
Autophagy-related gene 7 deficiency caused by miR-154-5p overexpression suppresses the cell viability and tumorigenesis of retinoblastoma by increasing cell apoptosis
Introduction
Retinoblastoma is a rare malignancy of the retina that accounts for 3% of all childhood cancers (1,2). Retinoblastoma most commonly affects very young children; children under the age of 2 make up two-thirds of cases diagnosed, and 95% of patients are diagnosed the before their fifth birthday. Clinically, two distinct forms of this disease exist. The first, bilateral or multifocal retinoblastoma, which comprises 25% of cases, is heritable and results from germline mutations of the RB1 gene. The second form, accounting for 75% of all cases, is unilateral or unifocal retinoblastoma, which is nonhereditary in 90% of cases (3).
The main symptoms of retinoblastoma are leukocoria and strabismus. If possible, these symptoms always necessitate clarification within 1 week to rule out retinoblastoma. Coats’ disease and persistent fetal vasculature are the most common differential diagnoses, along with other intraocular tumors, especially astrocytomas (4). However, the most common pseudo-retinoblastomas entities include Coats’ disease, persistent hyperplastic primary vitreous, and familial exudative vitreo-retinopathy (5).
With the development of wide-angle photography, high-resolution optical coherence tomography and autofluorescence, significant progress has been made in the diagnosis and treatment of retinoblastoma (6). Currently, global sparing therapies include selective ophthalmic artery chemotherapy, periocular carboplatin, focal consolidation, and intravitreal melphalan. Nevertheless, outcomes for patients with advanced retinoblastoma are still dire and can include enucleation or death (7). Therefore, there is a pressing need for new innovative and effective therapeutic strategies.
Epigenetic changes caused by abnormal DNA methylation and histone modification have been extensively explored in cervical cancer, and recent research has facilitated an in-depth understanding of non-coding RNAs, especially microRNAs (miRNAs) and long non-coding RNAs (lncRNAs) (8). miRNAs are important regulators of tumor cell proliferation, migration, invasion, and apoptosis (9). They have been shown to play an essential role in cervical cancer (10). A previous study described that miR-613 acts as a tumor suppressor in retinoblastoma through its inhibition of cell proliferation and invasion via the downregulation of E2F5 (11). Furthermore, in the human retinoblastoma cell line Y79, miR-140-5p was found to inhibit Y79 cell growth by blocking the c-Met/AKT/mTOR signaling pathway (12), while miR-204 was shown to suppress proliferation and to promote apoptosis of retinoblastoma cells by downregulating the expression of Bcl-2 and Sirt1 (13). Previous studies have suggested that miR-154-5p is involved in various types of cancer, including renal cell carcinoma (14), prostate cancer (15), glioblastoma (16), and lung cancer (17). Importantly, in prostate cancer cell lines miR-154-5p targets E2F5 to restrain tumor cell proliferation, migration, and invasion (15).
Autophagy-related gene 7 (ATG7) controls the assembly and expansion of autophagosomal membranes, which makes it essential for autophagy (18). In bladder cancer, ATG7 plays an oncogenic role through its promotion of cell invasion, metastasis, and stem-like properties (19). ATG7 augments cell death in colorectal cancer, which is inhibited by miR-106a (20). Deletion of ATG7 can inhibit the carcinogen-induced protumorigenic inflammatory microenvironment and epithelial tumorigenesis (21). Considering the findings of these earlier studies, we aimed to study the impact of miR-154-5p overexpression on ATG7’s role in cell viability and apoptosis by conducting in vitro experiments using the human retinoblastoma cell line Y79 and by establishing a tumor xenograft model in BALB/c athymic nude mice. The following article is presented in accordance with the ARRIVE reporting checklist (available at http://dx.doi.org/10.21037/atm-20-6009).
Methods
Cell culture
The Y79 cells line was derived from the Chinese Type Culture Collection. After culture in RPMI 1640 medium [containing 10% fetal bovine serum (FBS), 1×105 U/L penicillin and 1×105 U/L streptomycin], the Y79 cells were incubated with 5% carbon dioxide at 37 °C. The culture medium was replaced every 2 days, and cell passaging was performed every 3 to 4 days. Cells in the logarithmic growth phase were collected for use in the subsequent experiments. The RPMI 1640 medium, FBS, penicillin–streptomycin, and trypsin were purchased from Gibco, USA.
