
This article has an erratum available at: http://dx.doi.org/10.21037/atm-2022-34 the article has been update on 2022-09-05 at here.
miR-126-3p contributes to sorafenib resistance in hepatocellular carcinoma via downregulating SPRED1
Introduction
Since being approved for the treatment of hepatocellular carcinoma (HCC) by the U.S. Food and Drug Administration (FDA), sorafenib has become the standard treatment for patients in the advanced stages of the disease (1,2). Numerous clinical studies have shown that sorafenib prolongs the survival of patients with advanced HCC; however, due to primary or acquired drug resistance, the effectiveness of sorafenib is limited. A variety of factors have been shown to be closely correlated with sorafenib resistance, including epithelial-mesenchymal transition (EMT), hypoxia, cancer stem cell (CSC) generation, and activation of signaling pathways such as the PI3K/AKT, NF-κB, and MAPK pathways (3). Therefore, more efforts to clarify the mechanism of sorefenib resistance are called for to improve the effectiveness of sorafenib for a larger proportion of advanced HCC patients.
MicroRNAs (miRNAs) are short, conserved, noncoding RNAs that regulate gene expression at the post-transcriptional level. Recent studies have indicated that miRNAs possibly play critical roles in regulating sorafenib resistance in HCC through involvement in signaling pathways and other mechanisms (4,5). For instance, one study reported that miR-122 and miR-181a caused sorafenib resistance in HCC through regulating RAS/RAF/ERK signaling pathways, and another suggested that targeting miR-122 may improve the effectiveness of sorafenib in patients with HCC (6,7). Qiu et al. also showed that miR-16 could reverse sorafenib resistance by inhibiting HIF-1α-induced generation of cancer stem cells (8). Thus, the targeting of specific miRNAs may become an effective therapeutic method for overcoming sorafenib resistance.
miR-126-3p has been shown to be involved in the processes of inflammation and angiogenesis in numerous cancers, but it appears to have different effects in different cancers (9). Studies have demonstrated that miR-126-3p suppresses cell proliferation and metastasis in breast cancer and colorectal cancer (10,11). However, in leukemia, lung cancer, and HCC, miR-126-3p has been shown to contribute to tumor progression (12-14). It has also been reported that miR-126-3p plays a crucial role in regulating the response to anticancer therapy (15,16), but it is unclear if miR-126-3p is involved in sorafenib resistance in HCC.
Sprouty-related EVH1 domain-containing protein 1 (SPRED1), which is a widely known negative regulator of the Ras/Raf/ERK signaling pathway, has been shown to have a low expression in various tumor tissues (17,18). A recent study identified that SPRED1 loss was a driver of mucosal melanoma; knockdown of SPRED1 led to MAPK activation, increased cell proliferation, and promoted resistance to tyrosine kinase inhibitors (19). Another study demonstrated that SPRED1 was downregulated in HCC and that it negatively regulated HCC cell proliferation, invasion, and metastasis (20). Moreover, a recent study of HCC patients found that the mRNA level of SPRED1 was inversely correlated with that of miR-126-3p (21). Nevertheless, whether SPRED1 expression is related to resistance or sensitivity to sorafenib in HCC is still unknown.
In the present study, we found that miR-126-3p was significantly upregulated in HCC patients who were resistant to sorafenib. Bioinformatics analysis and dual luciferase reporter assay confirmed that SPRED1 was the target of miR-126-3p. We further demonstrated that miR-126-3p prompted sorafenib resistance by inhibiting SPRED1 expression and activating the ERK signaling pathway. We present the following article in accordance with the ARRIVE reporting checklist (available at http://dx.doi.org/10.21037/atm-20-2081).
Methods
Cell culture and reagents
Five HCC cell lines (HepG2, Hep3B, Huh-7, SK-HEP-1, and MHCC97H) were cultured in Dulbecco’s Modified Eagle Medium (DMEM; Gibco BRL, Grand Island, NY, USA) supplemented with 10% fetal bovine serum (FBS, Gibco BRL). Sorafenib was purchased from APExBIO Technology (TX, USA). The sorafenib reagent was dissolved in dimethyl sulfoxide (DMSO) at a dose of 100 µmol/L for storage, and was diluted to working concentrations with DMEM. SCH772984, an ERK inhibitor, was purchased from Selleck Chemicals (Houston, TX, USA) and used at concentration of 1 µmol/L, as recommended (22).
