
Genome-wide analysis of long non-coding RNA expression profile in lung adenocarcinoma compared to spinal metastasis
Introduction
Lung cancer is the leading cause of cancer-related death worldwide, comprising almost 20% of cancer-related deaths (1,2). More than 65% of lung carcinoma patients have local or disseminated metastatic disease (3,4). Tumor cells can metastasize locally to the lymph nodes and distant organs, including the skeleton. The rate of spinal metastasis (SM) is close to 50% in patients with lung cancer, followed by the ribs (27%), but rarely occurs in appendicular bones (<6%) (5,6). SM is viewed as a deleterious stage in patients, resulting in a series of skeleton-associated adverse events, especially paraplegia and lifelong urinary and fecal incontinence.
In recent years, increasingly more studies have verified that inappropriate gene expression due to genomic instability or alteration of epigenetic markers can play a major role in lung cancer metastasis (7,8). Consistent with the majority of malignant tumors, non-small cell lung cancer (NSCLC) metastasis is usually associated with a specific genetic chromosomal abnormality (9). However, SM pathogenesis of lung adenocarcinoma is still unclear. The aim of the present study was to investigate the expression profile of long non-coding RNAs (lncRNAs) and potential functions in SM of lung adenocarcinoma. In our study, we explored the expression and roles of lncRNAs in lung adenocarcinoma, which indicates its potential clinical applications as diagnostic or prognostic biomarkers, as well as therapeutic targets.
LncRNAs are defined as RNA molecules longer than 200 nucleotides that are unable to be translated into proteins, but can act as regulators of cancer signaling pathways and biomarkers in predicting cancer prognosis (10). Although the function of lncRNAs is still uncertain, some lncRNAs have been confirmed to be involved in functions involved in regulating gene expression, especially in some serious carcinomas, at the gene level of post-transcriptional processing, maturation, and transport (11). LncRNAs do not code for proteins, yet increasingly more evidence has revealed their involvement in the development of cancer metastasis, such as ovarian cancer (12), gastric cancer (13), and osteosarcoma (14). However, few lncRNAs have been reported to be important regulators of metastasis of lung adenocarcinoma. HOX transcript antisense RNA and brain cytoplasmic RNA 1 have been recently and widely studied lncRNAs associated with lung cancer (15,16). However, the definitive role of lncRNAs with regard to lung adenocarcinoma and its SM tissue is not well understood.
In the present study, we investigated the different expression profiles of lncRNAs and mRNAs in lung adenocarcinoma and SM tissue by using genome-wide microarray. Ten differentially expressed lncRNAs were then verified by quantitative reverse-transcriptase polymerase chain reaction (qRT-PCR). Gene function was explored by gene ontology (GO) and pathway analysis. Our study demonstrated that the aberrant expression of lncRNAs may be involved in inducing SM of lung adenocarcinoma. In addition, exploring the changes of the lncRNA expression profile may provide a new method for the diagnosis and prevention of lung cancer metastasis.
We present the following article in accordance with the MDAR reporting checklist (available at http://dx.doi.org/10.21037/atm-20-7046).
Methods
Patients and tissue samples
All 10 tissue specimens (see Table S1) were obtained from the Department of Surgery, Zhongshan Hospital, Fudan University (Shanghai, China). All tumor tissues were snap-frozen in liquid nitrogen and were immediately put into a −80 °C freezer. Pathology confirmed the diagnosis of both lung adenocarcinoma and SM. Five pairs of samples were used for the microarray analysis, while the other five pairs were used for qRT-PCR. In order to distinguish noise and true results, the inclusion criteria of this study were very strict, in which comprehensive pathological type, TNM staging and immunohistochemistry factors were considered to remove background interference values as much as possible. No patients had received chemotherapy or radiotherapy, or any other cancer treatment prior to surgery. Participants gave informed consent before taking part. All procedures performed in this study were conformed to the provisions of the Declaration of Helsinki (as revised in 2013) and were approved by the Ethics Committee of Zhongshan Hospital, Fudan University (number: Y2019-085).