Cell transfection
miR-154-5p mimic and pcDNA-ATG7 were purchased from GenePharma (Shanghai, China). Lipofectamine 2000 reagent (Invitrogen, Waltham, MA, USA) was used to transfect Y79 cells with miR-154-5p mimic or pcDNA-ATG7 in accordance with the manufacturer’s instructions.
qRT-PCR
For the quantitative reverse transcription polymerase chain reaction (qRT-PCR), TRIzol was used to extract total RNA from Y79 cells and nude mouse tumor tissues, and the absorbance was measured with a 721 type Nanodrop spectrophotometer (A260/A280, Mettler-Toledo, Switzerland). Next, following the manufacturer’s instructions, the PrimeScript RT reagent Kit (TakaRa, Dalian, China) and 2 SYBR Premix Ex Taq™ II Kit (TakaRa, Dalian, China) were employed to perform cDNA synthesis and PCR amplification, respectively. The reaction conditions were set as 94 °C pre-denaturing for 5 minutes, and then denaturing at 94 °C for 30 seconds, annealing at 55 °C for 30 seconds, extension at 72 °C for 30 seconds, and amplification for 35 cycles. Finally, extension was carried out at 72 °C for 10 minutes and the sample was stored at 4 °C. The relative mRNA expression levels were calculated using the 2–ΔΔCt method.
Western blot assay
According to the manufacturer’s protocol, RIPA lysis buffer (Beyotime Institute of Biotechnology, Shanghai, China) was employed to extract protein from cells. Protein concentrations were measured using a BCA Protein Assay Kit obtained from Beyotime Institute of Biotechnology Co., Ltd. (Shanghai, China). Next, 10% dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) was employed to separate the proteins, which were subsequently transferred onto polyvinylidene difluoride (PVDF) membranes (Merck Millipore) through electro-blotting. Then, 5% skim milk was used to block the membranes for 1 hour in tris-buffered saline with Tween (TBST), after which the membranes were incubated with primary antibody rabbit anti-ATG7 antibody (1:1,000, #8558, Cell Signaling), anti-p21 antibody (1:1,000, #2947, Cell Signaling), anti-Survivin (1:1,000, #2808, Cell Signaling), anti-E-cadherin (1:1,000, #3195, Cell Signaling), N-cadherin (1:1,000, #13116, Cell Signaling), VEGF (1:1,000, #9698, Cell Signaling), and Vimentin (1:1,000, #5741, Cell Signaling). After incubation overnight at 4 °C, the membranes were incubated for a second time with goat anti-rabbit IgG horseradish peroxidase-conjugated secondary antibodies for 1 hour at 37 °C. The band densities were analyzed with an automatic digital gel image analysis system (Bio-Rad CFX-96, CA, USA).
Luciferase assay
Lipofectamine 2000 was used to transfect Y79 cells with miR-154-5p mimic, which was followed by co-transfection with ATG7-wild type (wt) or ATG7-mutant (mut). The luciferase assay was analyzed using a dual-luciferase reporter assay system (Promega Corporation, Madison, WI, USA). After 48 hours, the cells were collected and examined for luciferase activity as previously described (22).
Cell grouping
Y79 cells were transfected with miR-NC or miR-154-5p. At the same time, Anti-miR-NC and Anti-miR-154-5p were transfected into Y79 cells alone, respectively. In addition, pcDNA and pcDNA-ATG7 were transfected into Y79 cells alone, respectively. miR-154-5p mimic and pcDNA-ATG7 alone or combination were transfected into Y79 cells. The control group, miR-154-5p group, ATG7 group and miR-154-5p + ATG7 group were performed for subsequent experiment.
Cell counting Kit-8 assay
Y79 cell proliferation was analyzed by performing a Cell Counting Kit-8 assay in accordance with the manufacturer’s protocol. Y79 cells were seeded in 96-well plates at a density of 5×104 cells/well in 100 µL Dulbecco’s Modified Eagle Medium (DMEM) medium supplemented with 10% FBS. Following incubation for 24, 48, 72, and 96 hours, CCK-8 reagent (10 µL) was added to each well, and the cells were cultured for 1 hour in 5% CO2 (Thermo Fisher Scientific, Carlsbad, CA, USA). A microplate reader (Bio-Rad, CA, USA) was used to measure the absorbance rate at 450 nm.
Colony formation assay
Y79 cells in the control group, control group, miR-154-5p group, ATG7 group, and miR-154-5p + ATG7 group were pretreated with phosphate-buffered saline (PBS), and then cultured for approximately 14 days. The cells were fixed in cold methanol-glacial acetic acid and then subjected to staining with crystal violet.
