three repeated experiments. The relative change represents the difference between various experimental group and the control group. The negative binomial distribution test is used to test the significance (P value) of the difference between the two groups.
Results
Analysis of the DEGs in the soleus muscle during the weightlessness-induced muscle atrophy process
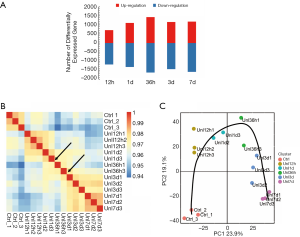
RNA samples from the soleus muscle were subjected to RNA-seq analysis, and many DEGs were obtained. Compared with that in the control group, at 12, 24, and 36 hours, three days, and seven days after hindlimb unloading, the expression levels of 712, 1,109, 1,433, 1,162, and 1,182 genes were upregulated in soleus muscle, respectively, and 1,186, 1,324, 1,632, 1,446, and 1,596 genes were downregulated, respectively (Figure 1A). Notably, at 36 hours after hindlimb unloading, the number of DEGs significantly increased, showing that the atrophy process might be widely activated at this time point. With the Pearson correlation heatmaps, 24 and 36 hours after and 36 hours and three days after hindlimb unloading, two-node transitions could be observed in the expression profile of DEGs. There were one sample deviating at 24 hours and one sample deviating at 36 hours after hindlimb unloading, therefore, they were excluded from subsequent analyses (Figure 1B). PCA results showed significant transitions in DEGs 36 hours after hindlimb unloading (Figure 1C). The Pearson correlation heatmaps and PCA results suggested that within seven days after hindlimb unloading, three different transcriptional phases occurred: 12–24 hours after hindlimb unloading (transcriptional phase I), 36 hours after hindlimb unloading (transcriptional phase II), and 3–7 days after hindlimb unloading (transcriptional phase III).
Analysis of the biological processes and signaling pathways of upregulated genes during the process of weightlessness-induced muscle atrophy
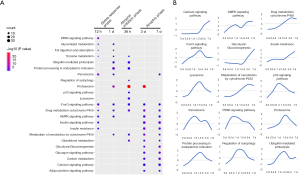
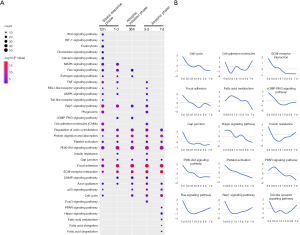
GO and KEGG analyses were performed to determine the biological processes and signaling pathways associated with the DEGs. Figures 2,3 show the enriched biological processes and signaling pathways, respectively, in which the upregulated genes participate at different time points after hindlimb unloading. During the transcriptional phase I (12–24 hours after hindlimb unloading), biological processes including “response to hypoxia,” “response to endoplasmic reticulum stress,” “cellular response to dexamethasone stimulus,” “response to glucocorticoid,” and “oxidation-reduction” were enriched. The biological processes “positive regulation of the apoptotic process,” “protein ubiquitination,” and “response to denervation involved in the regulation of muscle adaptation” were also enriched during transcriptional phase I. The signaling pathways enriched at this phase include “Drug metabolism-cytochrome P450”, “Metabolism of xenobiotics by cytochrome P450”, “Protein processing in the endoplasmic reticulum,” “Peroxisome,” and “PPAR signaling pathway.” These data indicate that oxidative stress, endoplasmic reticulum (ER) stress and cell apoptosis were activated suggesting that the transcriptional phase I primarily involved the activation of stress responses; therefore, this phase was defined as the stress response phase. During transcriptional phase II (36 hours after hindlimb unloading), the biological processes including “cellular response to lipopolysaccharide,” “proteasome-mediated ubiquitin-dependent protein catabolic process,” “autophagosome assembly,” “positive regulation of the apoptotic process,” “protein ubiquitination,” “response to oxidative stress,” and “oxidation-reduction process” were enriched. The signaling pathways enriched during this phase mainly included “Drug metabolism-cytochrome P450”, “Metabolism of xenobiotics by cytochrome P450”, “Ubiquitin mediated proteolysis,” “Lysosome,” “Proteasome,” “p53 signaling pathway,” and “FoxO signaling pathway.” The proteolysis-related “ubiquitin-proteasome system” and “autophagy-lysosomal system” were activated during this phase, accompanied by the appearance of inflammatory responses and further exacerbation of the oxidative stress response, suggesting that this phase was the atrophic initiation phase. During transcriptional phase III (3–7 days after hindlimb unloading), biological processes including “positive regulation of fast-twitch skeletal muscle fiber contraction,” “glycolysis,” “aging,” “insulin resistance,” “cellular response to tumor necrosis factor,” “response to hypoxia” and “oxidation-reduction process” were enriched; the signaling pathways enriched at this phase mainly included “drug metabolism-cytochrome P450”, “Metabolism of xenobiotics by cytochrome P450”, “Calcium signaling pathway” and “Glycolysis/Gluconeogenesis.” During transcriptional phase III, biological processes and pathways, including slow-to-fast muscle conversion and aging, were activated, with oxidative stress and proteolysis continued to be activated, suggesting that this phase was the atrophic phase.

