
Whole-exome sequencing identifies prognostic mutational signatures in gastric cancer
Introduction
Gastric cancer (GC) is one of the most common malignant tumors worldwide, having the highest incidence in Eastern Asia, particularly in China and Japan (1). The 5-year survival rate of advanced GC is approximately 29.3%, as GC is prone to relapses and metastasis (2). At present, surgery and chemotherapy are still the main treatment options for GC, despite the increasing importance of newer generation cancer therapies such as targeted therapies and immune checkpoint inhibition.
GC is a heterogeneous disease, and individual patients often exhibits distinct genetic and molecular profiles. The advent of next-generation sequencing has rapidly expanded our knowledge of the genetic basis of this disease, and several studies have helped to uncover potential therapeutic targets. Whole-exome sequencing (WES) selectively sequences all the exons or coding regions of the genome. It can reveal approximately 85% of known disease-related variants by sequencing less than 2% of the genome. Comprehensive molecular analysis, including WES, of 295 GCs recently led to a new classification system of GC into four distinct subtypes, characterized by Epstein-Barr viral (EBV) infection, microsatellite instability (MSI), high aneuploid and chromosomal instability (CIN), and stable genome and diffuse histology (3).
Different populations have some different molecular markers of gastric cancer. For example, Hispanic/Latino patients have a significantly larger proportion of genomically stable tumor subtype and a high rate of CDH1 germline variants compared with Asian and White patients (4). WES analysis of 74 GC patients from China showed a high concordance with TCGA and other studies on GC (5). In the same study, PTPRT was significantly associated with metastasis of GC, and mutations in MACF1, CDC27, HMCN1, CDH1 and PDZD2 were moderately enriched in peritoneal metastasis samples (5). Recently, several molecular classifications of GC have been proposed (3). Biomarkers to predict response to immune checkpoint inhibitors and combination therapy have been vigorously investigated (6). Although some studies have been conducted on molecular biomarkers, patients with advanced GC are still unable to benefit from targeted therapies, and there are currently no markers available for secondary diagnosis.
Thus, it is important to identify markers with prognostic and clinical value, for example oncogenes that promote GC metastasis or response to targeted therapies. In this study, we performed WES analysis and molecular characterization on 38 GC patients with a goal to identify new prognostic markers and potential therapeutic targets. We also compared our results with The Cancer Genome Atlas stomach adenocarcinoma (TCGA-STAD) database. We present this article in accordance with the MDAR reporting checklist (available at http://dx.doi.org/10.21037/atm-20-6620).
Methods
Patients and tissue samples
Eligible GC patients were retrospectively identified from the pathology biobank at our institution (Changzheng Hospital, Shanghai, China). Thirty-eight patients (26 metastasis and 12 non-metastasis) were included in the study. All procedures performed in this study involving human participants were in accordance with the Declaration of Helsinki (as revised in 2013). All patients provided written consent and the study protocol was reviewed and approved by the Ethics Committee of the hospital (NO. 2020SL039). Formalin-fixed paraffin-embedded (FFPE) samples and matched peripheral blood samples were collected from the institutional biobank. Patients were treated by surgical resection and first-line chemotherapy were retrospectively included for further survival analysis. Twenty-two patients had undergone surgery. The most commonly used chemotherapy regimen was Tegafur Gimeracil and Oteracil Potassium (n=11), followed by oxaliplatin‐based regimen (n=5). Clinical records, including initial age of diagnosis, sex, pathological type, relapse and metastasis were obtained from hospital medical records.
DNA extraction, library preparation and whole-exome sequencing
Tumor DNA was extracted from FFPE preserved tissue samples using a MagMAX FFPE DNA/RNA Ultra kit (cat# A31881, ThermoFisher), and the paired germline DNA was extracted from peripheral whole blood using a Maxwell RSC blood DNA kit (cat# AS1400, Promega). DNA was sheared with a Covaris L220 sonicator and hybridized to the probes using an Agilent SureSelect XT Human All Exon V5 kit (cat# 5190-6209, Agilent, Santa Clara, CA, USA) for exome enrichment. Captured exome DNA was PCR-amplified, end-repaired, and attached to the adapters and barcode using the SureSelect XT HS and Low Input Library Preparation Kit for Illumina (cat# G9704, Agilent) according to manufacturer’s specifications to prepare the sequencing libraries. The libraries were sequenced on an Illumina NovaSeq-6000 Sequencing System to generate 150x150-bp paired-end reads. The image analysis and base calling were performed using the Illumina onboard RTA3 program with default parameters.