Bioinformatics analysis
Gene Expression Omnibus (GEO) is a public database containing high-throughput gene expression data, gene chips analyses, and microarray data. We conducted a search of the GEO database using the keywords “microRNA” “sorafenib” and “HCC”, and identified the dataset, GSE56059, which was then analyzed with the Limma package (version: 3.40.2) in R software. Differentially expressed miRNAs were analyzed between sorafenib nonresponse HCC patients [progressive disease (PD)] and sorafenib response HCC patients [partial response (PR) or stable disease (SD)] (23). Fold-change (FC) between the two groups was calculated for each gene. Then, the differentially expressed genes were filtered for P<0.05 and FC >1.5. Next, candidate target genes of miR-126-3p were identified using three online programs: TargetScan 7.2 (http://www.targetscan.org) (24), miRDB (http://www.mirdb.org), and StarBase v2.0 (http://starbase.sysu.edu.cn) (25).Table S1 shows the predicted target genes that overlapped in the three programs.
Cell transfection
SPRED1 small interfering (siRNA), and miR-126-3p mimics and inhibitors were designed and synthesized by RiboBio (Guangzhou, China). The plasmid vector pCDH-MSCV-MCS-EF1-copGFP-T2A-Puro carrying the full-length complementary DNA of human SPRED1 was purchased from GenePharma (Shanghai, China). Lentivirus microRNA inhibitor of miR-126-3p was obtained from Sigma-Aldrich (Germany) and then cloned into the pLKO.1-puro vector. The 293T cell line was then transfected with either SPRED1 overexpression plasmid, lenti-miR-126-3p plasmid, or the blank plasmid together with assistant plasmids (psPAX2 and pMD2.G) to produce viral particles. Viral supernatants were collected and transfected into HCC cells. The infected cells were selected with 2 µg/mL of puromycin. The siRNAs, plasmids, and microRNA mimics and inhibitors were transfected into HCC cells using Lipofectamine2000 (Invitrogen, USA) according to the manufacturer’s instructions. The sequences of the siRNAs and microRNA mimics and inhibitors are listed in Table S2.
Quantitative reverse transcription polymerase chain reaction (qRT-PCR)
Total RNA was isolated using TRIzol reagent (Takara, Japan) and reversed transcribed into cDNA using PrimerScript RT Master (Takara). Gene expressions were examined using SYBR Premix Ex Taq (Takara) by quantitative reverse transcription polymerase chain reaction (qRT-PCR). The primers sequences of target genes are listed in Table S3.
Cell viability and cell proliferation assays
To assess cell viability, HepG2 and MHCC97H cells were seeded onto a 96-well plate at a density of 1×104 cells/well. The cells were treated with various concentrations of sorafenib for 48 hours. To determine cell proliferation, HepG2 and MHCC97H cells were seeded onto a 96-well plate at a density of 2×103 cells/well. The cells were then divided into different groups and cultured for 24, 48, or 72 hours. After washing the cells twice with phosphate-buffered saline (PBS), 100 µL of DMEM containing 10 µL of Cell Counting Kit-8 (CCK8) reagent was added to each well. After incubation for 1 hour at 37 °C, the absorbance was measured at 450 nm using an automated enzyme-linked immunosorbent assay (ELISA) plate reader.
EdU proliferation assay
The EdU assay was performed using a BeyoClick™ EdU Cell Proliferation Kit with Alexa Fluor 488 (Beyotime, Jiangsu, China) according to the manufacturer’s protocol. Briefly, HCC cells were seeded onto a 24-well plate and treated for 48 hours. After that, the cells were incubated with 10 µmol/L EdU diluted in DMEM for a further 2 hours. Then, the cells were fixed with 4% paraformaldehyde for 20 min and permeabilized with 0.3% Triton X-100 for 20 min at room temperature. After being washed three times with PBS, the cells were incubated with 0.5 mL of Click Reaction Mixture for 30 min. Subsequently, the cells were stained with Hoechst (1:1,000) for 2 min and observed under a fluorescence microscope (Olympus, Tokyo, Japan). The numbers of EdU-positive cells were counted in three random fields of view per slide, and the percentage of EdU-positive cells was calculated.
Dual luciferase reporter assay
Wild-type and mutational SPRED1 mRNA 3’-UTR luciferase reporter gene plasmids were constructed by GenePharma Inc (Shanghai, China). Briefly, 293T cells were cotransfected with pSPRED1-3’UTR-WT/miR-126-3p mimics or pSPRED1-3’UTR-MUT/miR-126-3p mimics using Lipofectamine-2000 in 96-well plates. After 48 hours, luciferase assay was performed using a Dual-Luciferase Reporter Gene Assay Kit (Beyotime, China) according to the manufacturer’s instructions. Renilla luciferase served as a control reporter for normalization, and the relative luciferase activity was calculated as the ratio of the firefly and Renilla luciferase activities.