Analysis of biological processes and signaling pathways of downregulated genes during the process of weightlessness-induced muscle atrophy
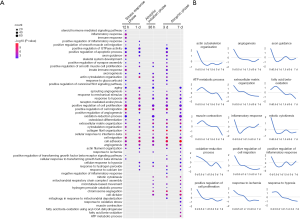
Figures 4,5 show the enriched biological processes and signaling pathways, respectively, in which the downregulated genes participate at different time points after hindlimb unloading. During the transcriptional phase, I, biological processes including “mitotic cytokinesis,” “angiogenesis,” “actin cytoskeleton organization,” “axon guidance,” “positive regulation of cell migration,” and “positive regulation of cell proliferation” were enriched. Signaling pathways including “Cell cycle,” “Focal adhesion,” “Gap junction,” “Rap1 signaling pathway”, “Protein digestion and absorption,” and the “PI3K-Akt signaling pathway” were enriched. These data indicate the stress response phase (transcriptional phase I) mainly involved inhibiting cell proliferation and angiogenesis inhibition after hindlimb unloading. During transcriptional phase II, biological processes including “chromosome segregation,” “mitochondrial respiratory chain complex I assembly,” and “osteoblast differentiation” were enriched, and signaling pathways including “PI3K-Akt signaling pathway”, “ECM-receptor interaction,” and “cGMP-PKG signaling pathway” were enriched. During transcriptional phase III, biological processes including “muscle contraction,” “fatty acid beta-oxidation,” and “ATP metabolic process” were enriched, and signaling pathways including “Hippo signaling pathway,” “Platelet activation,” “PI3K-Akt signaling pathway”, “ECM-receptor interaction” and “Fatty acid metabolism” were enriched. These data indicated that during transcriptional phases II–III after hindlimb unloading (atrophic initiation phase and atrophic phase), the processes related to energy metabolism were extensively inhibited, and cell proliferation and angiogenesis were constitutively inhibited.
Analysis of different functions of DEGs using the IPA database
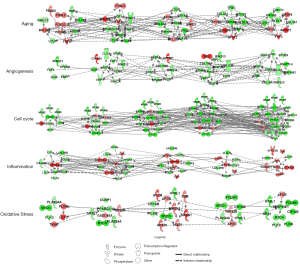
The IPA database platform was used to analyze DEGs’ enriched biological processes, and the genes related to 5 biological functions, i.e., aging, angiogenesis, cell cycle, immune response, and oxidative stress process of muscular atrophy, which play a key role in in the process of denervated muscle atrophy were identified. Molecular networks at different time points were further used to construct associations and analyze key regulatory molecules’ regulatory mechanisms. Figure 6 shows that muscle atrophy-induced aging-like changes during transcriptional phase III involved primarily in IGFBP1 and IGFBP5. Twenty-four hours after hindlimb unloading, downregulated angiogenesis-related genes, including VEGFA, KDR, FGF1, NRP1, and ANGPT2, significantly increased. Cell cycle-related genes showed more obvious trends during the muscle atrophy process; many cell cycle-related genes were downregulated across time. NFKBIA regulated inflammatory response-associated genes during the early phase of muscular atrophy, and then, NFKBIA, JAK2, and STAT5B together mediated inflammation responses. The number of genes involved in oxidative stress increased 12 to 36 hours after hindlimb unloading, including GADD45A and Cdkn1a, which promote reactive oxygen species (ROS) production (0.01<P<0.05, not shown in the figure), and a considerable number of antioxidant genes, including UCP3, HMOX1, NQO1, GPX3, GSR, and RXN1. The genes involved in regulating ROS genes after weightlessness included Nfe212, HDAC4, TGFBR2, SIRT3, and SIRT5 (1.5< FC <2, not shown in the figure). These genes were jointly involved in the cellular oxidative stress response after hindlimb unloading.