Microarray hybridization
RNA samples from each group were then used to generate biotinylated cRNA targets for the SBC human ceRNA array V1.0. Biotinylated cRNA targets were then hybridized with the slides. After hybridization, slides were scanned on the Agilent microarray scanner (Agilent Technologies, Santa Clara, CA, USA). Microarray experiments were performed by following the protocol of Agilent Technologies Shanghai Biotechnology.
Gene function analysis
We used the Database for Annotation, Visualization, and Integrated Discovery (http://david.abcc.ncifcrf.gov/) v6.7, and abnormally expressed genes were entered into the database, which utilized GO to identify biologic functions in the gene profile. The Kyoto Encyclopedia of Genes and Genomes (KEGG) was used to analyze the potential effects of these genes in tumor pathways (17,18). The lower the P value, the greater the statistical significance; the recommended cutoff for the P value was 0.05.
LncRNA target prediction
We aimed to identify the target genes of abnormally expressed lncRNAs via cis- or trans-regulatory functions. All the expressed lncRNAs incorporating all the differences were selected for target prediction following previous study (19). First, the algorithm searched for target genes in cis. Using the genome browser (http://genome.ucsc.edu/), genes transcribed within a 10 kbp window upstream or downstream of lncRNAs were regarded as cis target genes. Second, the effects of lncRNA combination on mRNA molecules were based on the algorithm of mRNA sequence complementarity and RNA double-chain energy prediction. BLAST software (http://www.ncbi.nlm.nih.gov/BLAST/) was used for first-round screening. Finally, trans-acting target genes were searched using RNAplex software.
qRT-PCR
Total RNA was extracted by TRIzol (Invitrogen, Carlsbad, CA, USA). RT-PCR analyses were conducted using the QuantiNovaTM SYBR Green PCR Kit (Qiagen, Hilden, Germany). qPCR data collection was conducted using a thermocycler (Thermo-ABI 7500; Applied Biosystems, Waltham, MA, USA). The Ct value was calculated based on the ΔΔCt method. Fold change of gene expression ratios were calculated using the 2−ΔΔCt method. The primers used are listed in Table 1.
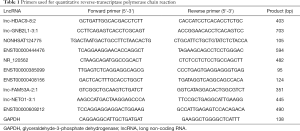
Full table
Statistical analysis
All data were presented as mean ± standard deviation. The standards for screening differentially expressed lncRNAs and mRNAs were a fold change >2 and a threshold P<0.05. All statistical analyses were carried out using SPSS statistical software version 20.0 (SPSS UK, Woking, UK).
Results
Differentially expressed lncRNAs and mRNAs
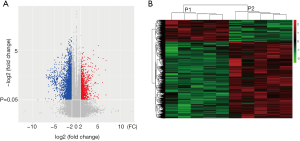
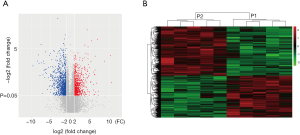
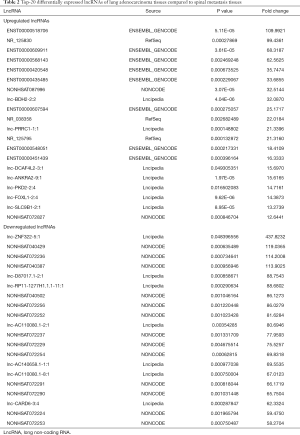
To profile differentially expressed lncRNAs between lung adenocarcinoma tissue and SM tissue, we analyzed a genome-wide result of lncRNA and mRNA expression in lung adenocarcinoma and matched SM. According to these microarray results, we found that 3,345 lncRNAs were differentially expressed (fold change >2, P<0.05) between lung adenocarcinoma and SM. Of these, 761 lncRNAs were upregulated and 2,584 lncRNAs were downregulated (Figure 1A). Heatmap analysis was used to divide all tissues into groups based on their expression levels. We were then able to draw conclusions about the relationships between them (Figure 1B). The top 20 differentially expressed lncRNAs are listed in Table 2. According to the same standard lncRNA, we found that 1,740 mRNAs were differentially expressed between lung adenocarcinoma and SM tissues (fold change >2, P<0.05). Of these, 801 mRNAs were upregulated and 939 mRNAs were downregulated (Figure 2). The top 20 differentially expressed mRNAs are listed in Table 3.