Identification of somatic and germline variations and copy numbers
After removing adapters and low-quality reads, the reads were aligned to NCBI human genome reference assembly hg19 using the Burrows-Wheeler Aligner (BWA) alignment algorithm. Further processing was performed using the Genome Analysis Toolkit (GATK, version 3.5), including the GATK Realigner Target Creator to identify regions that required realignment. The MuTect algorithm was applied to identify candidate somatic single nucleotide variants (SNVs) in tumors in comparison with the matched control blood sample from each patient. SNV annotation was performed using ANNOVAR. Rare germline variants with ≤0.05% allele frequency were identified using Varscan in the single-sample mode. Somatic copy number variations (CNVs) were called using ExomeCNV (7). Recurrent focal and broad CNV alterations were identified using GISTIC2.0 (8).
We identified germline variants using GATK HaplotypeCaller with default parameters, then filtered the variants with the parameter allelic depth (AD) ≥5 for alternative alleles and AD ≤2% for rare variants in the population frequency databases gnomAD and 1,000 g (2015aug version). We further selected germline variants related to tumor susceptibility based on the 135 tumor susceptibility genes recommended by the National Comprehensive Cancer Network (NCCN) guidelines.
Tumor mutation burden and MSI evaluation
The tumor mutation burden (TMB) score was defined by the total number of somatic nonsynonymous mutations (NSM), which was determined by comparing sequence data between tumor tissues and matched blood samples using a previously described method (9). All autosomal microsatellite tracts containing 1–5 bp repeating subunits and five or more repeats in GRCh37/hg19 were identified using MISA (http://pgrc.ipk-gatersleben.de/misa/misa.html). An MSI score (number of unstable microsatellite sites/total valid sites) of <1% was defined as low microsatellite instability (MSI-L), a score of ≤1% to <3.5% was defined as medium microsatellite instability (MSI-M) and a score of ≥3.5% was defined as high microsatellite instability (MSI-H).
Statistical analysis
All statistical analyses were performed using R (https://cran.r-project.org) or SPSS software (version 25.0; SPSS, Chicago, IL). Contingency tables were analyzed using Fisher’s exact test (the total number of cases, <40). All statistical tests were two-tailed, and P<0.05 indicated a significant difference.
Results
Patient survival, metastasis and recurrent somatic variants
The basic patient clinical and mutational burden characteristics are shown in Table 1. The median age of patients was 57 (range, 36–71) years, and the majority of patients were male 28 (73.7%). Metastasis occurred in 26 (68.4%) patients. The median TMB of all the 38 samples was 107 (range, 20–542) and the median MSI was 0.01 (range, 0–1.43). Five patients were classified as TMB-H (TMB >300) and no patients were classified as MSI-H (all MSI scores <3.5%). Twenty-nine patients in this study had a clear record of their tumor location. Among them, 16 patients had tumors in the distal stomach, 8 in the proximal stomach, and 5 in the middle stomach. Among the 38 patients, 29 have used chemotherapy and 22 have undergone surgery. The survival period of chemotherapy is better (P=0.064, HR=0.114), but the survival period of surgery is not significantly prolonged (P=0.546, HR=0.436).