Discussion
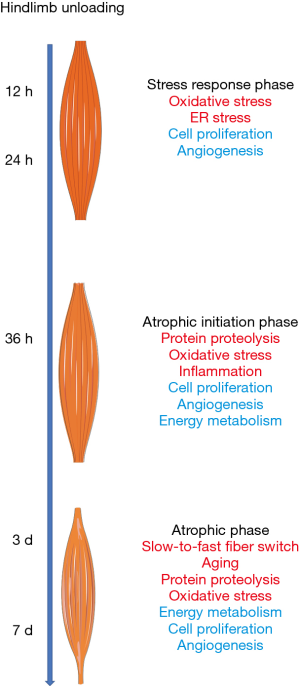
This study investigated gene expression changes in skeletal muscle at 12, 24, 36 hours, three days, and seven days after hindlimb unloading. We found a considerable number of DEGs in the soleus muscle of the unloaded hindlimb. From 12 hours–7 days after hindlimb unloading, the number of upregulated and downregulated genes in soleus muscle showed time-dependent changes. Notably, 36 hours after hindlimb unloading, the number of DEGs significantly increased, which indicated it was a critical time point for changes in gene expression patterns in soleus muscle. Most genes became activated at this time point, thus, the atrophic process was started. Pearson correlation heatmaps and PCA are used to analyze the gene expression profile of the unloaded soleus muscle of the hindlimb. The seven days’ time course of hindlimb unloading was divided into three transcriptional phases. KEGG analysis and GO annotation indicated that during transcriptional phase I, stress responses were activated, and cell proliferation and angiogenesis were inhibited; during transcriptional phase II, proteolytic-related processes were activated, accompanied by a certain degree of inflammatory response; and during transcriptional phase III, slow-to-fast muscle conversion, as well as aging and other processes, were activated, and energy metabolism was significantly inhibited. With these characteristics, transcriptional phases I, II, and III were sequentially appointed as the “stress response phase,” “atrophic initiation phase,” and “trophic phase.”
Oxidative stress first attracted our attention during the stress response phase. Our previous study has shown that the first phase of denervated muscle atrophy is the oxidative stress phase and that intervention with antioxidant drugs can effectively relieve muscular atrophy (16,19,20). It has been shown that a considerable amount of ROS is produced during the process of weightlessness-induced muscle atrophy and that the production of ROS is closely related to muscle atrophy (30,36,37). The cytochrome P450 enzyme is a marker of oxidative stress and the source of cellular ROS (38). During the stress response phase, the P450-associated pathway is activated, and the PPAR signaling pathway, which participates in the regulation of ROS production and degradation, is also activated. To note, some genes related to response to hypoxia were up-regulated, while some genes were down-regulated in the same biological process. As different gene groups showed different time-dependent expression changes, they exerted different functional effects on muscle atrophy. IPA was used to analyze further the DEGs associated with oxidative stress in muscular atrophy. In addition to the genes that induce ROS production (including GADD45a), the expression of many antioxidant genes (including Gpx3) was upregulated during the stress response. The number of DEGs related to oxidative stress increased during the atrophic initiation phase and persistently presented until the atrophic phase, with HOMX1 as the core regulator. Recent studies have shown that endoplasmic reticulum stress (ERS) plays an important role in maintaining myocyte homeostasis in healthy and disease-related conditions (39,40). It has been reported that PERK and XBP1 play important regulatory roles in the ERS pathway in cancer and starvation-induced muscle atrophy (41,42). Our study found that 24 hours after weightlessness, ERS was significantly activated, and the expression of XBP1 and its downstream genes was upregulated. In the stress response phase, unlike tibialis anterior atrophy caused by denervation, after hindlimb unloading, cells’ responses in the soleus muscles to dexamethasone and glucocorticoids were significantly activated. Literature reports have shown that glucocorticoid expression significantly increases during the early phase after weightlessness, resulting in multiple organ responses, including muscle atrophy, bone loss, and immune organ atrophy (43-45). Notably, the soleus muscle, as a slow muscle, is not sensitive to glucocorticoids (46). However, glucocorticoids have a strong anti-inflammatory effect, and the inflammatory response was inhibited during the stress response phase, especially 12 hours after hindlimb unloading. Consistent with previous studies, our analysis also showed the expression of cell cycle arrest genes increased. Many cell