Flow cytometry assay
Apoptosis of Y79 cells was detected via flow cytometry. As previously described (23), suspended cells (about 3×106/mL per sample) were collected into a 10 mL sterile centrifuge tube. Then, the cells were washed with incubation buffer and centrifuged before resuspension and incubation in the dark at room temperature for 10–15 minutes. After that, the cells were centrifuged, incubated again to wash, and then resuspended. Staining of the cells was carried out with Annexin V-FITC and propidium iodide (PI), respectively. Lastly, a FACS Aria cell sorter (BD Biosciences) was used to analyze the sample. Data were analyzed with Flow Jo (Tree Star, Ashland, OR). The Annexin V-FITC apoptosis detection kit (C1062S) was bought from the Shanghai Biyuntian Biotechnology Institute.
Nude mouse subcutaneous xenograft model
All experiments involving animals were conducted in accordance with the National Institutes of Health Laboratory Animal Care and Use Guidelines. The Ethics Committee of Experimental Animal Management and Animal Welfare of West China Hospital of Sichuan University reviewed and approved all animal experiments (code: No. 20190327). BALB/c nude mice (female, 5–6 weeks old, weight 18.36±3.21 g) were supplied by Chengdu Dashuo Biotechnology Co. Untreated Y79 cells in the logarithmic growth phase and Y79 cells transfected with miR-154-5p mimic were suspended in medium, and the cell concentration was adjusted to 5×106/mL. To study the effects of miR-154-5p on tumor growth, the mice were administered a subcutaneous injection of 5.0×106/mL Y79 cells via the right flank close to the hind leg. After the tumors had reached a volume of approximately 100 mm3, the texts began. The mice were randomly divided into two groups: (I) control group and (II) miR-154-5p group.
Immunohistochemical staining
Xylene was used to deparaffinize sections of formalin-fixed, paraffin-embedded tumor tissues. After deparaffinization, the tissue sections were rehydrated with gradient concentrations of ethanol and then incubated overnight with rabbit anti-p21 antibody (1:50, #2947, Cell Signaling) and anti-Survivin (1:400, #2808, Cell Signaling) (Cell Signaling, Danvers, MA, USA). The corresponding goat anti-rabbit IgG horseradish peroxidase-conjugated secondary antibodies were incubated at room temperature for 1 hour. An Olympus DX51 fluorescence microscope (Olympus, Tokyo, Japan) was used to visualize the sections, and analysis was conducted with image6.0 software.
Terminal-deoxynucleotidyl transferase mediated nick end labeling (TUNEL) staining
A TUNEL apoptosis detection kit (C1091) was obtained from the Shanghai Biyuntian Biotechnology Institute. Part of the tumor tissues were taken to make paraffin sections. After routine deparaffinization, the tissues were immersed in 0.1 M pH 6.0 citric acid buffer, microwaved for 5 minutes, and washed twice with PBS. Then, the sections were placed in TUNEL reaction solution in a wet box at 37 °C for 1 hour in the dark, after which they were washed three times with PBS. The slides were stained with TUNEL using a POD kit for in situ cell death detection (Roche, Penzberg, Germany). POD was added to the slices, and they were placed in a humid chamber at 37 °C for 30 minutes to react, then washed three times with PBS, and dried. DAB was added to react at room temperature for 10 minutes, and the sections were counterstained, dehydrated with gradient ethanol, clarified with xylene, and sealed with neutral glue. An optical microscope was used to observe the sections, and images from five randomly selected fields of view were taken and analyzed with image J software.
Statistical analysis
All experiments were independently repeated for a minimum of three times. All experimental data were expressed as mean ± standard deviation (SD). Differences between the two groups were analyzed with Student’s t-test. Statistical analyses were performed with SPSS 22.0 software (SPSS Inc.). P<0.05 was considered to indicate statistical significance.
Results
Expression levels of miR-154-5p and ATG7
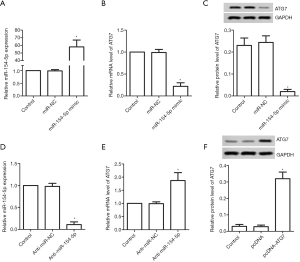
As shown in Figure 1A, Y79 cells were transfected with miR-NC or miR-154-5p mimic. The mRNA expression levels of miR-154-5p were elevated in the miR-154-5p mimic group compared to those in the control group (P<0.05); however, the mRNA and protein expression levels of the ATG7 group showed the opposite result (P<0.05, Figure 1B,C). After transfection with anti-miR-NC or anti-miR-154-5p, the mRNA expression levels of miR-154-5p in the anti-miR-154-5p group were observed to be decreased compared to those in the control group (P<0.05, Figure 1D). The mRNA expression levels of ATG7 in the anti-miR-154-5p group were increased compared to those in the control group (P<0.05, Figure 1E). Y79 cells were also transfected with pcDNA or pcDNA-ATG7. The protein expression levels of ATG7 in the pcDNA-ATG7 group were increased compared to those in the control group (P<0.05, Figure 1F).