Western blotting and immunohistochemical (IHC)
Western blotting and immunohistochemistry (IHC) assays were performed as described in our previous study (26). Antibody Sampler Kit (#9926) was purchased from Cell Signaling Technology (Danvers, MA, USA). The primary antibodies against SPRED1 and glyceraldehyde 3-phosphate dehydrogenase (GAPDH), and the horse-radish peroxidase (HRP)-labeled anti-rabbit secondary antibodies were obtained from ABclonal (Wuhan, China).
Animal experiments
Animal experiments were approved by the Bioethics Committee of Sun Yat-Sen University and were performed according to the National Institutes of Health guidelines for the use and care of laboratory animals (approval number: SYSU-IACUC-2019-B182). MHCC97H cells transfected with LV-vector or LV-miR-126-3p inhibitor (5×106) were injected subcutaneously into 4–6 weeks old BALB/C nude mice. Sorafenib was diluted in an oral vehicle containing Cremophor (Sigma), ethanol, and ddH2O at a ratio of 1:1:6. The mice injected with MHCC97H-LV-vector cells were then randomly divided into two groups: the control group (ddH2O, orally) and the sorafenib group (30 mg/kg/d, orally). The mice injected with MHCC97H-LV-miR-126-3p inhibitor were administered 30 mg/kg/d of sorafenib. Tumor volumes were measured every 3 days and were calculated using the following equation: V (mm3) = 0.5 × length × width2. Finally, the mice were sacrificed after 2 weeks of treatment, and the solid tumors were weighed and fixed in formaldehyde.
Statistical analysis
All data are presented as the mean ± standard deviation. Data were compared using Student’s t-test or two-way analysis of variance (ANOVA) with Bonferroni correction. All analyses were performed using IBM SPSS Statistics version 20.0 software. A P value of < 0.05 was considered to be statistically significant.
Results
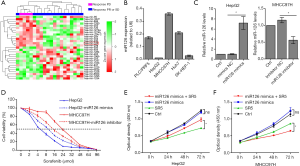
High expression of miR-126-3p was positively correlated with sorafenib resistance in HCC
Differentially expressed miRNAs were analyzed between sorafenib nonresponse HCC patients (PD) and sorafenib response HCC patients (PR or SD), and the results are shown in Figure 1. In our previous study, we demonstrated that miR-1226 was markedly downregulated in the PD group and could promote sorafenib sensitivity in HCC (27). Here, we found that miR-126-3p was highly expressed in the PD group. To determine whether miR-126-3p was involved in sorafenib sensitivity in HCC cells, we first used qRT-PCR to determine miR-126-3p expression levels in a panel of human HCC cell lines. The results showed that HepG2 cells exhibited the lowest expression of miR-126-3p, while MHCC97H cells exhibited the highest expression (Figure 1B). Figure 1D shows the effect of sorafenib on the viability of HepG2 and MHCC97H cells; the IC50 value of MHCC97H cells (22.54 µmol/L) was much higher than that of HepG2 cells (5.48 µmol/L). However, the IC50 value of MHCC97H cells transfected with miR-126-3p inhibitor decreased to 13.26 µmol/L, and the IC50 value of HepG2 cells transfected with miR-126-3p mimics increased to 9.03 µmol/L. Moreover, the CCK8 assay showed that overexpression of miR-126-3p significantly decreased the antiproliferative effect of sorafenib in HepG2 cells, while miR-126-3p inhibitor markedly enhanced the antitumor effect of sorafenib in MHCC97H cells (Figure 1E).
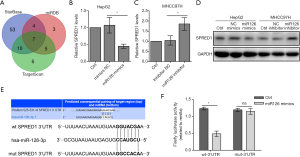
SPRED1 was a direct target of miR-126-3p
The direct target of miR-126-3p was predicted with the online programs TargetScan, miRDB, and StarBase v2.0. Seven genes (KANK2, ADAM9, SLC7A5, SPRED1, PLXNB2, ITGA6, and CRK) were predicted by all three programs (Figure 2A, Table S3). A recent study indicated that the mRNA level of SPRED1 was inversely correlated with that of miR-126-3p in HCC patients (21). Therefore, SPRED1 was selected for further experiments. The results of qRT-PCR and Western blotting assays showed that miR-126-3p mimics could inhibit the expression of SPRED1 in HepG2 cells, whereas miR-126-3p inhibitor markedly increased the expression of SPRED1 in MHCC97H cells (Figure 2B,C,D). Next, we performed a dual-luciferase reporter assay using reporter plasmid containing a wild-type or mutant sequence of 3’UTR region of SPRED1 (Figure 2E). The results showed that miR-126-3p mimic markedly reduced the firefly luciferase activity in the wild-type group; however, it had no effect on firefly luciferase activity in the mutant group (Figure 2F).