Full table
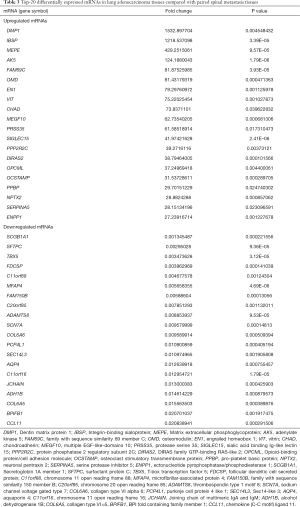
Full table
Function prediction of differentially expressed genes
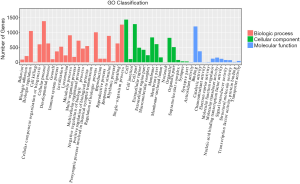
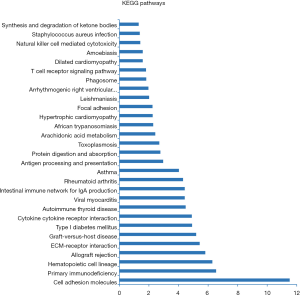
GO enrichment analysis of differentially expressed mRNAs was used to ascertain GOs with a higher confidence level in the differential expressed gene profiling (Figure 3). Enrichment provides a concept of the significant function, and as enrichment increases, the function of differentially expressed genes is more specific; in this way, target genes could be identified with more concrete functionality (20). Pathway analysis indicated that 29 pathways were significantly enriched among the differentially expressed transcripts (P<0.05). The most enriched pathway was cytokine–cytokine receptor interaction (P=1.74E−07), containing 48 genes. Many pathways are related to cancer, such as cell adhesion molecules (CAMs; associated with 42 genes), primary immunodeficiency (associated with 15 genes), focal adhesion (associated with 31 genes), type I diabetes mellitus (associated with 15 genes), extracellular matrix (ECM)-receptor interaction (associated with 23 genes), rheumatoid arthritis (associated with 22 genes), and T-cell receptor signaling pathway (associated with 18 genes) (Figure 4).
Analysis of lncRNA target prediction and function
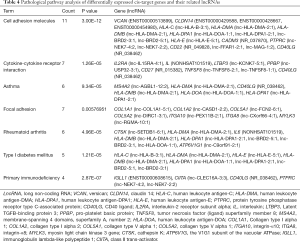
To identify whether dysregulated lncRNAs regulate genes and signaling pathways in SM of lung adenocarcinoma, we used target prediction programs to predict potential targets of lncRNAs in the database. In total, 9,458 differentially expressed lncRNAs were recognized to have cis or trans target genes. Of these, 2,317 lncRNAs targeted about 2,137 cis-genes, 7,141 lncRNAs had 2,932 trans-genes, and 448 lncRNAs had both cis and trans target genes. After expression profiling of mRNA, we integrated the 1024 cis target genes of lncRNA into differentially expressed mRNAs in the profile. We found that 463 of the differentially expressed cis-genes could be regulated by lncRNAs. Analysis using the Shbio enrichment system (http://enrich.shbio.com/) showed that the related tumor signaling pathways of these full target genes were enriched (Table 4). For example, cytokine-cytokine receptor interaction, CAMs, focal adhesion, and ECM-receptor interaction were the most enriched pathways linked with the differentially expressed genes.
