
A study on the pathogenesis of blood-heat psoriasis with transcriptome analysis
Introduction
Psoriasis, also known as “Baibi” or “Niupixuan” in traditional Chinese medicine, is a common chronic inflammatory skin disease affecting 2% of the world population. Psoriasis is clinically characterized by abnormal skin scales, erythema, and itching in the affected area. Psoriasis can occur systemically, but it is more common on the scalp and extremities. Damage to the affected skin sometimes induces new psoriasis lesions, known as the Koebner phenomenon. Patients with psoriasis have a higher probability of psoriatic arthritis, cardiovascular disease, and Crohn’s disease than normal subjects (1). Its incidence rate increases year by year, without significant difference in the incidence between men and women. However, the etiology of psoriasis has not been fully clarified. Therefore, psoriasis’ occurrence and development mechanism is needed further investigation to seek new therapeutic targets.
The theory of traditional Chinese hemo-modulation therapy for psoriasis has become widely accepted in clinical practice. In the criteria of diagnostic and therapeutic effects of diseases and syndromes in traditional Chinese medicine and differential diagnosis of symptoms in traditional Chinese medicine, psoriasis is divided into blood-heat type, blood-stasis type, and blood-dryness type. There are many differences among these three types in skin lesions, T lymphocytes activation, cytokines, genomics, proteomics, neuroendocrine-immune network, vascular endothelial growth factors, et al. in the peripheral blood and skin lesions (2,3).
The complex etiology of psoriasis leads to complex therapeutic targets and mechanisms (4). Omics technology, with high throughput, may quickly and comprehensively discover the potential biomarkers related to the mechanism of psoriasis and drug efficacy. This technology provides a new research direction for the potential molecular mechanism and signaling pathways in psoriasis’ pathogenesis and accelerates drug research in psoriasis treatment. At present, some valuable biomarkers and targets of psoriasis have been discovered in the studies of metabolomics, proteinomics, genomics, transcriptomics, and the gut microbiome.
Long non-coding RNAs (lncRNAs), with more than 200 nucleotides in length, are defined as a large group of transcripts not involved in encoding proteins. Existing studies have confirmed that lncRNAs are involved in regulating various biological processes (5-8), and it is confirmed to be involved in epigenetic and regulation of transcriptional and post-transcriptional levels (9-13). lncRNAs function in various aspects of gene expression regulation through different mechanisms, including RNA-DNA interaction, RNA-RNA base pairing, and RNA-protein interaction. For example, lncRNAs and microRNAs are usually interconnected and interact to regulate gene expression (14); lncRNAs regulate gene expression in specific diseases by directly binding to mRNAs (15). However, the potential role of lncRNAs in psoriasis patients is not clear.
Circular RNAs (CircRNAs), composed of hundreds to thousands of nucleotides, are a special class of non-coding circular RNAs formed in a closed circular single-strand structure. Their expression is tissue-specific, with high sequence conservation between species. The main function of circRNA is to bind and absorb RNA-binding proteins through miRNA response elements (MRE), which was previously mainly considered the product of transcription (16). With the development of sequencing technology, biostatistics, and experimental validation technology, some whole-transcriptome circRNA recognition software has been developed, including TopHat-Fusion, Segemehl, Mapice, CIRI, et al. finding a large number of circRNAs in cells, which may be related to diseases. Many studies have also found some circRNAs can be translated (17,18), but its relationship with psoriasis’ pathogenesis has not been studied.
Genome-wide association studies (GWAS) have developed rapidly in recent years, using genotyping chip technology primarily. Representatively, studies using immunochips (Illumina) have found 15 psoriasis-related genes (19), and some researchers have identified 65 causative genes using gene expression microarrays (Affymetrix) (20) through gene enrichment analysis. Researchers have also developed some exon microarrays to study mutations in the coding regions of genes and found some rare mutations and important mutations in coding regions (21). The studies using these microarrays are cheap and suitable for the study of large-scale samples. Some pathogenic genes have been found for these studies, and psoriasis’ pathology has been clarified to some extent.
This study is the first time to explore the differential expressions of mRNAs, lncRNAs, and circRNAs between patients with blood-heat psoriasis and normal subjects using high-throughput microarray. Bioinformatics was used to study the role of these differential expressions in psoriasis’ pathogenesis to understand the pathogenesis of blood-heat psoriasis further.
We present the following article in accordance with the MDAR reporting checklist (available at http://dx.doi.org/10.21037/atm-20-7137).