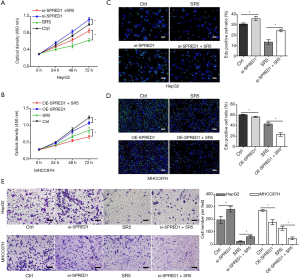
Upregulation of SPRED1 enhanced the antitumor effect of sorafenib against HCC
To determine whether SPRED1 was involved in sensitivity of HCC cells to sorafenib, we first determined the expression levels of SPRED1 in HepG2 and MHCC97H cells. The expression of SPRED1 was greater in the HepG2 cells than in the MHCC97H cells (Figure S1). The CCK-8 proliferation assay demonstrated that the downregulation of SPRED1 markedly impaired the antitumor effect of sorafenib in HepG2 cells (Figure 3A), whereas overexpression of SPRED1 enhanced the antitumor effect of sorafenib in MHCC97H cells (Figure 3B). The EdU assay demonstrated that sorafenib at 5 µmol/L significantly reduced the ratio of proliferating cells to 43% compared with the control in HepG2 cells. Conversely, the anti-tumor effect of sorafenib in HepG2 cells was markedly decreased after transfection with SPRED1 siRNA (Figure 3C). Additionally, sorafenib at 5 µmol/L could only reduce the ratio of proliferating cells to 71% compared with the control. However, in MHCC97H cells transfected with the SPRED1 overexpression plasmid, sorafenib decreased the proliferation inhibition rate to 38% compared with the control (Figure 3D). The Transwell migration and invasion assays consistently demonstrated that inhibiting SPRED1 markedly impaired the effect of sorafenib on the cell mobility and invasion ability of HepG2 cells, whereas upregulation of SPRED1 enhanced the effect of sorafenib on the cell mobility and invasion ability of MHCC97H cells (Figure 3E, Figure S1D,E).
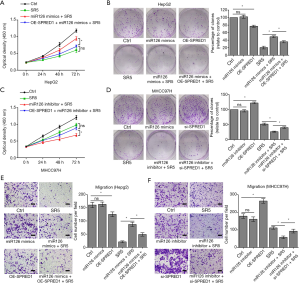
The effect of miR-126-3p on the sensitivity of HCC cells to sorafenib was dependent on the regulation of SPRED1 expression
To verify that miR-126-3p induced sorafenib resistance through downregulating SPRED1, rescue assays were performed. Consistent with the above-mentioned results, miR-126-3p u-regulation markedly impaired the effect of sorafenib in HepG2 cells. However, cotransfection with SPRED1 overexpression plasmid and miR-126-3p mimic could resensitize HepG2 cells to the antitumor effect of sorafenib (Figure 4A,B). In the MHCC97H cells, CCK8 and colony-formation assays both showed that downregulation of miR-126-3p increased the antitumor effect of sorafenib treatment, while cotransfection with SPRED1 siRNA and miR-126-3p inhibitor almost completely abrogated this increased sensitivity to sorafenib (Figure 4C,D). Furthermore, Transwell migration and invasion assays of HepG2 cells treated with sorafenib showed that the increased cell mobility and invasion ability induced by the overexpression of miR-126-3p was reversed by the upregulation of SPRED1 (Figure 4E, Figure S2A). Transwell assay also found similar results in MHCC97H cells (Figure 4F, Figure S2B). Collectively, these data suggested that miR-126-3p affects sorafenib sensitivity through regulating SPRED1 expression.