Full table
qRT-PCR validation
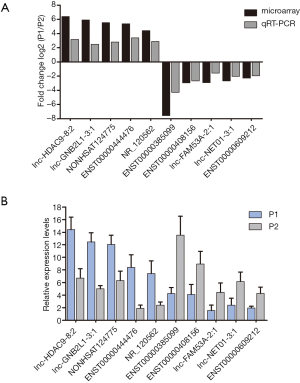
To verify the microarray data, we randomly chose 10 differentially expressed lncRNAs for qRT-PCR, of which TCONS_l2_00010365, n386477, n340790, lnc-LLPH-2:1, and NR_003225.2 were found to be upregulated, and lnc-PSD4-1:14, n335550, lnc-KCMF1-2:1, lnc-PLA2R1-1:1, and ENST00000422494.1 were found to be downregulated in our samples. The qRT-PCR results were consistent with the microarray data, and all 10 of the differentially expressed lncRNAs showed the same trend (P<0.05) (Figure 5).
Discussion
The incidence of bone metastasis has been increasing globally for several decades, with an increasingly greater number of lung cancer patients, especially those with SM. The prognosis of patients with lung carcinoma is generally promising because of advanced inspection and therapy, including surgical operations and chemotherapy (21,22). However, some limitations exist in current therapies, especially for some distant metastatic patients. Preventing cancer cells from metastasizing and invading is crucial for lung cancer patients.
In the past few decades, researchers have determined that SM of lung cancer is a complex biologic process characterized by various molecular abnormalities, such as the axis of stromal derived factor-1/chemokine (C-X-C motif) receptor 4 (CXCR4) (23), and the activation of the receptor activator of the NF-κB (RANK) ligand (RANKL) and Parathyroid Hormone-Related Protein (PTHrP) (24,25). However, the specific pathogenesis of this process still remains unclear.
LncRNAs were previously regarded as transcriptional noise or junk, but are now, conversely, attracting many scientists to investigate their potential role in tumorigenesis (26-28). Similar to mRNAs, they may act as oncogenes or anti-oncogenes via regulating transcriptional processes directly, or via alteration of the expression of related genes (29). However, to the best of our knowledge, there have been no specific studies on the expression profile of lncRNA microarray in SM of lung adenocarcinoma to date.
With the advent of high-throughput lncRNA microarrays, we can explore the expression of a large number of lncRNAs and mRNAs synchronously, leading to increased understanding of the overall transcriptome in cancer or normal cells (30). In our research, we first used microarray technology to evaluate the genome-wide lncRNA and mRNA expression profiles in lung adenocarcinoma tissue compared with matched SM tissue, and detected their possible functions by utilizing GO and KEGG pathway analysis. We found that there were significant numbers of differentially expressed lncRNAs and mRNAs between lung adenocarcinoma and SM tissue. To further confirm the validity of the microarray data, we independently measured lncRNA expression level in lung adenocarcinoma and SM samples by qRT-PCR. The microarray data were consistent with the qRT-PCR results.
To date, many studies have focused on the effect of lncRNAs in non-small cell lung cancer related to carcinoma tissue and para-carcinoma tissue (31,32), but to the best of our knowledge, no studies have focused on distant metastatic sites, especially common sites. For example, metastasis-associated lung adenocarcinoma transcript 1 was not only shown to promote proliferation of lung tumor cells, but also induce the epithelial-mesenchymal transition to migrate to distant sites, such as the brain and liver (33). In the microarray study, several differentially expressed levels of lncRNA and mRNA were in accordance with those reported in the literature, such as NR_125795, which can promote doxorubicin resistance in osteosarcoma by increasing the expression of the transcription factor Forkhead box protein C2, with a 21-fold change in lung adenocarcinoma tissues compared with SM tissues (34).