Methods
General information
A total of ten patients with blood-heat psoriasis (case group) diagnosed in our hospital and ten healthy volunteers (normal group) received a physical examination in our hospital between August 2017, and June 2018 were included. There were eight males and two females in the case group, aged 19–44 years, with an average age of 29.2 years, while in the normal group, there were five males and five females, aged 24–30 years, with an average age of 26 years. Peripheral venous blood was collected from all patients and healthy subjects for high-throughput microarray. All procedures performed in this study involving human participants were in accordance with the Declaration of Helsinki (as revised in 2013). The study was approved by the Ethics Committee of Beijing Hospital of Traditional Chinese Medicine (approval no. 2017BL-071) and informed consent was taken from all the patients.
Instruments and reagents
TRIzol; RNeasy Total RNA isolation kit; RNeasy mini kit; Agilent Bioanalyzer 2100; Sino Human ceRNA array V3.0; Agilent microarray scanner; isopropanol; absolute ethanol.
Study method
High-throughput microarray to screen differential expression
The RNA from whole blood was isolated using RNeasy Total RNA isolation kit and TRIzol isolation reagent and purified using an RNeasy mini kit. The RNA integrity number (RIN) of all RNAs was detected using Agilent Bioanalyzer 2100 to check RNA integrity. Then, the RNA samples were generated into biotin-labeled cRNA targets using Sino Human ceRNA array V3.0. Biotin-labeled cRNA targets were mixed on the slides and then scanned using an Agilent microarray scanner. The scanned data were extracted using Feature Extraction software 10.7, and the original data were normalized in with the quantile using the R package “limma.”
Statistical analysis
Differential expression analysis of the samples
Microarray data were used to compare and analyze differential expression in the same transcript between two samples. There were two evaluation criteria. One was the fold change of the expression level of the same transcript in two samples. The other was a false discovery rate (FDR); the P value for each transcript was calculated, followed by FDR control for multiple P value testing. By default, P2-fold were considered the differentially expressed genes. The results were displayed as scatter plots and volcano plots.
Cluster analysis of gene expression patterns
Hierarchical clustering was performed on the transcripts with differential expression. Cluster analysis can calculate the distance and similarity between samples. The distance matrix was constructed, and the two closest categories were combined into a new category; the above steps were continuously repeated to calculate the distance between the new category and the current categories until there is only one category. The direct correlation of samples was calculated by the expression of the selected differential transcripts. In general, the samples in the same category appear in the same cluster through hierarchical clustering and transcripts in the same cluster with similar biological functions (22). The results were displayed as heat maps.
Gene ontology (GO) and KEGG enrichment analysis for the genes with differential expression
GO databases were classified according to biological processes, cell components, and molecular functions, in which genes were involved. For genes with differential expression, GO functional enrichment analysis was performed to elucidate the differences between samples at the gene function level, and pathway enrichment analysis was carried out to further understand the biological functions of genes. The main public pathway database is currently KEGG (23).
Analysis of protein-protein interaction (PPI) networks and submodules of mRNAs with differential expression
PPI network analysis of differentially expressed mRNAs in psoriasis was performed using STRING 10.0 software. The PPI network was constructed using Cytoscape software, and the degree value of each protein node was calculated to select the core genes.
Results
Screening results of mRNAs, lncRNAs, and circRNAs with differential expression in psoriasis
The peripheral venous blood from ten patients with blood-heat psoriasis and ten normal controls were collected for high-throughput microarray. The mRNAs, lncRNAs, and circRNAs with differential expression were screened and shown in the scatter plots and volcano plots (Figure 1).
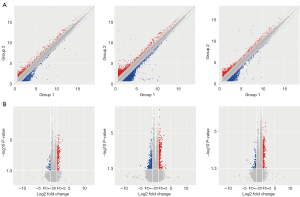
Fold change, and P value was used to select the mRNAs, lncRNAs, and circRNAs with significant expression differences, with conditions P2-fold or Table 1.

Full table
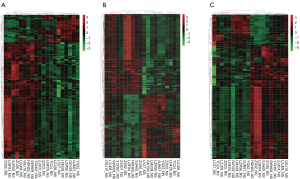
Heat maps and hierarchical cluster analysis of mRNAs, lncRNAs, and circRNAs
Hierarchical clustering was performed on the transcripts with differential expression. Red showed highly expressed transcripts, green showed lowly expressed transcripts, and the black part showed no significant difference. The same cluster samples appeared in the same cluster through hierarchical clustering and the transcripts in the same cluster with similar biological functions (Figure 2). Compared with that in the normal group, significant differences in gene mRNAs, lncRNAs, and circRNAs in the case group were identified.