cycle regulatory genes’ expression decreased during muscle atrophy, showing significant inhibition of cell proliferation, continuing until seven days after hindlimb unloading (28). Skeletal muscle is composed of terminally differentiated multinucleated muscle fibers. Although muscle loss occurs during muscle atrophy, muscle fiber atrophy is due primarily to reduced size caused by protein loss. Therefore, the inhibition of cell proliferation after hindlimb unloading might occur in muscle satellite cells. In addition to the cell cycle, angiogenesis-related genes (including VEGFA and KDR) and genes involved in sprouting angiogenesis (including NRP1 and ANGPT) were downregulated. The soleus muscle is a slow muscle composed primarily of oxidized type I fibers, with a particularly rich capillary network, the capillary network is vital for maintaining soleus muscle function (47). The downregulation of angiogenesis-related genes after hindlimb unloading further worsens muscular atrophy.
Multiple biological processes and signaling pathways in 2 major systems related to proteolysis, “Ubiquitin-Proteasome System” and “Autophagy-Lysosomal System,” were extensively activated during the atrophic initiation phase, and the activation of proteolysis was significantly advanced rather than four days after hindlimb unloading previously reported in the literature (28). This is due primarily to the broader early time points, that is, only one day and four days, used in previous studies of DEGs in rats after hindlimb unloading. Like this issue, our previous studies showed that muscle atrophy started 3–7 days after denervation, however, time points between 24 hours and three days were lacking (16). During this stage, the positive regulation of cells via LPS activation and inflammatory responses was significantly upregulated compared to that at 12 hours, indicating that inflammatory responses were activated, to a certain degree, during the atrophic initiation phase. We found the JAK/STAT pathway started to be constitutively activated 24 hours after hindlimb unloading. Although inflammation caused by hindlimb unloading occurred later than 24 hours, that inflammation was significantly activated after denervating muscle atrophy. There was an upregulation trend from 36 hours. The upregulated genes in the trophic phase were slow-to-fast muscle conversion- and aging-related genes. Consistent with earlier studies, our data showed during the muscle atrophy process, the soleus muscle fiber type changed from oxidized type I to glycolysis type II. Consistent with fiber-type conversion, glycolysis also significantly increased three days after hindlimb unloading (28). Like denervated muscle atrophy, we found that upregulated genes participated in the aging process, including insulin-like growth factor-binding protein 5 (IGFBP5) and IGF binding protein 1 (IGFBP1). Aging-associated insulin resistance was significantly upregulated during the atrophic phase. It is well-known that insulin resistance plays a role in protein synthesis (48). The downregulated genes in the atrophic initiation and atrophic phases were involved in mitochondrial respiration, fatty-acid β-oxidation, and protein synthesis, indicating that energy metabolism during weightlessness-induced muscle atrophy was significantly inhibited, that is consistent with those for other models of skeletal muscle atrophy (49).
It should be mentioned that there are some limitations. The proposed functional consequences of the gene expression changes identified tested in vivo or in vitro are warranted. Another drawback of our study was that lncRNA and microRNA were not investigated. Detection of lncRNA and microRNA will support the whole transcriptome survey for obtaining more comprehensive information.
Hindlimb unloading induces skeletal muscles’ disuse, leading to the generation of oxidative stress and other stress responses in muscle cells, resulting in inflammation responses and proteolysis. As proteolytic reactions continue to be activated, energy metabolism is reduced, and muscle atrophy occurs (Figure 7). Notably, the atrophic initiation phase, that is, 36 hours after hindlimb unloading, might be a critical time point for weightlessness-induced muscle atrophy. Our results may help understand the molecular mechanisms underlying weightlessness-induced skeletal muscle atrophy and develop promising therapeutic strategy to combat muscle loss.