In the present study, we discovered that those differentially expressed genes participated in many different biochemical processes and signaling pathways. A number of pathways related to cancer development and metastasis were ascertained. For instance, cytokine-cytokine receptor interaction and CAMs were found to be 2 of the most enriched pathways. In addition, focal adhesion (as discussed above) and primary immunodeficiency, which have been reported in the literature, are specifically associated with lung carcinoma invasion and metastasis (35,36). These results also confirm the reliability of our microarray data.
Although increasingly more lncRNAs are being discovered, very few of them have been linked to specific biologic cell functions. A large-scale prediction of lncRNA function in the mammalian genome has been reported, based on the transcription of lncRNAs that affect the expression of its flanking coding genes (30). LncRNAs may be co-expressed and affect the function of neighboring coding genes (37,38). In our study, we attempted to use pathway analysis to associate lncRNAs that were upregulated or downregulated with their adjacent genes (cis target genes) that were differentially expressed, and we uncovered specific signaling pathways that were in accordance with transcripts. The most enriched pathway was cytokine-cytokine receptor interaction, including 7 differentially expressed target genes (Table 4). CD22, human leukocyte antigen-DMA (HLA-DMA), and CD40LG were three common molecules involved in cancers pathways cancers. These molecules were predicted to be cis-regulated by the dysregulated lnc-MAG-1:2 (chr19: 35319261-35347355, 28094bp), lnc-HLA-DMA-2:1 (chr6: 32948613-32969094, 20481bp), and NR_049828 (chr19: 35319261-35347355, 28094bp), respectively. In addition, we randomly verified 10 lncRNAs by qRT-PCR in the samples of patients which were significantly dysregulated in lung adenocarcinoma SM (Figure 5). Therefore, lnc-MAG-1:2, lnc-HLA-DMA-2:1, and NR_049828 may play a crucial role in the pathological signal pathways in SM through CD22, HLA-DMA, and CD40LG, although identification of the exact mechanism remains to be elucidated. However, our study indicates several exact directions for future research.
To the best of our knowledge, the present study is the first to screen and analyze the lncRNA expression profile of lung adenocarcinoma and SM on a large scale. Large numbers of lncRNAs and mRNAs were found to be differentially expressed in SM tissue compared with lung adenocarcinoma tissue, which may be involved in the biologic process related to metastasis of lung adenocarcinoma through target genes. Our study provides a theoretical basis for the functional and mechanistic research of lncRNAs related to SM of lung adenocarcinoma, which facilitate comprehensively to research the competitive crosstalk among RNA transcripts and potential therapeutic target for lung cancer. Subsequently, we may develop targeted drugs for lung cancer according to differentially expressed lncRNAs and mRNAs.
Acknowledgments
Funding: This work was sponsored by Natural Science Foundation of China (grant number 81572629 and 81772855).
Footnote
Reporting Checklist: The authors have completed the MDAR reporting checklist. Available at http://dx.doi.org/10.21037/atm-20-7046
Data Sharing Statement: Available at http://dx.doi.org/10.21037/atm-20-7046