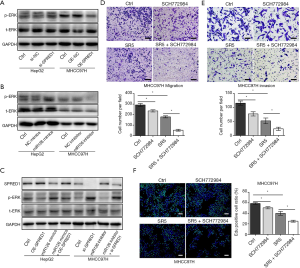
miR-126-3p/SPRED1 influenced sorafenib sensibility by modulating the ERK signaling pathway
Studies have shown that the Ras/Raf/MAPK signaling pathway is the key target of sorafenib and responsible for its antitumor effects (28). Therefore, we investigated the effect of miR-126-3p on this signaling pathway. Western blotting analysis demonstrated that inhibition of SPRED1 induced ERK phosphorylation in HepG2 cells, while overexpression of SPRED1 markedly reduced the phospho-ERK (p-ERK) levels in MHCC97H cells (Figure 5A). Moreover, our findings also revealed that upregulation of miR-126-3p markedly increased p-ERK in HepG2 cells, while the downregulation of miR-126-3p significantly decreased p-ERK in MHCC97H cells (Figure 5B). We also performed rescue assays to investigate the role of miR-126-3p/SPRED1/p-ERK signaling pathway. The results showed that miR-126 mimics inhibited the expression of SPRED1 and increased the p-ERK levels in HepG2 cells, while simultaneously, the overexpression of SPRED1 impaired the increase of p-ERK levels caused by miR-126 mimics (Figure 5C). Moreover, treatment of HepG2 and MHCC97H cells with a combination of sorafenib and SCH772984 (an ERK inhibitor) demonstrated that SCH772984 (1 µmol/L) strongly and synergistically enhanced the anti-tumor effect of sorafenib (Figure 5D,E,F). These results indicate that the regulation of miR-126-3p/SPRED1 influences the p-ERK levels, and synergistic inhibition of p-ERK markedly enhances the anti-tumor effect of sorafenib in HCC cells.
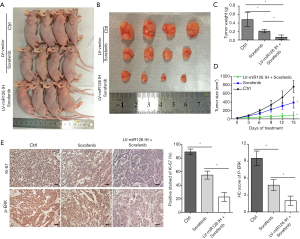
Downregulation of miR-126-3p promoted the antitumor effects of sorafenib in vivo
We constructed murine xenograft tumor models with MHCC97H-LV-vector and MHCC97H-LV-miR-126-3p inhibitor cells, and then treated the mice with vehicle control or sorafenib for 2 weeks (Figure 6). The results showed that sorafenib combined with miR-126-3p inhibition markedly inhibited tumor growth in the xenograft mice compared with the control and sorafenib alone groups (Figure 6D). The mean tumor weight in the sorafenib alone group was 43.6% of that in the control group, and the mean tumor weight in the sorafenib combined with miR-126-3p inhibition group was 13.9% of that in the control group (Figure 6C). Also, IHC analysis revealed that sorafenib combined with miR-126-3p inhibition markedly decreased the positive staining of Ki-67 and the level of p-ERK compared with the control and sorafenib groups (Figure 6E).
Discussion
Sorafenib is the first-line treatment for patients with advanced HCC. However, for a large proportion of patients, drug resistance makes sorafenib ineffective. In this study, we explored the role of miRNAs in the regulation of sorafenib resistance. For the first time we confirmed that miR-126-3p is positively correlated with sorafenib resistance in HCC patients.
A number of studies have indicated that dysregulation of miR-126-3p affects various biologic behaviors in tumor cells, including cell proliferation, migration, and tumor self-renewal (9,13,29). However, the function of miR-126-3p in HCC is controversial, and it is still unknown whether its expression is related to sorafenib sensitivity. Some studies have demonstrated that miR-126-3p is upregulated in HCC compared with metastatic liver tumors. High levels of circulating miR-126-3p are considered to be a promising noninvasive diagnostic biomarker for hepatitis B virus (HBV)—related or hepatitis C virus (HCV)—related HCC (12,30,31). Nevertheless, other studies have reported that a low expression of miR-126-3p is positively correlated with HCC recurrence after liver transplantation, and that miR-126-3p has an inhibitory effect on cell proliferation and induces apoptosis of HCC cells through targeting SOX2 (32,33). miRNA–mRNA interactions suggest that the same miRNA could have multipletargets, and the same mRNA could be targeted by mulpitlemiRNAs. Thus, a possible explanation for the paradoxical role of miR-126-3p in tumors might be that distinct target genes and distinct signaling pathways are involved, which gives rise to different biological consequences. Studies also have shown that miR-126-3p has both oncogenic and suppressive capabilities in leukemia (13,34). In the current study, in vitro and in vivo experiments demonstrated that HCC cells with a high expression of miR-126-3p are more resistant to sorafenib treatment. To the best of our knowledge, this is the first study to report that miR-126-3p is associated with sorafenib resistance in HCC.