Acknowledgments
Funding: This work was supported by the Natural Science Foundation of Jiangsu Province (Grant No. BK202012095) and the National Natural Science Foundation of China (Grant No. 81901933, 82072160).
Footnote
Reporting Checklist: The authors have completed the ARRIVE reporting checklist. Available at http://dx.doi.org/10.21037/atm-20-7400
Data Sharing Statement: Available at http://dx.doi.org/10.21037/atm-20-7400
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/atm-20-7400). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. The experiments involving animals were conducted following the animal care guidelines of Nantong University and ethically approved by the Jiangsu Administration Committee for Experimental Animals (No. 20180305-004).
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Bodine SC. Disuse-induced muscle wasting. Int J Biochem Cell Biol 2013;45:2200-8. [Crossref] [PubMed]
- Segales J, Perdiguero E, Serrano AL, et al. Sestrin prevents atrophy of disused and aging muscles by integrating anabolic and catabolic signals. Nat Commun 2020;11:189. [Crossref] [PubMed]
- Demontis GC, Germani MM, Caiani EG, et al. Human Pathophysiological Adaptations to the Space Environment. Front Physiol 2017;8:547. [Crossref] [PubMed]
- Ebner N, Elsner S, Springer J, et al. Molecular mechanisms and treatment targets of muscle wasting and cachexia in heart failure: an overview. Curr Opin Support Palliat Care 2014;8:15-24. [Crossref] [PubMed]
- Rudrappa SS, Wilkinson DJ, Greenhaff PL, et al. Human Skeletal Muscle Disuse Atrophy: Effects on Muscle Protein Synthesis, Breakdown, and Insulin Resistance-A Qualitative Review. Front Physiol 2016;7:361. [Crossref] [PubMed]
- Scicchitano BM, Dobrowolny G, Sica G, et al. Molecular Insights into Muscle Homeostasis, Atrophy and Wasting. Curr Genomics 2018;19:356-69. [Crossref] [PubMed]
- Scicchitano BM, Faraldi M, Musaro A. The Proteolytic Systems of Muscle Wasting. Recent Adv DNA Gene Seq 2015;9:26-35. [Crossref] [PubMed]
- Bodine SC, Baehr LM. Skeletal muscle atrophy and the E3 ubiquitin ligases MuRF1 and MAFbx/atrogin-1. Am J Physiol Endocrinol Metab 2014;307:E469-84. [Crossref] [PubMed]
- Baehr LM, West DWD, Marshall AG, et al. Muscle-specific and age-related changes in protein synthesis and protein degradation in response to hindlimb unloading in rats. J Appl Physiol (1985) 2017;122:1336-50. [Crossref] [PubMed]
- Zhao J, Brault JJ, Schild A, et al. FoxO3 coordinately activates protein degradation by the autophagic/lysosomal and proteasomal pathways in atrophying muscle cells. Cell Metab 2007;6:472-83. [Crossref] [PubMed]
- Wu CL, Cornwell EW, Jackman RW, et al. NF-kappaB but not FoxO sites in the MuRF1 promoter are required for transcriptional activation in disuse muscle atrophy. Am J Physiol Cell Physiol 2014;306:C762-7. [Crossref] [PubMed]
- Weng J, Zhang P, Yin X, et al. The Whole Transcriptome Involved in Denervated Muscle Atrophy Following Peripheral Nerve Injury. Front Mol Neurosci 2018;11:69. [Crossref] [PubMed]
- Kruger K, Dischereit G, Seimetz M, et al. Time course of cigarette smoke-induced changes of systemic inflammation and muscle structure. Am J Physiol Lung Cell Mol Physiol 2015;309:L119-28. [Crossref] [PubMed]
- Fisher AG, Seaborne RA, Hughes TM, et al. Transcriptomic and epigenetic regulation of disuse atrophy and the return to activity in skeletal muscle. Faseb j 2017;31:5268-82. [Crossref] [PubMed]