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/atm-20-7046). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. All procedures performed in this study were conformed to the provisions of the Declaration of Helsinki (as revised in 2013) and were approved by the Ethics Committee of Zhongshan Hospital, Fudan University (number: Y2019-085). Participants gave informed consent before taking part.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Devesa SS, Bray F, Vizcaino AP, et al. lung cancer trends by histologic type: male:female differences diminishing and adenocarcinoma rates rising. Int J Cancer 2005;117:294-9. [Crossref] [PubMed]
- Siegel R, Naishadham D, Jemal A. Cancer statistics, 2013. CA Cancer J Clin 2013;63:11-30. [Crossref] [PubMed]
- Morgensztern D, Ng SH, Gao F, et al. Trends in stage distribution for patients with non-small cell lung cancer: a National Cancer Database survey. J Thorac Oncol 2010;5:29-33. [Crossref] [PubMed]
- Rami-Porta R, Eberhardt WEE. Clinical implications of the innovations in the primary tumour and metastasis of the 8(th) edition of the TNM classification for lung cancer. J Thorac Dis 2018;10:S2682-5. [Crossref] [PubMed]
- Sugiura H, Yamada K, Sugiura T, et al. Predictors of survival in patients with bone metastasis of lung cancer. Clin Orthop Relat Res 2008;466:729-36. [Crossref] [PubMed]
- Zheng XQ, Huang JF, Lin JL, et al. Incidence, prognostic factors, and a nomogram of lung cancer with bone metastasis at initial diagnosis: a population-based study. Transl Lung Cancer Res 2019;8:367-79. [Crossref] [PubMed]
- Goto T, Hirotsu Y, Mochizuki H, et al. Mutational analysis of multiple lung cancers: Discrimination between primary and metastatic lung cancers by genomic profile. Oncotarget 2017;8:31133-43. [Crossref] [PubMed]
- Illei PB, Belchis D, Tseng LH, et al. Clinical mutational profiling of 1006 lung cancers by next generation sequencing. Oncotarget 2017;8:96684-96. [Crossref] [PubMed]
- Guan ZB, Cao YS, Li Y, et al. Knockdown of lncRNA GHET1 suppresses cell proliferation, invasion and LATS1/YAP pathway in non small cell lung cancer. Cancer Biomark 2018;21:557-63. [Crossref] [PubMed]
- Wang M, Sun X, Wang H, et al. Long non-coding RNAs in non-small cell lung cancer: functions and distinctions from other malignancies. Transl Cancer Res 2019;8:2636-53. [Crossref]
- Schmitt AM, Chang HY. Long Noncoding RNAs in Cancer Pathways. Cancer Cell 2016;29:452-63. [Crossref] [PubMed]
- Xi J, Feng J, Zeng S. Long noncoding RNA lncBRM facilitates the proliferation, migration and invasion of ovarian cancer cells via upregulation of Sox4. Am J Cancer Res 2017;7:2180-9. [PubMed]
- Lin Z, Lai S, He X, et al. Decreased long non-coding RNA MTM contributes to gastric cancer cell migration and invasion via modulating MT1F. Oncotarget 2017;8:97371-83. [Crossref] [PubMed]
- Yang C, Wang G, Yang J, et al. Long noncoding RNA NBAT1 negatively modulates growth and metastasis of osteosarcoma cells through suppression of miR-21. Am J Cancer Res 2017;7:2009-19. [PubMed]
- Hu T, Lu YR. BCYRN1, a c-MYC-activated long non-coding RNA, regulates cell metastasis of non-small-cell lung cancer. Cancer Cell Int 2015;15:36. [Crossref] [PubMed]
- Loewen G, Jayawickramarajah J, Zhuo Y, et al. Functions of lncRNA HOTAIR in lung cancer. J Hematol Oncol 2014;7:90. [Crossref] [PubMed]
- Huang W, Sherman BT, Lempicki RA. Bioinformatics enrichment tools: paths toward the comprehensive functional analysis of large gene lists. Nucleic Acids Res 2009;37:1-13. [Crossref] [PubMed]
- Huang W, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc 2009;4:44-57. [Crossref] [PubMed]