Full table
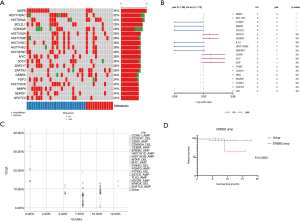
We first investigated the somatic mutation pattern of the patients (Figure 1). We compared the frequency of the above mutations between metastatic and non-metastatic patients (Figure 1A, Table S1). We found that ATAD3B, ARID1A, MGA, ZFHX3 and other mutations only appeared in metastatic patients, while ASTN1, HIST2H2AC, LRRC37A3, SAGE1, and AHNAK mutations only appeared in non-metastatic patients (Figure 1B). Among them, SAGE1, LRRC37A3, HIST2H2AC and ASTN1 were significant (Figure 1B, P<0.05). ATAD3B was also found to be associated with metastasis (Table S1, P=0.074). However, AHNAK2 and CDC27 were not found to be significantly biased in either group. The above results indicated that ATAD3B may be a key driving gene promoting GC metastasis.
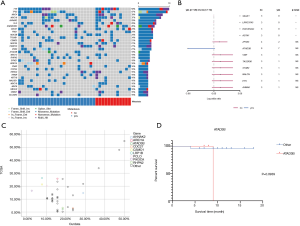
We then compared difference between our data and TCGA data. Three of the top 20 mutated genes, AHNAK2, CDC27, and ATAD3B, are novel in Chinese patients as they are absent in the top mutated genes in the TCGA-STAD cohort (Figure 1C), which were dominantly Caucasians and included 395 patients (3). Additionally, several top mutated genes in the TCGA-STAD cohort, such as ARID1A, CSMD1, and PIK3CA, have significantly lower frequencies in our data (Table 2). ATAD3B, a c-MYC and myogenin target gene, has been observed in different types of cancers and associated with cancer development and progression (10). We found that ATAD3B somatic mutation was associated with shorter overall survival (OS) (Figure 1D, P=0.0939). These discoveries suggested a strong trend towards progression and metastasis in our patients.
Full table
Association between TMB/MSI values and prevalent somatic mutations
While immune checkpoint inhibition (ICI) therapy such as anti PD-1/L1 antibodies have revolutionized cancer treatment, notably in melanoma, NSCLC and breast cancers, little progress has been made in GC. The response rate of GC to ICI is low if PD-L1 expression is used as the sole patient selection marker. High TMB and high instability in the microsatellite regions are emerging as selection biomarkers which have improved compatibility with ICI treatment (11). To test the possibility of using TMB and MSI as biomarkers in GCs, we stratified patients based on their TMB and MSI scores. Five patients were classified as TMB-H (with TMB score ≥300), and the rest of the patients were classified as TMB-L (TMB score <300). There was no obvious correlation between TMB score and metastasis in our data (P>0.05). Several genes, including AKAP9, PCLO, RELN and ASXL1, had significantly higher frequencies in the high TMB group than in the low TMB group (P<0.05) (Table S2). In the TCGA-STAD database, MSI-H accounted for 19.09% patients (12). However, no patient reached MSI-H in our cohort.
Rare germline variants
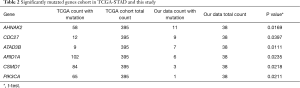
Germline variants are an important source of carcinogenesis but they are not well studied in GC due to previous technical difficulties of calling germline variants confidently. We used an improved germline variant calling procedure on GC patients (13). The 38 patients of our study had rare tumor susceptibility-related germline variants with a median of 5 and range from 1 to 10. The top 5 most frequent germline variants included AR, POLE, ATM, BRCA2, and ALK (Figure 2A).
We analyzed their association with patient survival time. POLE showed the strongest association (P=0.010, Figure 2B). It is noteworthy that patients with and without germline variations on POLE had markedly different survival outlooks within the follow up period, which was 20 months (Figure 2B). By comparing with the reference genome, we found 6 missense single nucleotide polymorphisms (SNPs) and 1 frame shift insertion in POLE, each occurring in 1 patient (Figure 2C). The SNPs included p.A2239V, p.A31S, p.K101E, p.A992T, p.A1943V, and p.A2180V, and have been reported in the NCBI dbSNP database except for p.A1943V. One of the SNPs, p.A992T, is located in the catalytic subunit A domain and this change may disrupt the catalytic function of the polymerase. The frame shift insertion was p.F1513fs. It has not been reported previously and the functional consequence of this change needs to be examined. These results indicate that POLE germline mutations may be suitable biomarkers for GC phenotypes.