- Ma W, Zhang R, Huang Z, et al. PQQ ameliorates skeletal muscle atrophy, mitophagy and fiber type transition induced by denervation via inhibition of the inflammatory signaling pathways. Ann Transl Med 2019;7:440. [Crossref] [PubMed]
- Shen Y, Zhang R, Xu L, et al. Microarray Analysis of Gene Expression Provides New Insights Into Denervation-Induced Skeletal Muscle Atrophy. Front Physiol 2019;10:1298. [Crossref] [PubMed]
- Wu C, Tang L, Ni X, et al. Salidroside Attenuates Denervation-Induced Skeletal Muscle Atrophy Through Negative Regulation of Pro-inflammatory Cytokine. Front Physiol 2019;10:665. [Crossref] [PubMed]
- Ma W, Zhang R, Huang Z, et al. PQQ ameliorates skeletal muscle atrophy, mitophagy and fiber type transition induced by denervation via inhibition of the inflammatory signaling pathways. Ann Transl Med 2019;7:440. [Crossref] [PubMed]
- Huang Z, Fang Q, Ma W, et al. Skeletal Muscle Atrophy Was Alleviated by Salidroside Through Suppressing Oxidative Stress and Inflammation During Denervation. Front Pharmacol 2019;10:997. [Crossref] [PubMed]
- Qiu J, Fang Q, Xu T, et al. Mechanistic Role of Reactive Oxygen Species and Therapeutic Potential of Antioxidants in Denervation- or Fasting-Induced Skeletal Muscle Atrophy. Front Physiol 2018;9:215. [Crossref] [PubMed]
- Xu T, Yang X, Wu C, et al. Pyrroloquinoline quinone attenuates cachexia-induced muscle atrophy via suppression of reactive oxygen species. J Thorac Dis 2018;10:2752-9. [Crossref] [PubMed]
- Qiu J, Yang X, Wang L, et al. Isoquercitrin promotes peripheral nerve regeneration through inhibiting oxidative stress following sciatic crush injury in mice. Ann Transl Med 2019;7:680. [Crossref] [PubMed]
- Shen Y, Zhang Q, Huang Z, et al. Isoquercitrin Delays Denervated Soleus Muscle Atrophy by Inhibiting Oxidative Stress and Inflammation. Front Physiol 2020;11:988. [Crossref] [PubMed]
- Gambara G, Salanova M, Ciciliot S, et al. Gene Expression Profiling in Slow-Type Calf Soleus Muscle of 30 Days Space-Flown Mice. PLoS One 2017;12:e0169314. [Crossref] [PubMed]
- Gambara G, Salanova M, Ciciliot S, et al. Microgravity-Induced Transcriptome Adaptation in Mouse Paraspinal longissimus dorsi Muscle Highlights Insulin Resistance-Linked Genes. Front Physiol 2017;8:279. [Crossref] [PubMed]
- Zhang X, Trevino MB, Wang M, et al. Impaired Mitochondrial Energetics Characterize Poor Early Recovery of Muscle Mass Following Hind Limb Unloading in Old Mice. J Gerontol A Biol Sci Med Sci 2018;73:1313-22. [Crossref] [PubMed]
- Fernandez-Gonzalo R, Tesch PA, Lundberg TR, et al. Three months of bed rest induce a residual transcriptomic signature resilient to resistance exercise countermeasures. Faseb j 2020;34:7958-69. [Crossref] [PubMed]
- Stevenson EJ, Giresi PG, Koncarevic A, et al. Global analysis of gene expression patterns during disuse atrophy in rat skeletal muscle. J Physiol 2003;551:33-48. [Crossref] [PubMed]
- Globus RK, Morey-Holton E. Hindlimb unloading: rodent analog for microgravity. J Appl Physiol (1985) 2016;120:1196-206. [Crossref] [PubMed]
- Qaisar R, Karim A, Elmoselhi AB. Muscle unloading: A comparison between spaceflight and ground-based models. Acta Physiol (Oxf) 2020;228:e13431. [Crossref] [PubMed]
- Zhang J, Li Y, Li G, et al. Identification of the optimal dose and calpain system regulation of tetramethylpyrazine on the prevention of skeletal muscle atrophy in hindlimb unloading rats. Biomed Pharmacother 2017;96:513-23. [Crossref] [PubMed]