Targeting the relationship between miR-154-5p and ATG7
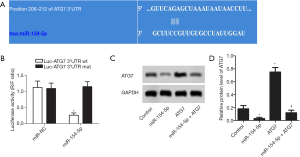
As shown in Figure 2A, miR-154-5p contained a complementary element to the three prime untranslated region (3’-UTR) of ATG7. ATG7 was predicted to be a potential target of miR-154-5p. A dual-luciferase reporter assay was carried out using Y79 cells, and luciferase activity was found to be obviously suppressed in Y79 cells cotransfected with wild-type ATG7 3'-UTR and miR-154-5p, as shown in Figure 2B (P<0.05). As shown in Figure 2C,D, the protein expression levels of ATG7 in the miR-154-5p group were decreased compared to those in the control group (P<0.05) but were increased in the ATG7 group (P<0.05). Meaningfully, the levels of ATG7 were low in the miR-154-5p + ATG7 group contrasted with ATG7 group (P<0.05). These findings indicated that miR-154-5p could negatively regulate the expression of ATG7 via interaction with the 3’-UTR of ATG7 mRNA.
miR-154-5p overexpression inhibits Y79 cells proliferation via targeting of ATG7
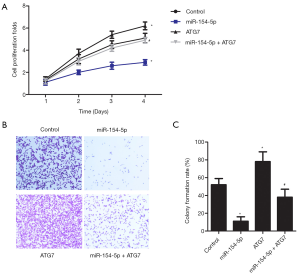
Cell proliferation was assessed in the cell groups by conducting a CCK-8 assay (Figure 3A). The colony formation rate in the miR-154-5p group was significantly reduced compared to that of the control group (P<0.05), while an obvious increase was observed in the ATG7 group (P<0.05). They were significantly lower in the ATG7 group than in the miR-154-5p + ATG7 group (P<0.05). Cell growth in the different groups was assessed by performing a colony formation assay, and the results are shown in Figure 3B and C. Compared with those in the control group, the cell proliferation rate in the miR-154-5p group was reduced (P<0.05); however, the ATG7 group showed the opposite result (P<0.05). Moreover, the rate of cell proliferation in the ATG7 group was significantly lower than that in the miR-154-5p + ATG7 group (P<0.05). These results suggested that miR-154-5p overexpression could inhibit Y79 cell proliferation via targeting of ATG7.
miR-154-5p overexpression promotes Y79 cells apoptosis via targeting of ATG7
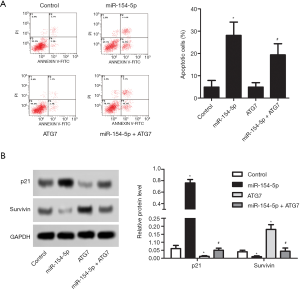
Flow cytometry was conducted to detect apoptosis in the different cell groups, and Figure 4A shows the results. The cell apoptosis rate in the miR-154-5p group was increased compared to that in the control group (P<0.05); however, the opposite result was observed in the ATG7 group (P<0.05). The rate of cell apoptosis in the miR-154-5p + ATG7 group was higher than that in the ATG7 group, and the difference was statistically significant (P<0.05). Western blot was carried out to detect the protein levels of p21 and Survivin in the different cell groups, and Figure 4B shows the results. Compared with the control group, the protein expression levels of p21 were obviously increased in the miR-154-5p group (P<0.05) but were decreased in the ATG7 group (P<0.05). However, the level of p21 in the miR-154-5p + ATG7 group was significantly higher than that in the ATG7 group (P<0.05). Further, the protein expression level of Survivin was found to be downregulated in the miR-154-5p group (P<0.05) but upregulated in the ATG7 group (P<0.05). However, the levels of Survivin were significantly lower in the miR-154-5p + ATG7 group than in the ATG7 group (P<0.05). These results suggested that miR-154-5p overexpression could inhibit Y79 cells apoptosis via targeting of ATG7.