Next, we compared the differences of germline variants between the metastatic and non-metastatic groups (Table S3), but no significant differences in germline variants were found. The frequencies of POLE were 8.3% (1/12) and 23.1% (6/26) in the non-metastatic and metastatic groups respectively. This is suggestive of a correlation between POLE germline variants and metastasis, but does not reach statistical significance most likely due to the limited sample size.
Aberrations in somatic copy number alteration
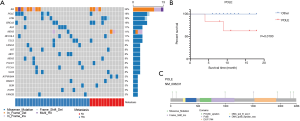
We analyzed CNVs in the 38 GC patients between tumor tissue and peripheral blood. The most significant recurrent, arm-level gains occurred on chromosomal arm 20q, and the most significant recurrent, arm-level loss occurred on chromosomal arm 4q (Figure 3A). A gain on 16p was found to be associated with shorter overall survival (OS) (Figure 3B, P=0.0143). A loss on 17p was also found to be associated with shorter OS (Figure 3C, P=0.0939, HR=8.00), and involved five patients.
At the cytoband level, we found that amplification of 16p11.2, 19q13.32 and 19q13.33, and deletion of 2q24.3, 3p22.2, 3p22.3, 9p21.3, 9p24.1, 9q31.1, 11q11 and 12p13.33 were associated with shorter OS (P<0.05) (Table S4).
We compared the CNVs of the cytobands in the metastatic and non-metastatic groups. Among them, 18q12.1, 4q21.1, 4q13.3, 4q31.3 and 4p12 only appeared in the metastatic group, and were found to be associated with metastasis (Figure 3D, P<0.05).
Notably, the 9p24.1 chromosomal region contains CD274/PD-L1, PD-L2 and JAK2 genes, which are all important to tumor killing and immune checkpoints. It has been reported that amplification of the 9p24.1 chromosomal region and the CD274/PD-L1 gene is an important mechanism for increased PD-L1 expression, which may predict the response to PD-1/PD-L1 targeted therapy (14). However, in our study, deletion of 9p24.1 was associated with shorter OS (Figure 3E, P=0.0376). In a recent study, high frequency 9p24.1 deletion occurred in the post chemotherapy setting but the significance is unclear, requiring further studies (15). Additionally, we found 16p11.2 amplification in 5 (13.2%) patients (Figure 3F, P=0.0066). This is the first time 16p11.2 amplification has been reported in GC. Genes located at 16p11.2 include TP53TG3, MIR762, UBE2MP1 and ZNF771. These four genes were also amplified in the cases of GC with amplification at 16p11.2. TP53TG3 is a novel TP53‐inducible gene. It has been reported to play an important role in the TP53-mediated signaling pathway (16). Amplification of 16p11.2 and associated TP53TG3 may be a reflection of disrupted TP53 signaling pathway in GC.
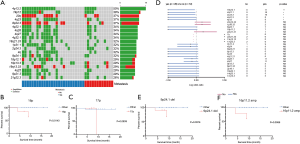
To further analyze the contribution of CNVs from somatic mutations to metastasis, we compared SCNAs between metastasis patients and non-metastasis patients (Figure 4A). The results showed that among the non-metastasis patients, the amplification of BCL11B and MNX1 was significant, whilst among the metastasis patients, the amplification of MMP9, PTPN1 and SS18L1 was significant (Figure 4B, Table S5, P<0.05). These results suggest that metastasis may result from the increase in effective copy number of driver oncogenes.
We compared our somatic copy number alteration (SCNA) analysis with the TCGA-STAD database which is comprised predominantly of European descendants. The frequencies of the top SCNA genes largely overlapped in the two cohorts (Figure 4C, Table S6). For example, the cancer driver genes CCNE1 and ERBB2/HER2 had significant copy number gains in both this study and the TCGA cohorts. We found that patients with ERBB2 amplification had a shorter 1-year survival rate (83.3% vs. 96.9% in wildtype patients, HR =5.0, P=0.246) but this was not statistically significant, perhaps due to the limited sample size (Figure 4D). One gene with a significant difference in distribution between our study cohort and the TCGA cohort was HIST1H3B. It was amplified in 10.5% cases in this study, while being deleted in 0.2% cases in the TCGA cohort.