- Han L, Zhang K, Shi Z, et al. LncRNA pro fi le of glioblastoma reveals the potential role of lncRNAs in contributing to glioblastoma pathogenesis. Int J Oncol 2012;40:2004-12. [PubMed]
- Schlitt T, Palin K, Rung J, et al. From gene networks to gene function. Genome Res 2003;13:2568-2576. [Crossref] [PubMed]
- Aberle DR, Adams AM, Berg CD, et al. Reduced lung-cancer mortality with low-dose computed tomographic screening. N Engl J Med 2011;365:395-409. [Crossref] [PubMed]
- Steger V, Walles T, Kosan B, et al. Trimodal therapy for histologically proven N2/3 non-small cell lung cancer: mid-term results and indicators for survival. Ann Thorac Surg 2009;87:1676-83. [Crossref] [PubMed]
- Salcedo R, Oppenheim JJ. Role of chemokines in angiogenesis: CXCL12/SDF-1 and CXCR4 interaction, a key regulator of endothelial cell responses. Microcirculation 2003;10:359-70. [Crossref] [PubMed]
- Jones DH, Nakashima T, Sanchez OH, et al. Regulation of cancer cell migration and bone metastasis by RANKL. Nature 2006;440:692-6. [Crossref] [PubMed]
- Zong JC, Wang X, Zhou X, et al. Gut-derived serotonin induced by depression promotes breast cancer bone metastasis through the RUNX2/PTHrP/RANKL pathway in mice. Oncol Rep 2016;35:739-48. [Crossref] [PubMed]
- Eades G, Zhang YS, Li QL, et al. Long non-coding RNAs in stem cells and cancer. World J Clin Oncol 2014;5:134-41. [Crossref] [PubMed]
- Gibb EA, Brown CJ, Lam WL. The functional role of long non-coding RNA in human carcinomas. Mol Cancer 2011;10:38. [Crossref] [PubMed]
- Bi YY, Shen G, Quan Y, et al. Long noncoding RNA FAM83H-AS1 exerts an oncogenic role in glioma through epigenetically silencing CDKN1A (p21). J Cell Physiol 2018;233:8896-907. [Crossref] [PubMed]
- Moran VA, Perera RJ, Khalil AM. Emerging functional and mechanistic paradigms of mammalian long non-coding RNAs. Nucleic Acids Res 2012;40:6391-400. [Crossref] [PubMed]
- Wang L, Fu D, Qiu Y, et al. Genome-wide screening and identification of long noncoding RNAs and their interaction with protein coding RNAs in bladder urothelial cell carcinoma. Cancer Lett 2014;349:77-86. [Crossref] [PubMed]
- Xu G, Chen J, Pan Q, et al. Long noncoding RNA expression profiles of lung adenocarcinoma ascertained by microarray analysis. PLoS One 2014;9:e104044. [Crossref] [PubMed]
- Yang J, Lin J, Liu T, et al. Analysis of lncRNA expression profiles in non-small cell lung cancers (NSCLC) and their clinical subtypes. Lung Cancer 2014;85:110-5. [Crossref] [PubMed]
- Shen L, Chen L, Wang Y, et al. Long noncoding RNA MALAT1 promotes brain metastasis by inducing epithelial-mesenchymal transition in lung cancer. J Neurooncol 2015;121:101-8. [Crossref] [PubMed]
- Zhang CL, Zhu KP, Ma XL. Antisense lncRNA FOXC2-AS1 promotes doxorubicin resistance in osteosarcoma by increasing the expression of FOXC2. Cancer Lett 2017;396:66-75. [Crossref] [PubMed]
- Ohs I, Ducimetiere L, Marinho J, et al. Restoration of Natural Killer Cell Antimetastatic Activity by IL12 and Checkpoint Blockade. Cancer Res 2017;77:7059-71. [Crossref] [PubMed]
- Wei Y, Kim TJ, Peng DH, et al. Fibroblast-specific inhibition of TGF-beta1 signaling attenuates lung and tumor fibrosis. J Clin Invest 2017;127:3675-88. [Crossref] [PubMed]
- Dinger ME, Amaral PP, Mercer TR, et al. Long noncoding RNAs in mouse embryonic stem cell pluripotency and differentiation. Genome Res 2008;18:1433-45. [Crossref] [PubMed]
- Ponjavic J, Oliver PL, Lunter G, et al. Genomic and transcriptional co-localization of protein-coding and long non-coding RNA pairs in the developing brain. PLoS Genet 2009;5:e1000617. [Crossref] [PubMed]
(English Language Editor: R. Scott)