miR-154-5p overexpression promotes Y79 cells migration via targeting of ATG7
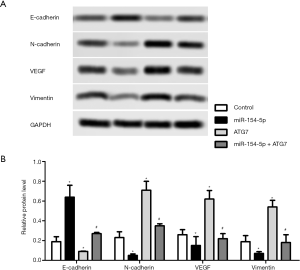
Western blot was carried out to detect the protein expression levels of E-cadherin, N-cadherin, VEGF, and Vimentin in the different groups of cells, and the results are shown in Figure 5A and B. The protein expression levels of E-cadherin in the miR-154-5p group were obviously increased compared to those in the control group (P<0.05), while the ATG7 group showed the opposite result (P<0.05). Moreover the levels of E-cadherin in the miR-154-5p + ATG7 group were significantly higher than those in the ATG7 group (P<0.05). In addition, the protein expression levels of N-cadherin, VEGF, and Vimentin were downregulated in the miR-154-5p group (P<0.05), but upregulated in the ATG7 group (P<0.05). However, the levels of N-cadherin, VEGF, and Vimentin in the miR-154-5p + ATG7 group were significantly lower than those in the ATG7 group (P<0.05). These results indicated that miR-154-5p overexpression could inhibit Y79 cells migration via targeting of ATG7.
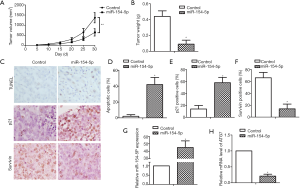
Effect of miR-154-5p overexpression in nude mice model
Figure 6A and B show the volume and weight of the mouse xenograft tumors results after 30 days. On day 30, the tumor volume and weight of the miR-154-5p group mice was obviously decreased compared to that of the control mice (P<0.05). Figure 6C shows the results of TUNEL staining and immunohistochemistry of mouse tumor tissue samples. Compared with that in the control group, the cell apoptosis rate in the miR-154-5p group was increased (P<0.05, Figure 6D), and the miR-154-5p group also had a significantly increased number of P21-positive cells (P<0.05, Figure 6E). The number of cells that stained positive for Survivin was significantly lower in the miR-154-5p group compared with the control group (P<0.05, Figure 6F). Compared with those in the control group, the expression levels of miR-154-5p in the miR-154-5p group were upregulated (P<0.05); in contrast, the levels of ATG7 expression were downregulated (P<0.05) (Figure 6G,H).
Discussion
p21 was the first CKI gene to be discovered. Overactivation of p21 can block cell proliferation and lead to disorders of the cell cycle (24). The associations of rs2279744 and rs937283 in the MDM2 gene, and p21 rs1801270 polymorphism, with the development of retinoblastoma have been demonstrated previously (25). In retinoblastoma, lncRNA H19 binds to the miR-17-92 cluster to inhibit its suppressive effect on p21 (26).
Survivin, an evolutionarily conserved eukaryotic protein, is crucial for cell division and can inhibit cell death. Normally it is only expressed in actively proliferating cells, but is upregulated in most, if not all cancers (27). Survivin inhibition by targeted siRNA suppresses the proliferation, growth, invasion, migration and induced apoptosis of retinoblastoma Y79 cells in vitro. In addition, Survivin inhibition by targeted shRNA suppresses in vivo orthotopic tumors and liver metastasis in BALB/c nude mice (28). In the research, the protein expression levels of p21 in the miR-154-5p group was obviously increased, but was decreased in the ATG7 group. In addition, the protein expression level of Survivin was downregulated in the miR-154-5p group, but upregulated in the ATG7 group. In vivo, the number of p21-positive cells was significantly increased in the miR-154-5p group compared with the control group. The number of Survivin-positive cells was significantly downregulated in the miR-154-5p group contrasted with the control group.
E-cadherin mediates cell-cell adhesion, integrin mediates cell-matrix contact, and regulates tumor invasion and metastasis (29). Lower expression of E-cadherin leads to loss of contact inhibition and is associated with enhanced cell motility and advanced cancer (30). In addition, N-cadherin functions as an invasion suppressor and is up-regulated in most carcinomas (31,32). Vascular endothelial growth factor (VEGF) represents a growth factor with important pro-angiogenic activity, having a mitogenic and an anti-apoptotic effect on endothelial cells, increasing the vascular permeability, promoting cell migration, etc. (33). Previous research have exhibited that VEGF overexpression is significantly associated with the risk of retinoblastoma. Besides, the results suggested that VEGF overexpression have a crucial effect on the optic nerve involvement and differentiation of retinoblastoma (34). The intermediate filament vimentin is required for cells to transition from the epithelial state to the mesenchymal state and migrate as single cells (35). Vimentin is a cytoskeletal intermediate filament protein that is expressed in mesenchymal cells and cancer cells during the epithelial-mesenchymal transition (36). In the research, the protein expression levels of E-cadherin in the miR-154-5p group was obviously increased, but was decreased in the ATG7 group contrasted with the control group. However, the level of E-cadherin in the miR-154-5p + ATG7 group was significantly higher than in the ATG7 group. In addition, the protein expression level of N-cadherin, VEGF and Vimentin was downregulated in the miR-154-5p group, but upregulated in the ATG7 group. However, the level of N-cadherin, VEGF and Vimentin in miR-154-5p + ATG7 group was significantly lower than in the ATG7 group.