Discussion
We used WES analysis to reveal the distinct mutational landscape of 38 GC patients and provided a comprehensive analysis of somatic and germline alterations. We compared our study with the TCGA-STAD cohort. We found an overall similarity between the two cohorts but highlighted some significant differences. For example, we discovered some previously unreported mutation sites, such as AHNAK2, CDC27, and ATAD3B in addition to commonly mutated genes that were also reported in TCGA-STAD, including TP53, CSMD3, ARID1A, and KMT2C. AHNAK2 and CDC27 mutations have been reported to be closely linked to progression of patients with unresectable metastatic GC, and to tumorigenesis and progression by supporting epithelial-mesenchymal transition (EMT) and gaining tumor cell-like properties (17,18). CDC27 was reported to be associated with a higher risk of peritoneal metastasis and poor survival in GC (19), suggesting that patients with this mutation may benefit from immunotherapy.
We found that somatic mutation of ATAD3B was related to tumor metastasis, which may provide opportunities for future study. ATAD3B is a negative regulator of ATAD3A and may function as an adaptor of mitochondrial homeostasis and metabolism in hESCs and cancer cells (20). A study revealed that ATAD3A increased breast cancer metastasis through its interaction with GPR78 and the metastasis promoting protein WASF3 (21). ATPase family AAA domain-containing protein 3 proteins A and B (ATAD3A and ATAD3B) are crucial for normal mitochondrial-ER interactions and are fundamental to the processes underlying mitochondrial biogenesis. ATAD3B supports mitochondrial stemness properties through negative regulation of ATAD3A function (22). ATAD3A has been identified to be a chemo-resistance factor in prostate cancer (23), cervical cancer (24), lung cancer (25) and glioma (26). Two compounds have been identified to decrease ATAD3A expression. They are calphostin C, an inhibitor of PKC, and resveratrol, and may hold potential to treat GC (27).
We also discovered certain rare germline alterations that were associated with GC survival or metastasis, including POLE, FANCM, and PDGFRA. Recent studies have shown that people with POLE germline mutations are susceptible to gastrointestinal tumors (28). POLE encodes the catalytic subunit of DNA polymerase epsilon, the primary DNA polymerase in the base excision repair (BER) pathway (29-31). Defects in the DNA polymerase epsilon complex would lead to mismatch repair (MMR) deficiency. MMR deficient cells usually have many DNA mutations, which may lead to colorectal cancer and other types of gastrointestinal cancer. According to a recent study, somatic and germline mutations in the exonuclease domain of the POLE protein are important carcinogenic drivers (32). Another study also showed the importance of screening POLE/POLD1 germline and somatic variants in unexplained MSI-H and MMR-deficient tumors (33). Our analyses showed that POLE germline mutations could be effective molecular markers for predicting survival and metastasis.
Additionally, we analyzed CNV in patients and revealed both reported and novel potential biomarkers of immunotherapy in GC. It is worth noting that the 9p24.1 amplicon includes PD-L1, PD-L2, and JAK2, and has been reported in both GC and in lymphomas (34). In our study, we found a correlation between 9p24.1 amplification and prognosis and metastasis, and may ultimately be an indicator for immunotherapy.
The cancer driver genes CCNE1 and ERBB2 were found to be amplified in both our study and the TCGA cohort, and the frequency of HIST1H3B amplification was significantly higher. ERBB2 amplification may provide value in the development of GC therapy since trastuzumab, an antibody against HER2 (also known as ERBB2), has been introduced in GC therapy (35). ERBB2/HER2 is a member of the epidermal growth factor receptor (EGFR) family. It occasionally occurs in metastatic GC and plays a role in the metastatic processes of some GCs (36). CCNE1 was reported to be significantly associated with liver metastasis (37), having implications for the targeting of cell cycle deregulation and therapeutic cyclin-dependent kinase (CDK) inhibition (38). HIST1H3B encodes histone variant H3.1. It has a significant impact on the regulation of gene transcription and DNA methylation in pediatric diffuse intrinsic pontine glioma (39). The significance of HIST1H3B CNA in GC remains to be further studied.