GO and KEGG enrichment analysis of mRNAs, lncRNAs, and circRNAs with differential expression
GO and KEGG enrichment analysis of mRNAs with differential expression
The results of GO and KEGG enrichment analysis of mRNAs with differential expression are shown in Figure 3. The GO enrichment analysis showed mRNAs with differential expression were mainly involved in biological processes including cellular process, biological regulation, single-organism process, and regulation of the biological process involved in cellular components including cell part and organelle, and involved in molecular functions including binding. The mRNAs with differential expression are mainly enriched in the formation of ribosomes, protein localization to the endoplasmic reticulum, SRP-dependent cotranslational protein targeting the membrane, and proteins targeting estrogen receptors, indicating that these biological processes were closely related to the pathogenesis of psoriasis. KEGG pathway enrichment analysis revealed 30 biological pathways with enrichment of differentially expressed mRNAs (Figure 3D). Most pathways take part in the regulation of immune response and lipid metabolism, as well as translation. The pathways with significant enrichment are ribosome, oxidative phosphorylation, cytokine-cytokine receptor interaction, non-alcoholic fatty liver disease (NAFLD), and NF-kappa B signaling pathway.
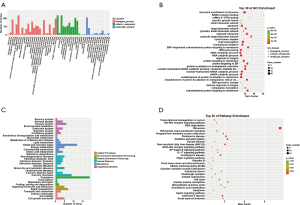
GO and KEGG enrichment analysis of cis target genes of lncRNAs with differential expression
The results of GO and KEGG enrichment analysis of cis target genes of lncRNAs with differential expression are shown in Figure 4. The GO enrichment analysis showed that cis target genes of lncRNAs with differential expression were primarily involved in biological processes, including cellular process, biological regulation, single-organism process, and regulation of the biological process. Cis target genes of lncRNAs with differential expression were mainly enriched in membrane coat, negative regulation of protein binding, response to nutrients, regulation of cell cycle checkpoints, and negative regulation of binding, indicating that these biological processes were closely related to the pathogenesis of psoriasis. KEGG pathway enrichment analysis revealed 30 biological pathways with enrichment of cis target genes of lncRNAs with differential expression (Figure 3D). Most pathways took part in the regulation of immune response and signal transduction, as well as metabolism. The pathways with significant enrichment are Salmonella infection, Ribrine, Rap1 signaling pathway, Platelet activation, Endocrine, and other calcium absorption factor-regulated reosomes.
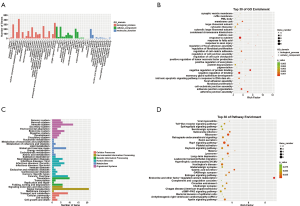
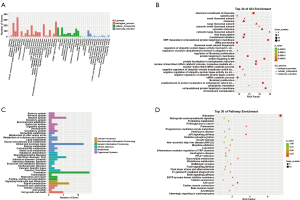
GO and KEGG enrichment analysis of trans target genes of lncRNAs with differential expression
The results of GO and KEGG enrichment analysis of trans target genes of lncRNAs with differential expression are shown in Figure 5. The GO enrichment analysis showed that trans target genes of lncRNAs with differential expression were mainly involved in biological processes including cellular process, biological regulation, single-organism process, and the metabolic process involved in cellular components, including organelle involved in molecular functions including binding and catalysis. Trans target genes of lncRNAs with differential expression were mainly enriched in aspartic endopeptidase activity, intramembrane cleavage, cytoplasmic exosomes (RNase complexes), regulation of amyloid fibril formation, maintenance of protein location in the nucleus, and mitochondrial RNA processing, indicating that these biological processes were closely related to the pathogenesis of psoriasis. KEGG pathway enrichment analysis revealed 30 biological pathways with enrichment of trans target genes of lncRNAs with differential expression (Figure 5D). Most pathways took part in the regulation of immune response and signal transduction, as well as metabolism. The pathways with significant enrichment were the p53 signaling pathway, Platinum drug resistance, Lysosome, Tryptophan metabolism, Valine, leucine, and isoleucine degradation.