Increasing evidence indicates that microRNAs can serve as oncogenes in cancers, though this depends on their target genes and context. For example, miR-145 restrains the proliferation of human retinoblastoma Y79 cells by downregulating the expression of Insulin-like growth factor 1 receptor (IGF-1R) (37). An miR-21 inhibitor suppresses cell proliferation, migration, and invasion via the PTEN/PI3K/AKT signal pathway in the progression of retinoblastoma (38), and miR-124 inhibits the proliferation and invasion of human retinoblastoma cell lines Y79 and SO-RB50 by targeting STAT3 (39). Previous studies have suggested that miR-154-5p contributes to various types of cancer, including renal cell carcinoma (14), prostate cancer (15), glioblastoma (16), nasopharyngeal carcinoma (40) and lung cancer (17). Interesting, lncRNA small nucleolar RNA host gene 11 (SNHG11) functions as an oncogene in glioma and promotes proliferation, invasion, and migration via epithelial-mesenchymal transition by targeting miR-154-5p (41). Up-regulating of miRNA-154-5p holds back from tumorigenesis of osteosarcoma (42), and miR-154-5p inhibits cell invasion and migration via inhibiting the expressive levels of Kinesin family member 14 (KIF14) in nasopharyngeal carcinoma (40). In the study, overexpression of miR-154-5p could inhibited Y79 cells proliferation and migration, as well as promoted Y79 cells apoptosis.
Overexpression of ATG7 is a vital for bladder cancer tumorigenic growth in vitro and in vivo through targeting of the TS2/miRNA-196b/Forkhead box other 1(FoxO1)/p27 axis (43). CD44s is a necessary downstream regulator of ATG7 for the formation, invasion and lung metastasis of bladder cancer (19). ATG7 deficiency can lead to intestinal dysbiosis, and this microbiota is necessary for the anti-cancer response (44). In addition, ATG7 deficiency causes a stress response in tumor cells, accompanied by metabolic defects, AMPK activation, and p53-mediated cell cycle arrest, but not in normal tissues (44). ATG7-deficient prostate tumors display evidence of endoplasmic reticulum stress, suggesting that autophagy promotes prostate tumorigenesis via management of protein homeostasis (45). In the current research, ATG7 overexpression induced Y79 cells proliferation and migration, as well as restrained apoptosis. According to previous research, miR-154 functions as a tumor suppressor in bladder cancer by directly targeting ATG7 (46). In the present research, miR-154-5p overexpression inhibited Y79 cells proliferation and migration, as well as promoted Y79 cells apoptosis via targeting ATG7, providing a basis to explore the functions of miRNAs on the treatment of retinoblastoma.
Acknowledgments
Funding: None.
Footnote
Reporting Checklist: The authors have completed the ARRIVE reporting checklist. Available at http://dx.doi.org/10.21037/atm-20-6009
Data Sharing Statement: Available at http://dx.doi.org/10.21037/atm-20-6009
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/atm-20-6009). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. All animal experiments were performed in light of the National Institutes of Health Laboratory Animal Care and Use Guidelines and were supported by West China Hospital of Sichuan University. The Ethics Committee of Experimental Animal Management and Animal Welfare of West China Hospital of Sichuan University reviewed and approved. Code: No. 20190327.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Rao R, Honavar SG. Retinoblastoma. Indian J Pediatr 2017;84:937-44. [Crossref] [PubMed]
- Malkani I, Warrier RP, Yu LC, et al. Retinoblastoma: a review. Indian J Pediatr 1993;60:227-36. [Crossref] [PubMed]