CNVs of several genes including MMP9, PTPN1, and SS18L1 were found to be significantly related to metastasis. Matrix metalloproteinase 9 (MMP9) encodes a type IV and V collagen degradation enzyme, and is involved in IL-8-induced mobilization of hematopoietic progenitor cells from bone marrow. MMP9 was reported to be associated with invasion and metastasis of GC by degrading the extracellular matrix (ECM) and basement membrane barriers (40). PTPN1 belongs to the protein tyrosine phosphatase (PTP) family. PTPN1 promoted proliferation, colony formation and migration, while decreasing apoptosis of cancer cells through activating extracellular signal-regulated kinase 1/2 (41). PTPN1 has also been implicated with MMP9 in the pathways of cell growth control and response to interferon stimulation (41).
We further analyzed the correlation between somatic mutations and TMB with both patient survival and tumor metastasis to test their value as biomarkers in patients with metastatic GC. Currently, PD-1/PDL-1, MSI-H, and TMB have been used as predictive markers to identify GC patients who would benefit from immunotherapy (42), but none fully satisfies clinical use. More robust markers are still needed for patients to gain clear benefits from precision therapies.
In conclusion, we discovered several new molecular markers of GC, which may potentially predict survival and metastasis, and may provide guidance for clinical treatment. However, more rigorous test of their clinical value is required. Our data also suggested that high-risk GC may be driven by rare germline variants or copy number changes during tumor evolution.
Acknowledgments
We thank the patients and their family members for their supports of this study.
Funding: None.
Footnote
Reporting Checklist: The authors have completed the MDAR reporting checklist. Available at http://dx.doi.org/10.21037/atm-20-6620
Data Sharing Statement: Available at http://dx.doi.org/10.21037/atm-20-6620.
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/atm-20-6620). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. All procedures performed in this study involving human participants were in accordance with the Declaration of Helsinki (as revised in 2013). The study was approved by Shanghai Changzheng Hospital (NO. 2020SL039) and informed consent was taken from all the patients.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Torre LA, Bray F, Siegel RL, et al. Global cancer statistics, 2012. CA Cancer J Clin 2015;65:87-108. [Crossref] [PubMed]
- Zubarayev M, Min EK, Son T. Clinical and molecular prognostic markers of survival after surgery for gastric cancer: tumor-node-metastasis staging system and beyond. Transl Gastroenterol Hepatol 2019;4:59. [Crossref] [PubMed]
- Cancer Genome Atlas Research Network. Comprehensive molecular characterization of gastric adenocarcinoma. Nature 2014;513:202-9. [Crossref] [PubMed]
- Wang SC, Yeu Y, Hammer STG, et al. Hispanic/Latino Patients with Gastric Adenocarcinoma Have Distinct Molecular Profiles Including a High Rate of Germline. Cancer Res 2020;80:2114-24. [Crossref] [PubMed]
- Chen C, Shi C, Huang X, et al. Molecular Profiles and Metastasis Markers in Chinese Patients with Gastric Carcinoma. Sci Rep 2019;9:13995. [Crossref] [PubMed]
- Mukherjee S, Hochwald SN. Pembrolizumab in advanced gastric cancer: pioneering or prosaic? Ann Transl Med 2019;7:S3. [Crossref] [PubMed]
- Sathirapongsasuti JF, Lee H, Horst BA, et al. Exome sequencing-based copy-number variation and loss of heterozygosity detection: ExomeCNV. Bioinformatics 2011;27:2648-54. [Crossref] [PubMed]
- Mermel CH, Schumacher SE, Hill B, et al. GISTIC2.0 facilitates sensitive and confident localization of the targets of focal somatic copy-number alteration in human cancers. Genome Biol 2011;12:R41. [Crossref] [PubMed]
- Chalmers ZR, Connelly CF, Fabrizio D, et al. Analysis of 100,000 human cancer genomes reveals the landscape of tumor mutational burden. Genome Med 2017;9:34. [Crossref] [PubMed]
- Hubstenberger A, Labourdette G, Baudier J, et al. ATAD 3A and ATAD 3B are distal 1p-located genes differentially expressed in human glioma cell lines and present in vitro anti-oncogenic and chemoresistant properties. Exp Cell Res 2008;314:2870-83. [Crossref] [PubMed]
- Samstein RM, Lee CH, Shoushtari AN, et al. Tumor mutational load predicts survival after immunotherapy across multiple cancer types. Nat Genet 2019;51:202-6. [Crossref] [PubMed]
- Bonneville R, Krook MA, Kautto EA, et al. Landscape of Microsatellite Instability Across 39 Cancer Types. JCO Precis Oncol 2017;2017:PO.17.00073.