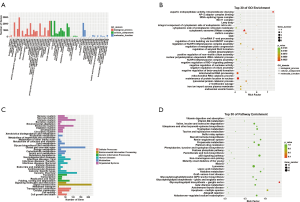
GO and KEGG enrichment analysis of circRNA target genes with differential expression
The results of GO and KEGG enrichment analysis of circRNA target genes with differential expression are shown in Figure 6. The GO enrichment analysis showed circRNA target genes with differential expression were mainly involved in biological processes including cellular process, biological regulation, single-organism process and regulation of the biological process, cellular components including cells, and molecular dynamics functions as binding. CircRNA target genes with differential expression were mainly enriched in the structural composition of ribosomes, cytoplasmic ribosomes, SRP-dependent cotransproteins targeting the membrane, and cotransproteins targeting the membrane, indicating that these biological processes were related to the pathogenesis of psoriasis. KEGG pathway enrichment analysis revealed 30 biological pathways with enrichment of circRNA target genes with differential expression (Figure 6D). Most of the pathways regulate immune response and signal transduction, metabolism, translation processes, cell growth, and apoptosis. The pathways with significant enrichment were the p53 signaling pathway, Proteasome, Cell cycle, Glycerolipid metabolism, Oocyte meiosis.
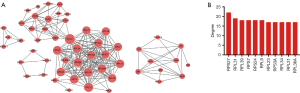
Network plot of mRNAs with differential expression
To obtain the core differentially expressed genes in differentially expressed mRNAs, the PPI network plot was performed in Figure 7A and revealed the top 11 core differentially expressed genes (RPS27, RPL31, RPL39, RPS7, RPS24, RPL9, RPL23, RPS3A, RPL34, RPL21, and RPL36A) according to the level of node degree (degree). The degree exceeded 15, indicating a close correlation between proteins (Figure 7B).
Discussion
Psoriasis is a chronic inflammatory disease characterized by keratinocyte proliferation and inflammatory cell infiltration. In traditional Chinese medicine, psoriasis is divided into blood-heat type, blood-dryness type, and blood-stasis type. The blood-heat type is common in psoriasis’s progressive stage, characterized by the rapid development of skin lesions covered with more scales, erythema, and severe itching, mainly isomorphic reactions. Blood-heat psoriasis is treated with clearing heat, cooling blood, activating blood, and Liangxuehuoxue decoction. Blood-dryness psoriasis is common in the quiescent and regression stage, with a protracted course, less new skin lesions, some skin lesions disappeared. Blood-dryness psoriasis is appropriate to be treated with nourishing blood, nourishing yin, moistening dryness, and Liangxuejiedu decoction was applied. The blood-stasis type is common in the quiescent stage, with prolonged skin lesions in patchy and dark red, covered with thick scales. Blood-stasis psoriasis is appropriate to be treated with activating blood, absorbing clots, promoting qi, and Huoxuesanyu decoction was applied.
At present, psoriasis is generally recognized as a disease involving multiple pathways; the discovered signaling pathways related to psoriasis include Janus kinase-signal transduction and activator of transcription (JAK-STAT) signaling pathway, mitogen-activated protein kinase (MAPK) signaling pathway, PI3K/Akt signaling pathway, nuclear transcription factor kappa B (NF-KappaB) pathway (24,25). However, the signaling pathways involved are complex, which is still to be further studied. Currently, psoriasis diagnosis primarily depends on clinical manifestations and pathological examination, with low sensitivity and specificity. Therefore, researchers continue to find biological markers for their diagnosis and find the corresponding therapeutic targets.
Existing studies have shown that lncRNA plays a key role in the pathogenesis and development of many diseases, especially tumors and autoimmune-related diseases (26,27). Psoriasis has a significant genetic susceptibility. lncRNAs are involved in psoriasis’ pathogenesis (28). In the study of lncRNA and psoriasis, including PRINS, TINCR, and RNA-MSX2P1, these RNAs are crucial in psoriasis behavior’s biological regulation (29-31). However, the correlation of psoriasis and the abnormal expression of lncRNA has not been fully clarified, and the regulatory role and mechanism of lncRNA in psoriasis need further in-depth study. Meanwhile, few studies on the mechanism by which circRNA plays in psoriasis also need our further study.