- Rodriguez-Galindo C, Orbach DB, VanderVeen D. Retinoblastoma. Pediatric clinics of North America 2015;62:201-23. [Crossref] [PubMed]
- Bornfeld N, Lohmann D, Bechrakis NE, et al. Retinoblastoma. Ophthalmologe 2020;117:389-402. [Crossref] [PubMed]
- Ghassemi F, Bazvand F, Makateb A. Lesions Simulating Retinoblastoma at a Tertiary Care Center. J Ophthalmic Vis Res 2015;10:316-9. [Crossref] [PubMed]
- Villegas VM, Hess DJ, Wildner A, et al. Retinoblastoma. Curr Opin Ophthalmol 2013;24:581-8. [Crossref] [PubMed]
- Chen Y, Henson ES, Xiao W, et al. Tyrosine kinase receptor EGFR regulates the switch in cancer cells between cell survival and cell death induced by autophagy in hypoxia. Autophagy 2016;12:1029-46. [Crossref] [PubMed]
- Fang J, Zhang H, Jin S. Epigenetics and cervical cancer: from pathogenesis to therapy. Tumour Biol 2014;35:5083-93. [Crossref] [PubMed]
- Almeida MI, Reis RM, Calin GA. MicroRNA history: discovery, recent applications, and next frontiers. Mutat Res 2011;717:1-8. [Crossref] [PubMed]
- He Y, Lin J, Ding Y, et al. A systematic study on dysregulated microRNAs in cervical cancer development. Int J Cancer 2016;138:1312-27. [Crossref] [PubMed]
- Zhang Y, Zhu X, Zhu X, et al. MiR-613 suppresses retinoblastoma cell proliferation, invasion, and tumor formation by targeting E2F5. Tumour Biol 2017;39:1010428317691674. [Crossref] [PubMed]
- Liao Y, Yin X, Deng Y, et al. MiR-140-5p suppresses retinoblastoma cell growth via inhibiting c-Met/AKT/mTOR pathway. Biosci Rep 2018;38:BSR20180776. [Crossref] [PubMed]
- Ding J, Lu X. Expression of miR-204 in pediatric retinoblastoma and its effects on proliferation and apoptosis of cancer cells. Oncol Lett 2018;16:7152-7. [Crossref] [PubMed]
- Lin C, Li Z, Chen P, et al. Oncogene miR-154-5p regulates cellular function and acts as a molecular marker with poor prognosis in renal cell carcinoma. Life Sci 2018;209:481-9. [Crossref] [PubMed]
- Zheng Y, Zhu C, Ma L, et al. miRNA-154-5p Inhibits Proliferation, Migration and Invasion by Targeting E2F5 in Prostate Cancer Cell Lines. Urol Int 2017;98:102-10. [Crossref] [PubMed]
- Wang X, Sun S, Tong X, et al. MiRNA-154-5p inhibits cell proliferation and metastasis by targeting PIWIL1 in glioblastoma. Brain Res 2017;1676:69-76. [Crossref] [PubMed]
- Huang J, Wu J, Li Y, et al. Deregulation of serum microRNA expression is associated with cigarette smoking and lung cancer. Biomed Res Int 2014;2014:364316. [Crossref] [PubMed]
- Xiong J. Atg7 in development and disease: panacea or Pandora’s Box? Protein Cell 2015;6:722-34. [Crossref] [PubMed]
- Zhu J, Huang G, Hua X, et al. CD44s is a crucial ATG7 downstream regulator for stem-like property, invasion, and lung metastasis of human bladder cancer (BC) cells. Oncogene 2019;38:3301-15. [Crossref] [PubMed]
- Hao H, Xia G, Wang C, et al. miR-106a suppresses tumor cells death in colorectal cancer through targeting ATG7. Med Mol Morphol 2017;50:76-85. [Crossref] [PubMed]
- Qiang L, Sample A, Shea CR, et al. Autophagy gene ATG7 regulates ultraviolet radiation-induced inflammation and skin tumorigenesis. Autophagy 2017;13:2086-103. [Crossref] [PubMed]
- Ji Q, Xu X, Xu Y, et al. miR-105/Runx2 axis mediates FGF2-induced ADAMTS expression in osteoarthritis cartilage. J Mol Med (Berl) 2016;94:681-94. [Crossref] [PubMed]
- Ferretti AC, Hidalgo F, Tonucci FM, et al. Metformin and glucose starvation decrease the migratory ability of hepatocellular carcinoma cells: targeting AMPK activation to control migration. Sci Rep 2019;9:2815. [Crossref] [PubMed]
- Sherr CJ, McCormick F. The RB and p53 pathways in cancer. Cancer Cell 2002;2:103-12. [Crossref] [PubMed]
- Cao Q, Wang Y, Song X, et al. Association between MDM2 rs2279744, MDM2 rs937283, and p21 rs1801270 polymorphisms and retinoblastoma susceptibility. Medicine 2018;97:e13547. [Crossref] [PubMed]