- McKenna A, Hanna M, Banks E, et al. The Genome Analysis Toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res 2010;20:1297-303. [Crossref] [PubMed]
- Panarese I, De Vita F, Ronchi A, et al. Predictive biomarkers along gastric cancer pathogenetic pathways. Expert Rev Anticancer Ther 2017;17:417-25. [Crossref] [PubMed]
- Gupta S, Vanderbilt CM, Cotzia P, et al. Next-Generation Sequencing-Based Assessment of JAK2, PD-L1, and PD-L2 Copy Number Alterations at 9p24.1 in Breast Cancer: Potential Implications for Clinical Management. J Mol Diagn 2019;21:307-17. [Crossref] [PubMed]
- Ng CC, Koyama K, Okamura S, et al. Isolation and characterization of a novel TP53-inducible gene, TP53TG3. Genes Chromosomes Cancer 1999;26:329-35. [Crossref] [PubMed]
- Wang M, Li X, Zhang J, et al. AHNAK2 is a Novel Prognostic Marker and Oncogenic Protein for Clear Cell Renal Cell Carcinoma. Theranostics 2017;7:1100-13. [Crossref] [PubMed]
- Xin Y, Ning S, Zhang L, et al. CDC27 Facilitates Gastric Cancer Cell Proliferation, Invasion and Metastasis via Twist-Induced Epithelial-Mesenchymal Transition. Cell Physiol Biochem 2018;50:501-11. [Crossref] [PubMed]
- Wu R, Li Q, Wu F, et al. Comprehensive Analysis of CDC27 Related to Peritoneal Metastasis by Whole Exome Sequencing in Gastric Cancer. Onco Targets Ther 2020;13:3335-46. [Crossref] [PubMed]
- Merle N, Feraud O, Gilquin B, et al. ATAD3B is a human embryonic stem cell specific mitochondrial protein, re-expressed in cancer cells, that functions as dominant negative for the ubiquitous ATAD3A. Mitochondrion 2012;12:441-8. [Crossref] [PubMed]
- Teng Y, Ren X, Li H, et al. Mitochondrial ATAD3A combines with GRP78 to regulate the WASF3 metastasis-promoting protein. Oncogene 2016;35:333-43. [Crossref] [PubMed]
- Baudier J. ATAD3 proteins: brokers of a mitochondria-endoplasmic reticulum connection in mammalian cells. Biol Rev Camb Philos Soc 2018;93:827-44. [Crossref] [PubMed]
- Huang KH, Chow KC, Chang HW, et al. ATPase family AAA domain containing 3A is an anti-apoptotic factor and a secretion regulator of PSA in prostate cancer. Int J Mol Med 2011;28:9-15. [PubMed]
- Chen TC, Hung YC, Lin TY, et al. Human papillomavirus infection and expression of ATPase family AAA domain containing 3A, a novel anti-autophagy factor, in uterine cervical cancer. Int J Mol Med 2011;28:689-96. [PubMed]
- Fang HY, Chang CL, Hsu SH, et al. ATPase family AAA domain-containing 3A is a novel anti-apoptotic factor in lung adenocarcinoma cells. J Cell Sci 2010;123:1171-80. [Crossref] [PubMed]
- You WC, Chiou SH, Huang CY, et al. Mitochondrial protein ATPase family, AAA domain containing 3A correlates with radioresistance in glioblastoma. Neuro Oncol 2013;15:1342-52. [Crossref] [PubMed]
- Carter LG, D'Orazio JA, Pearson KJ. Resveratrol and cancer: focus on in vivo evidence. Endocr Relat Cancer 2014;21:R209-25. [Crossref] [PubMed]