To further investigate the mechanisms of the relationship between miR-126-3p and sorafenib, we identified putative target genes of miR-126-3p and confirmed that SPRED1 is a direct target of miR-126-3p. A recent study reported that SPRED1 mRNA expression was inversely correlated with miR-126-3p expression in HCC patients (21). Another study reported that SPRED1 could inhibit cell proliferation and migration in HCC (20). However, it was still unclear if SPRED1 is related to sorafenib sensitivity in HCC. Our results demonstrated that overexpression of SPRED1 markedly enhances the antitumor effect of sorafenib in HCC, and we further confirmed that miR-126-3p contributes to sorafenib resistance in HCC cells via inhibiting SPRED1 expression.
Because sorafenib is an inhibitor of multiple receptor tyrosine kinases (VEGFR-2, PDGFR-β, Flt-3, and Kit) and Raf kinases, it can inhibit the ERK signaling pathway in HCC (28,35). Previous studies have revealed that higher p-ERK levels are associated with increased time to progression (TTP) in HCC and are potentially a predictive biomarker for poor outcomes in patients with advanced HCC who are treated with sorafenib (36,37). Furthermore, a preclinical study also suggested that p-ERK might be a prognostic biomarker for HCC patients treated with the combination of sorafenib and MEK inhibitor (38). Studies have shown that SPRED1 could inhibit the ERK activation by forming a complex with Raf, and another study demonstrated that SPRED1 inhibits p-ERK levels through downregulating Ras-GTP levels (39,40). In the current study, we identified that upregulation of SPRED1 markedly decreases ERK phosphorylation and that the overexpression of miR-126-3p increases p-ERK levels via the downregulation of SPRED1 in HCC cells. Overall, our results indicate that miR-126-3p/SPRED1 influences the sensitivity of HCC to sorafenib by modulating ERK signaling pathway.
Conclusions
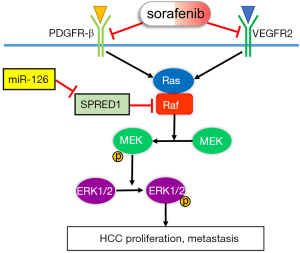
In conclusion, we systematically demonstrated that miR-126-3p contributes to sorafenib resistance in HCC via inhibiting SPRED1 and activating the ERK signaling pathway (Figure 7). Our findings provide preclinical evidence to support further study on the potential use of the miR-126-3p/SPRED1 axis as a predictive biomarker of sorafenib response in patients with HCC.
Acknowledgments
The authors thank J. Reynolds and J. Gray from AME Hong Kong office for their helpful English language editing.
Funding: This work was supported by grants from the National Natural Science Foundation of China (nos. 81972263, 81572398, and 81672419), and the Science and Technology Planning Project of Guangdong Province (nos. 2017A010105003, 2015A050502023, and 2016A020216010).
Footnote
Reporting Checklist: The authors have completed the ARRIVE reporting checklist. Available at http://dx.doi.org/10.21037/atm-20-2081
Data Sharing Statement: Available at http://dx.doi.org/10.21037/atm-20-2081
Peer Review File: Available at http://dx.doi.org/10.21037/atm-20-2081
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/atm-20-2081). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work, and the questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. Animal experiments were approved by the Bioethics Committee of Sun Yat-Sen University and were performed according to the National Institutes of Health guidelines for the use and care of laboratory animals (approval number: SYSU-IACUC-2019-B182).