- Zhang A, Shang W, Nie Q. Long non-coding RNA H19 suppresses retinoblastoma progression via counteracting miR-17-92 cluster. J Cell Biochem 2018;119:3497-509. [Crossref] [PubMed]
- Wheatley SP, Altieri DC. Survivin at a glance. J Cell Sci 2019;132:jcs223826. [Crossref] [PubMed]
- Liu K, Liu Y, Zhao G. Targeting survivin suppresses proliferation and invasion of retinoblastoma cells in vitro and in vivo. Int J Clin Exp Pathol 2017;10:9352-61. [PubMed]
- Canel M, Serrels A, Frame MC, et al. E-cadherin-integrin crosstalk in cancer invasion and metastasis. J Cell Sci 2013;126:393-401. [Crossref] [PubMed]
- Mendonsa AM, Na TY, Gumbiner BM. E-cadherin in contact inhibition and cancer. Oncogene 2018;37:4769-80. [Crossref] [PubMed]
- Derycke LD, Bracke ME. N-cadherin in the spotlight of cell-cell adhesion, differentiation, embryogenesis, invasion and signalling. Int J Dev Biol 2004;48:463-76. [Crossref] [PubMed]
- Cao ZQ, Wang Z, Leng P. Aberrant N-cadherin expression in cancer. Biomed Pharmacother 2019;118:109320. [Crossref] [PubMed]
- Melincovici CS, Boşca AB, Şuşman S, et al. Vascular endothelial growth factor (VEGF) - key factor in normal and pathological angiogenesis. Rom J Morphol Embryol 2018;59:455-67. [PubMed]
- Wu Q, Sun X, Zheng G. VEGF overexpression is associated with optic nerve involvement and differentiation of retinoblastoma: A PRISMA-compliant meta-analysis. Medicine 2018;97:e13753. [Crossref] [PubMed]
- Costigliola N, Ding L, Burckhardt CJ, et al. Vimentin fibers orient traction stress. 2017;114:5195-200.
- Trogden KP, Battaglia RA, Kabiraj P, et al. An image-based small-molecule screen identifies vimentin as a pharmacologically relevant target of simvastatin in cancer cells. FASEB J 2018;32:2841-54. [Crossref] [PubMed]
- Chen Z, Yang H, Nie Y, et al. miR-145 regulates the proliferation and apoptosis of Y79 human retinoblastoma cells by targeting IGF-1R. Int J Clin Exp Pathol 2018;11:4331-8. [PubMed]
- Gui F, Hong Z, You Z, et al. MiR-21 inhibitor suppressed the progression of retinoblastoma via the modulation of PTEN/PI3K/AKT pathway. Cell Biol Int 2016;40:1294-302. [Crossref] [PubMed]
- Liu S, Hu C, Wang Y, et al. miR-124 inhibits proliferation and invasion of human retinoblastoma cells by targeting STAT3. Oncol Rep 2016;36:2398-404. [Crossref] [PubMed]
- Chen J, Ma C. MiR-154-5p Suppresses Cell Invasion and Migration Through Inhibiting KIF14 in Nasopharyngeal Carcinoma. Onco Targets Ther 2020;13:2235-46. [Crossref] [PubMed]
- Geng YB, Xu C, Wang Y, et al. Long non-coding RNA SNHG11 promotes cell proliferation, invasion and migration in glioma by targeting miR-154-5p. Eur Rev Med Pharmacol Sci 2020;24:4901-8. [PubMed]
- Tian Q, Gu Y, Wang F, et al. Upregulation of miRNA-154-5p prevents the tumorigenesis of osteosarcoma. Biomed Pharmacother 2020;124:109884. [Crossref] [PubMed]
- Zhu J, Li Y, Tian Z, et al. ATG7 Overexpression Is Crucial for Tumorigenic Growth of Bladder Cancer In Vitro and In Vivo by Targeting the ETS2/miRNA196b/FOXO1/p27 Axis. Mol Ther Nucleic Acids 2017;7:299-313. [Crossref] [PubMed]
- Lévy J, Cacheux W, Bara MA, et al. Intestinal inhibition of Atg7 prevents tumour initiation through a microbiome-influenced immune response and suppresses tumour growth. Nat Cell Biol 2015;17:1062-73. [Crossref] [PubMed]
- Santanam U, Banach-Petrosky W, Abate-Shen C, et al. Atg7 cooperates with Pten loss to drive prostate cancer tumor growth. Genes Dev 2016;30:399-407. [Crossref] [PubMed]
- Zhang J, Mao S, Wang L, et al. MicroRNA-154 functions as a tumor suppressor in bladder cancer by directly targeting ATG7. Oncol Rep 2019;41:819-28. [PubMed]
(English Language Editors: K. Brown and J. Reynolds)