- Castellsagué E, Li R, Aligue R, et al. Novel POLE pathogenic germline variant in a family with multiple primary tumors results in distinct mutational signatures. Hum Mutat 2019;40:36-41. [Crossref] [PubMed]
- Sobol RW. Genome instability caused by a germline mutation in the human DNA repair gene POLB. PLoS Genet 2012;8:e1003086. [Crossref] [PubMed]
- Briggs S, Tomlinson I. Germline and somatic polymerase epsilon and delta mutations define a new class of hypermutated colorectal and endometrial cancers. J Pathol 2013;230:148-53. [Crossref] [PubMed]
- Hansen MF, Johansen J, Bjornevoll I, et al. A novel POLE mutation associated with cancers of colon, pancreas, ovaries and small intestine. Fam Cancer 2015;14:437-48. [Crossref] [PubMed]
- Ahn SM, Ansari AA, Kim J, et al. The somatic POLE P286R mutation defines a unique subclass of colorectal cancer featuring hypermutation, representing a potential genomic biomarker for immunotherapy. Oncotarget 2016;7:68638-49. [Crossref] [PubMed]
- Jansen AM, van Wezel T, van den Akker BE, et al. Combined mismatch repair and POLE/POLD1 defects explain unresolved suspected Lynch syndrome cancers. Eur J Hum Genet 2016;24:1089-92. [Crossref] [PubMed]
- Green MR, Monti S, Rodig SJ, et al. Integrative analysis reveals selective 9p24.1 amplification, increased PD-1 ligand expression, and further induction via JAK2 in nodular sclerosing Hodgkin lymphoma and primary mediastinal large B-cell lymphoma. Blood 2010;116:3268-77. [Crossref] [PubMed]
- Van Cutsem E, Sagaert X, Topal B, et al. Gastric cancer. Lancet 2016;388:2654-64. [Crossref] [PubMed]
- Lee JW, Soung YH, Kim SY, et al. ERBB2 kinase domain mutation in a gastric cancer metastasis. APMIS 2005;113:683-7. [Crossref] [PubMed]
- Kim B, Shin HC, Heo YJ, et al. CCNE1 amplification is associated with liver metastasis in gastric carcinoma. Pathol Res Pract 2019;215:152434. [Crossref] [PubMed]
- Etemadmoghadam D, George J, Cowin PA, et al. Amplicon-dependent CCNE1 expression is critical for clonogenic survival after cisplatin treatment and is correlated with 20q11 gain in ovarian cancer. PLoS One 2010;5:e15498. [Crossref] [PubMed]
- Wu G, Broniscer A, McEachron TA, et al. Somatic histone H3 alterations in pediatric diffuse intrinsic pontine gliomas and non-brainstem glioblastomas. Nat Genet 2012;44:251-3. [Crossref] [PubMed]
- Matsumura S, Oue N, Nakayama H, et al. A single nucleotide polymorphism in the MMP-9 promoter affects tumor progression and invasive phenotype of gastric cancer. J Cancer Res Clin Oncol 2005;131:19-25. [Crossref] [PubMed]
- Liu J, Luan W, Zhang Y, et al. HDAC6 interacts with PTPN1 to enhance melanoma cells progression. Biochem Biophys Res Commun 2018;495:2630-6. [Crossref] [PubMed]
- Kim ST, Cristescu R, Bass AJ, et al. Comprehensive molecular characterization of clinical responses to PD-1 inhibition in metastatic gastric cancer. Nat Med 2018;24:1449-58. [Crossref] [PubMed]