With the advent of the post-genomics era, gene chips and high-throughput sequencing have become mature and widely popularized, becoming important technologies in molecular biology research. High throughput sequencing has developed later than gene chips, it has become a hotspot in biological gene research due to its economy, efficiency, and rapid development (32). However, in this paper, for the first time, peripheral venous blood from ten patients with blood-heat psoriasis (eight males and two females) and ten healthy controls (five males and five females) were collected for high-throughput microarray to further clarify the mRNAs, lncRNAs, and circRNAs with abnormal expression in psoriasis for its stability and accuracy. The results showed 205 circRNAs, 393 lncRNAs, and 157 mRNAs with differential expression between the case and normal groups, indicating that many mRNAs, lncRNAs, and circRNAs with differential expression were in the blood-heat psoriasis patients and were involved in the occurrence and development of blood-heat psoriasis.
lncRNA has confirmed to be involved in many biological processes; it plays a critical role in epigenetics and transcription by cis- and trans- acting on transcriptional regulation and constitutes a network of ribonucleic acid complexes (33,34). Qiu et al. (35) found that the percentage of CD4+ T lymphocytes in the peripheral blood of patients with blood-heat psoriasis was higher than the blood-dryness and blood-stasis types, and there was no significant difference between the blood-dryness group and the blood-stasis group; no significant differences were identified in CD8+ and CD3+ T cells among the three groups. In the present paper, the enrichment analysis of the selected mRNAs, lncRNAs (cis and trans), and circRNAs with differential expression was performed to obtain the top 30 GO terms. The mRNAs, cis, and trans target genes of lncRNAs, and circRNA target genes with differential expression were mainly involved in biological processes including cellular processes, biological regulation, and regulation of the biological process, involved in cellular components including organelle, and involved in molecular functions including binding and catalysis. These RNAs lead to immune system disorders in psoriasis patients primarily through inducing the proliferation and division of lymphocytes and immune cells. Gu et al. (36) selected key biomarkers (9) from the metabolomics experiment results and enzymes and key proteins screened by the network database. Then, molecular docking was used to find the proteins and enzymes with high binding energy after calculating; the binding energy-weighted polypharmacological index was introduced to assess the correlation of pathways, and the result showed the PI3K-Akt signaling pathway and five other pathways might be the key pathways in the development of psoriasis. In our study, KEGG pathway enrichment analysis revealed 30 biological pathways with significant enrichment of differently expressed mRNAs, lncRNAs (cis and trans), and circRNAs. Most pathways involve regulating immune response and lipid metabolism, translation, cell growth, and apoptosis. Therefore, in patients with blood-heat psoriasis, the genes with differential expression damage the lipids and proteins that constitute the mitochondrial inner membrane and promote changes in mitochondrial inner membrane fluidity and permeability, thereby promoting apoptosis and causing metabolic disorders. Also, inflammatory factors and adipokines are closely related to the occurrence and development of psoriasis and metabolic syndrome and participate in regulating various pathways. The above results further reveal the close relationship between psoriasis and metabolic syndrome at the gene level. Further, the 11 core differentially expressed genes in psoriasis were obtained through a PPI network of mRNAs with differential expression according to the level of network nodes, including RPS27, RPL31, RPL39, RPS7, RPS24, RPL9, RPL23, RPS3A, RPL34, RPL21, and RPL36A, which are mainly involved in RNA formation.
Conclusions
It is the first time to collect the whole blood genes from patients with blood-heat psoriasis and normal subjects for high-throughput microarray. A total of 205 circRNAs, 393 lncRNAs, and 157 mRNAs were found to be differentially expressed in blood-heat psoriasis, showing that they are involved in the pathogenesis of blood-heat psoriasis. GO analysis of mRNAs, lncRNAs, and circRNAs with differential expression showed they are involved in cellular processes, biological regulation, single-organism processes, and regulation of biological process regulation, suggesting that these biological processes play an important role in the development of blood-heat psoriasis. On KEGG analysis, mRNAs, lncRNAs, and circRNAs with differential expression are enriched in 30 biological pathways, and most pathways are associated with autoimmune reactions and lipid metabolism, and signal transduction. The PPI network of mRNAs with differential expression reveals 11 core differential genes. Although with the limitations, including the small sample size, this is a pilot study to explore the differentially expressed transcripts, providing a preliminary basis for the preliminary exploration of mRNAs, lncRNAs, and circRNAs in blood-heat psoriasis. Meanwhile, this study investigates the material basis, molecular function, and biological process in the occurrence and development of blood-heat psoriasis at the gene level, providing a reference for the mechanism of its pathogenesis and subsequent studies. High-throughput analysis obtains rich data, and many valuable data still can be further studied.