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Cheng AL, Kang YK, Chen Z, et al. Efficacy and safety of sorafenib in patients in the Asia-Pacific region with advanced hepatocellular carcinoma: a phase III randomised, double-blind, placebo-controlled trial. Lancet Oncol 2009;10:25-34. [Crossref] [PubMed]
- Llovet JM, Montal R, Sia D, et al. Molecular therapies and precision medicine for hepatocellular carcinoma. Nat Rev Clin Oncol 2018;15:599-616. [Crossref] [PubMed]
- Zhu YJ, Zheng B, Wang HY, et al. New knowledge of the mechanisms of sorafenib resistance in liver cancer. Acta Pharmacol Sin 2017;38:614-22. [Crossref] [PubMed]
- Shah MY, Ferrajoli A, Sood AK, et al. microRNA Therapeutics in Cancer - An Emerging Concept. EBioMedicine 2016;12:34-42. [Crossref] [PubMed]
- Pratama MY, Pascut D, Massi MN, et al. The role of microRNA in the resistance to treatment of hepatocellular carcinoma. Ann Transl Med 2019;7:577. [Crossref] [PubMed]
- Xu Y, Huang J, Ma L, et al. MicroRNA-122 confers sorafenib resistance to hepatocellular carcinoma cells by targeting IGF-1R to regulate RAS/RAF/ERK signaling pathways. Cancer Lett 2016;371:171-81. [Crossref] [PubMed]
- Azumi J, Tsubota T, Sakabe T, et al. miR-181a induces sorafenib resistance of hepatocellular carcinoma cells through downregulation of RASSF1 expression. Cancer Sci 2016;107:1256-62. [Crossref] [PubMed]
- Qiu Y, Shan W, Yang Y, et al. Reversal of sorafenib resistance in hepatocellular carcinoma: epigenetically regulated disruption of 14-3-3eta/hypoxia-inducible factor-1alpha. Cell Death Discov 2019;5:120. [Crossref] [PubMed]
- Ebrahimi F, Gopalan V, Smith RA, et al. miR-126 in human cancers: clinical roles and current perspectives. Exp Mol Pathol 2014;96:98-107. [Crossref] [PubMed]
- Zhang Y, Yang P, Sun T, et al. miR-126 and miR-126* repress recruitment of mesenchymal stem cells and inflammatory monocytes to inhibit breast cancer metastasis. Nat Cell Biol 2013;15:284-94. [Crossref] [PubMed]
- Ebrahimi F, Gopalan V, Wahab R, et al. Deregulation of miR-126 expression in colorectal cancer pathogenesis and its clinical significance. Exp Cell Res 2015;339:333-41. [Crossref] [PubMed]
- Ghosh A, Ghosh A, Datta S, et al. Hepatic miR-126 is a potential plasma biomarker for detection of hepatitis B virus infected hepatocellular carcinoma. Int J Cancer 2016;138:2732-44. [Crossref] [PubMed]
- de Leeuw DC, Denkers F, Olthof MC, et al. Attenuation of microRNA-126 expression that drives CD34+38- stem/progenitor cells in acute myeloid leukemia leads to tumor eradication. Cancer Res 2014;74:2094-105. [Crossref] [PubMed]
- Donnem T, Lonvik K, Eklo K, et al. Independent and tissue-specific prognostic impact of miR-126 in nonsmall cell lung cancer: coexpression with vascular endothelial growth factor-A predicts poor survival. Cancer 2011;117:3193-200. [Crossref] [PubMed]
- Luo W, Yan D, Song Z, et al. miR-126-3p sensitizes glioblastoma cells to temozolomide by inactivating Wnt/beta-catenin signaling via targeting SOX2. Life Sci 2019;226:98-106. [Crossref] [PubMed]
- Caporali S, Amaro A, Levati L, et al. miR-126-3p down-regulation contributes to dabrafenib acquired resistance in melanoma by up-regulating ADAM9 and VEGF-A. J Exp Clin Cancer Res 2019;38:272. [Crossref] [PubMed]
- Kawazoe T, Taniguchi K. The Sprouty/Spred family as tumor suppressors: Coming of age. Cancer Sci 2019;110:1525-35. [Crossref] [PubMed]
- Jiang CF, Shi ZM, Li DM, et al. Estrogen-induced miR-196a elevation promotes tumor growth and metastasis via targeting SPRED1 in breast cancer. Mol Cancer 2018;17:83. [Crossref] [PubMed]
- Ablain J, Xu M, Rothschild H, et al. Human tumor genomics and zebrafish modeling identify SPRED1 loss as a driver of mucosal melanoma. Science 2018;362:1055-60. [Crossref] [PubMed]
- Yoshida T, Hisamoto T, Akiba J, et al. Spreds, inhibitors of the Ras/ERK signal transduction, are dysregulated in human hepatocellular carcinoma and linked to the malignant phenotype of tumors. Oncogene 2006;25:6056-66. [Crossref] [PubMed]