- Roberson PA, Shimkus KL, Welles JE, et al. A time course for markers of protein synthesis and degradation with hindlimb unloading and the accompanying anabolic resistance to refeeding. J Appl Physiol (1985) 2020;129:36-46. [Crossref] [PubMed]
- Marzuca-Nassr GN, Vitzel KF, Murata GM, et al. Experimental Model of HindLimb Suspension-Induced Skeletal Muscle Atrophy in Rodents. Methods Mol Biol 2019;1916:167-76. [Crossref] [PubMed]
- Li S, Xue C, Yuan Y, et al. The transcriptional landscape of dorsal root ganglia after sciatic nerve transection. Sci Rep 2015;5:16888. [Crossref] [PubMed]
- Wang H, Zhou Y, Cong M, et al. Comparative transcriptomic profiling of peripheral efferent and afferent nerve fibres at different developmental stages in mice. Sci Rep 2018;8:11990. [Crossref] [PubMed]
- Powers SK. Can antioxidants protect against disuse muscle atrophy? Sports Med 2014;44 Suppl 2:S155-65. [Crossref] [PubMed]
- Hu NF, Chang H, Du B, et al. Tetramethylpyrazine ameliorated disuse-induced gastrocnemius muscle atrophy in hindlimb unloading rats through suppression of Ca(2+)/ROS-mediated apoptosis. Appl Physiol Nutr Metab 2017;42:117-27. [Crossref] [PubMed]
- Tomasi ML, Ramani K, Ryoo M, et al. SUMOylation regulates cytochrome P450 2E1 expression and activity in alcoholic liver disease. Faseb j 2018;32:3278-88. [Crossref] [PubMed]
- Vainshtein A, Sandri M. Signaling Pathways That Control Muscle Mass. Int J Mol Sci 2020;21:4759. [Crossref] [PubMed]
- Bohnert KR, McMillan JD, Kumar A. Emerging roles of ER stress and unfolded protein response pathways in skeletal muscle health and disease. J Cell Physiol 2018;233:67-78. [Crossref] [PubMed]
- Bohnert KR, Goli P, Roy A, et al. The Toll-Like Receptor/MyD88/XBP1 Signaling Axis Mediates Skeletal Muscle Wasting during Cancer Cachexia. Mol Cell Biol 2019;39:e00184-19. [Crossref] [PubMed]
- Ebert SM, Monteys AM, Fox DK, et al. The transcription factor ATF4 promotes skeletal myofiber atrophy during fasting. Mol Endocrinol 2010;24:790-9. [Crossref] [PubMed]
- Yang J, Li J, Cui X, et al. Blocking glucocorticoid signaling in osteoblasts and osteocytes prevents mechanical unloading-induced cortical bone loss. Bone 2020;130:115108. [Crossref] [PubMed]
- Wang KX, Shi Y, Denhardt DT. Osteopontin regulates hindlimb-unloading-induced lymphoid organ atrophy and weight loss by modulating corticosteroid production. Proc Natl Acad Sci U S A 2007;104:14777-82. [Crossref] [PubMed]
- Steffen JM, Musacchia XJ. Disuse atrophy, plasma corticosterone, and muscle glucocorticoid receptor levels. Aviat Space Environ Med 1987;58:996-1000. [PubMed]
- Schakman O, Kalista S, Barbé C, et al. Glucocorticoid-induced skeletal muscle atrophy. Int J Biochem Cell Biol 2013;45:2163-72. [Crossref] [PubMed]
- Hendrickse P, Degens H. The role of the microcirculation in muscle function and plasticity. J Muscle Res Cell Motil 2019;40:127-40. [Crossref] [PubMed]
- Gao Y, Arfat Y, Wang H, et al. Muscle Atrophy Induced by Mechanical Unloading: Mechanisms and Potential Countermeasures. Front Physiol 2018;9:235. [Crossref] [PubMed]
- Ibebunjo C, Chick JM, Kendall T, et al. Genomic and proteomic profiling reveals reduced mitochondrial function and disruption of the neuromuscular junction driving rat sarcopenia. Mol Cell Biol 2013;33:194-212. [Crossref] [PubMed]
(English Language Editor: J. Chapnick)