Acknowledgments
Funding: This work was supported by National Natural Science Foundation of Beijing (grant number 7171003); Capital’s Funds for Health Improvement and Research (CFH2018-2-1162); the Beijing Traditional Chinese Medicine Science and Technology Development Fund Project (QN2018-21).
Footnote
Reporting Checklist: The authors have completed the MDAR reporting checklist. Available at http://dx.doi.org/10.21037/atm-20-7137
Data Sharing Statement: Available at http://dx.doi.org/10.21037/atm-20-7137
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/atm-20-7137). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. All procedures performed in this study involving human participants were in accordance with the Declaration of Helsinki (as revised in 2013). The study was approved by the Ethics Committee of Beijing Hospital of Traditional Chinese Medicine (approval No. 2017BL-071) and informed consent was taken from all the patients.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Nestle FO, Kaplan DH, Barker J. Psoriasis. N Engl J Med 2009;361:496-509. [Crossref] [PubMed]
- Boehncke WH, Schön MP. Psoriasis. Lancet 2015;386:983-94. [Crossref] [PubMed]
- Yue T, Ji M, Qu H, et al. Comprehensive analyses of long non-coding RNA expression profiles by RNA sequencing and exploration of their potency as biomarkers in psoriatic arthritis patients. BMC Immunol 2019;20:28. [Crossref] [PubMed]
- Lowes MA, Bowcock AM, Krueger JG. Pathogenesis and therapy of psoriasis. Nature 2007;445:866-73. [Crossref] [PubMed]
- Wang M, Sun X, Wang H, et al. Long non-coding RNAs in non-small cell lung cancer: functions and distinctions from other malignancies. Transl Cancer Res 2019;8:2636-53. [Crossref]
- Leti F, Legendre C, Still CD, et al. Altered expression of MALAT1 lncRNA in nonalcoholic steatohepatitis fibrosis regulates CXCL5 in hepatic stellate cells. Transl Res 2017;190:25-39.e21. [Crossref] [PubMed]
- Vennin C, Adriaenssens E. Long non-coding RNA and messenger RNA-the meeting of two worlds. J Thorac Dis 2018;10:544-6. [Crossref] [PubMed]
- Li LJ, Zhao W, Tao SS, et al. Comprehensive long non-coding RNA expression profiling reveals their potential roles in systemic lupus erythematosus. Cell Immunol 2017;319:17-27. [Crossref] [PubMed]
- Fatica A, Bozzoni I. Long non-coding RNAs: new players in cell differentiation and development. Nat Rev Genet 2014;15:7-21. [Crossref] [PubMed]
- Luo Z. Regulatory mechanisms of long non-coding RNAs. Oncology and Translational Medicine 2019;005:147-51.
- Yong Z, Ying Z, Hua SL, et al. Long noncoding RNAs: emerging players in cardiac electrical and structural remodeling. Chinese Journal of Pharmacology Toxicology 2019. Available online: http://en.cnki.com.cn/Article_en/CJFDTotal-YLBS201910096.htm
- Zhao XY, Lin JD. Long Noncoding RNAs: A New Regulatory Code in Metabolic Control. Trends Biochem Sci 2015;40:586-96. [Crossref] [PubMed]
- Xie Y, Yao J, Zhang X, et al. A panel of extracellular vesicle long noncoding RNAs in seminal plasma for predicting testicular spermatozoa in nonobstructive azoospermia patients. Hum Reprod 2020. [Crossref] [PubMed]
- Wang Z, Li B, Li Y, et al. Identification and characterization of long noncoding RNA in Paulownia tomentosa treated with methyl methane sulfonate. Physiol Mol Biol Plants 2018;24:325-34. [Crossref] [PubMed]
- Zhang Y, Pitchiaya S, Cieślik M, et al. Analysis of the androgen receptor-regulated lncRNA landscape identifies a role for ARLNC1 in prostate cancer progression. Nat Genet 2018;50:814-24. [Crossref] [PubMed]
- Wu DM, Wen X, Han XR, et al. Role of Circular RNA DLEU2 in Human Acute Myeloid Leukemia. Mol Cell Biol 2018;38:e00259-18. [Crossref] [PubMed]