- Ji JS, Xu M, Song JJ, et al. Inhibition of microRNA-126 promotes the expression of Spred1 to inhibit angiogenesis in hepatocellular carcinoma after transcatheter arterial chemoembolization: in vivo study. Onco Targets Ther 2016;9:4357-67. [Crossref] [PubMed]
- Sun WJ, Huang H, He B, et al. Romidepsin induces G2/M phase arrest via Erk/cdc25C/cdc2/cyclinB pathway and apoptosis induction through JNK/c-Jun/caspase3 pathway in hepatocellular carcinoma cells. Biochem Pharmacol 2017;127:90-100. [Crossref] [PubMed]
- Peveling-Oberhag J, Doring C, Hartmann S, et al. Feasibility of global miRNA analysis from fine-needle biopsy FFPE material in patients with hepatocellular carcinoma treated with sorafenib. Clin Sci (Lond) 2015;128:29-37. [Crossref] [PubMed]
- Agarwal V, Bell GW, Nam JW, et al. Predicting effective microRNA target sites in mammalian mRNAs. Elife 2015;4:e05005. [Crossref] [PubMed]
- Li JH, Liu S, Zhou H, et al. starBase v2.0: decoding miRNA-ceRNA, miRNA-ncRNA and protein-RNA interaction networks from large-scale CLIP-Seq data. Nucleic Acids Res 2014;42:D92-7. [Crossref] [PubMed]
- Tan W, Luo X, Li W, et al. TNF-α is a potential therapeutic target to overcome sorafenib resistance in hepatocellular carcinoma. EBioMedicine 2019;40:446-56. [Crossref] [PubMed]
- Chen X, Tan W, Li W, et al. miR-1226-3p Promotes Sorafenib Sensitivity of Hepatocellular Carcinoma via Downregulation of DUSP4 Expression. J Cancer 2019;10:2745-53. [Crossref] [PubMed]
- Liu L, Cao Y, Chen C, et al. Sorafenib blocks the RAF/MEK/ERK pathway, inhibits tumor angiogenesis, and induces tumor cell apoptosis in hepatocellular carcinoma model PLC/PRF/5. Cancer Res 2006;66:11851-8. [Crossref] [PubMed]
- Zhang B, Nguyen LXT, Li L, et al. Bone marrow niche trafficking of miR-126 controls the self-renewal of leukemia stem cells in chronic myelogenous leukemia. Nat Med 2018;24:450-62. [Crossref] [PubMed]
- Ali HEA, Abdel Hameed R, Effat H, et al. Circulating microRNAs panel as a diagnostic tool for discrimination of HCV-associated hepatocellular carcinoma. Clin Res Hepatol Gastroenterol 2017;41:e51-62. [Crossref] [PubMed]
- Khairy A, Hamza I, Shaker O, et al. Serum miRNA Panel in Egyptian Patients with Chronic Hepatitis C Related Hepatocellular Carcinoma. Asian Pac J Cancer Prev 2016;17:2699-703. [PubMed]
- Zhao C, Li Y, Zhang M, et al. miR-126 inhibits cell proliferation and induces cell apoptosis of hepatocellular carcinoma cells partially by targeting Sox2. Hum Cell 2015;28:91-9. [Crossref] [PubMed]
- Chen H, Miao R, Fan J, et al. Decreased expression of miR-126 correlates with metastatic recurrence of hepatocellular carcinoma. Clin Exp Metastasis 2013;30:651-8. [Crossref] [PubMed]
- Li Z, Chen P, Su R, et al. Overexpression and knockout of miR-126 both promote leukemogenesis. Blood 2015;126:2005-15. [Crossref] [PubMed]
- Nagel C, Armeanu-Ebinger S, Dewerth A, et al. Anti-tumor activity of sorafenib in a model of a pediatric hepatocellular carcinoma. Exp Cell Res 2015;331:97-104. [Crossref] [PubMed]
- Negri FV, Dal Bello B, Porta C, et al. Expression of pERK and VEGFR-2 in advanced hepatocellular carcinoma and resistance to sorafenib treatment. Liver Int 2015;35:2001-8. [Crossref] [PubMed]
- Zhang Z, Zhou X, Shen H, et al. Phosphorylated ERK is a potential predictor of sensitivity to sorafenib when treating hepatocellular carcinoma: evidence from an in vitro study. BMC Med 2009;7:41. [Crossref] [PubMed]
- Wang C, Jin H, Gao D, et al. Phospho-ERK is a biomarker of response to a synthetic lethal drug combination of sorafenib and MEK inhibition in liver cancer. J Hepatol 2018;69:1057-65. [Crossref] [PubMed]
- Suzuki M, Morita R, Hirata Y, et al. Spred1, a Suppressor of the Ras-ERK Pathway, Negatively Regulates Expansion and Function of Group 2 Innate Lymphoid Cells. J Immunol 2015;195:1273-81. [Crossref] [PubMed]
- Stowe IB, Mercado EL, Stowe TR, et al. A shared molecular mechanism underlies the human rasopathies Legius syndrome and Neurofibromatosis-1. Genes Dev 2012;26:1421-6. [Crossref] [PubMed]
(English Language Editors: J. Reynolds and J. Gray)