- Hansen TB, Jensen TI, Clausen BH, et al. Natural RNA circles function as efficient microRNA sponges. Nature 2013;495:384-8. [Crossref] [PubMed]
- Li X, Yang L, Chen LL. The Biogenesis, Functions, and Challenges of Circular RNAs. Mol Cell 2018;71:428-42. [Crossref] [PubMed]
- Tsoi LC, Spain SL, Knight J, et al. Identification of 15 new psoriasis susceptibility loci highlights the role of innate immunity. Nat Genet 2012;44:1341-8. [Crossref] [PubMed]
- Chen W, Xie K, Liu X, et al. Identification of key pathways and genes in psoriasis via gene microarray analysis. Mol Med Rep 2016;13:2327-37. [Crossref] [PubMed]
- Zuo X, Sun L, Yin X, et al. Whole-exome SNP array identifies 15 new susceptibility loci for psoriasis. Nat Commun 2015;6:6793. [Crossref] [PubMed]
- Li J, Li X, Feng X, et al. A novel target convergence set based random walk with restart for prediction of potential LncRNA-disease associations. BMC Bioinformatics 2019;20:626. [Crossref] [PubMed]
- Liu YZ, Zhang L, Roy-Engel AM, et al. Carcinogenic effects of oil dispersants: A KEGG pathway-based RNA-seq study of human airway epithelial cells. Gene 2017;602:16-23. [Crossref] [PubMed]
- Zhao H, Mitra N, Kanetsky PA, et al. A practical approach to adjusting for population stratification in genome-wide association studies: principal components and propensity scores (PCAPS). Stat Appl Genet Mol Biol 2018. [Crossref] [PubMed]
- Johansen C, Rittig AH, Mose M, et al. STAT2 is involved in the pathogenesis of psoriasis by promoting CXCL11 and CCL5 production by keratinocytes. PLoS One 2017;12:e0176994. [Crossref] [PubMed]
- Guttman M, Rinn JL. Modular regulatory principles of large non-coding RNAs. Nature 2012;482:339-46. [Crossref] [PubMed]
- Ricciuti B, Mencaroni C, Paglialunga L, et al. Long noncoding RNAs: new insights into non-small cell lung cancer biology, diagnosis and therapy. Med Oncol 2016;33:18. [Crossref] [PubMed]
- Harden JL, Krueger JG, Bowcock AM. The immunogenetics of Psoriasis: A comprehensive review. J Autoimmun 2015;64:66-73. [Crossref] [PubMed]
- Sonkoly E, Bata-Csorgo Z, Pivarcsi A, et al. Identification and characterization of a novel, psoriasis susceptibility-related noncoding RNA gene, PRINS. J Biol Chem 2005;280:24159-67. [Crossref] [PubMed]
- Széll M, Danis J, Bata-Csörgő Z, et al. PRINS, a primate-specific long non-coding RNA, plays a role in the keratinocyte stress response and psoriasis pathogenesis. Pflugers Arch 2016;468:935-43. [Crossref] [PubMed]
- Danis J, Göblös A, Bata-Csörgő Z, et al. PRINS Non-Coding RNA Regulates Nucleic Acid-Induced Innate Immune Responses of Human Keratinocytes. Front Immunol 2017;8:1053. [Crossref] [PubMed]
- Lacarte MG. miRNAs as predictive biomarkers of obesity and weight loss: epigenetic regulation and target gene identification. 2018. Available online: https://hdl.handle.net/10171/56410
- Tsoi LC, Iyer MK, Stuart PE, et al. Analysis of long non-coding RNAs highlights tissue-specific expression patterns and epigenetic profiles in normal and psoriatic skin. Genome Biol 2015;16:24. [Crossref] [PubMed]
- Yuan DM, Ma J, Fang WB. Identification of non-coding RNA regulatory networks in pediatric acute myeloid leukemia reveals circ-0004136 could promote cell proliferation by sponging miR-142. Eur Rev Med Pharmacol Sci 2019;23:9251-8. [PubMed]
- Qiu B. The Relationship of Peripheral Blood T Cell Subsets with Different TCM Syndromes of Psoriasis Vulgaris. Chinese Medicine Modern Distance Education of China 2014. Available online: http://en.cnki.com.cn/Article_en/CJFDTotal-ZZYY201411015.htm
- Gu J, Li L, Wang D, et al. Deciphering metabonomics biomarkers-targets interactions for psoriasis vulgaris by network pharmacology. Ann Med 2018;50:323-32. [Crossref] [PubMed]
(English Language Editor: J. Chapnick)


