
Comparative expression analysis of PD-1, PD-L1, and CD8A in lung adenocarcinoma
Introduction
Lung cancer is the leading cause of death in men and the second leading cause of death in women worldwide, with 2.1 million new cases and 1.8 million deaths predicted in 2018 (1,2). Lung cancer survival is poor, and the 5-year overall survival (OS) for lung cancer has been between 7% and 18% in different countries over the last two decades (3). Non-small cell lung cancer (NSCLC) is a subgroup of lung cancer, and the major histologic type of NSCLC is lung adenocarcinoma (LUAD), which accounts for 40% of all lung cancers (4). Because of the lack of typical symptoms at early stages, most patients with lung cancer are diagnosed at a late stage (1). The benefits of surgical resection are limited in patients with metastatic NSCLC, and biotherapies such as targeted therapy and immunotherapy are regarded as first-line treatments for advanced NSCLC (5).
Immunotherapy is short for immune checkpoint therapy, which consists of the inhibition of immune checkpoint proteins to reduce the effect of the tumor microenvironment and suppress host antitumor immunity (6). The programmed cell death protein 1 (PD-1)/programmed cell death-ligand 1 (PD-L1) pathway is one of the main targets of cancer immunotherapy. The PD-1 pathway is associated with tumor immune escape, and the overexpression of PD-1 is detected in many tumors including those of melanoma, renal cell carcinoma, breast cancer, and NSCLC (7-10). PD-1 is detected at high levels in elements of the tumor microenvironment such as in tumor-infiltrating lymphocytes (TILs), and the ligands of PD-1, especially PD-L1, may be induced by PD-1 during the exhaustion of TILs on tumor cells (11,12). Overexpression of PD-1 and its ligand inhibit T cell activation and result in adaptive immune resistance.
Although PD-1 pathway inhibitors such as nivolumab and pembrolizumab are generally used in antitumor strategies including the treatment of advanced LUAD, the roles of PD-1 pathway-related factors in the LUAD tumor microenvironment remain unclear (13,14). In clinical trials of pembrolizumab for the treatment of NSCLC, PD-L1 expression was not identified as a predictive factor for discriminating responders from non-responders, and other factors from the tumor microenvironment may affect the prognosis of patients receiving PD-1 pathway immunotherapy (15). As members of TILs, CD8-positive cytolytic T cells (CD8+ CTLs) are recruited during tumor immune responses and play a crucial role in immunotherapy, and the genetic expression of CD8A may be a measurable indicator of immune cell infiltration (16). A previous study for pan-cancer have shown that high expression of PD-L1 with high status of CD8A might be a good predictive factor for the response to anti-PD-1/PD-L1 therapy (17). The Cancer Genome Atlas (TCGA) is a database of human tumors established through large-scale genome sequencing. The mRNA expression levels of target genes and clinical factors, and the prognosis of patients are recorded in this large, publicly available database (18). Previous studies have revealed microenvironment according to PD-1 pathway in LUAD, however exact values for defining expression situation were still limited. In this study, we first investigated the optimal cutoff value for identifying CD8A, PD-1, and PD-L1 high or low expression, and analyzed the relationship between the mRNA expression levels of these three biomarkers of the PD-1 pathway and the clinical factors of LUAD based on a large data derived from TCGA database. We present the following article in accordance with the REMARK reporting checklist (available at http://dx.doi.org/10.21037/atm-20-6486).
Methods
Study population
In this study, we used level 3 data derived from TCGA database (https://cancergenome.nih.gov/), which is freely available to the public. Clinical information and mRNA expression data of LUAD patients obtained by RNA-sequencing of TCGA tumor samples were downloaded for analysis. Clinical information included diagnosis, age, sex, primary site of tumor, tumor stage, tumor-mutation-node (TMN) status, vital status, and OS rates. Cases lacking PD-1, PD-L1, or CD8A mRNA expression data were excluded. Finally, 484 cases were enrolled in the study. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013).
Characteristics of patients
For further analyses, some variables were divided into sub-groups. Based on the optimal cutoff value, the expression of PD-1, PD-L1, and CD8A was classified as high expression or low expression. Patients aged <65 years were included in the younger patient group and those aged ≥65 years were included in the elderly patient group. Regarding TNM stage, as defined by the seventh edition of the American Joint Committee on Cancer, only T1, T1a, and T1b were defined as early T stage, whereas T2, T3, and T4 were defined as advanced T stage. Patients with N0 stage were included in the lymph node-negative group, and those with N1, N2, or N3 stage were included in the lymph node-positive group. M0 stage was classified into a non-metastatic group, whereas other M stages were classified into a distant-metastasis group. Regarding clinical stage, stage I was considered as early stage, whereas stages higher than stage I were considered as advanced stage.
Statistical analyses
The optimal cutoff values for PD-1, PD-L1, and CD8A were determined by Cutoff Finder, which is an R software engineered, web-based system designed by Budczies et al. (http://molpath.charite.de/cutoff/) (19). The chi-square or Fisher’s exact test was used to compare categorical variables, and the results are expressed as numbers and percentages of patients. Survival curves for each variable were evaluated using the Kaplan-Meier method and compared using the log-rank test. A logistic regression model was used to identify risk factors in univariate analysis. Variables in the univariate analysis with a significance of probability (P) value <0.05 were included in the multivariate analysis using the Cox proportional hazards regression model. All statistical analyses were performed using basic and the survival package of R 3.5.0 (http://www.r-project.org). A P value <0.05 was considered to have statistical significance.
Results
Patient characteristics
A total of 484 cases were enrolled in the current study according to the selection process. The median age was 66 years (range, 38–88 years), and most of the patients were elderly (n=265, 54.8%) and women (n=258, 53.3%). Most of the patients had advanced T stage (n=320, 66.1%). The majority of cases had no record of nodal metastasis (n=313, 64.7%), or distant metastasis (n=323, 66.7%). The median follow-up time was 22.34 months (range, 2.04–238.11 months). The baseline characteristics of the 484 patients included are shown in Table 1.
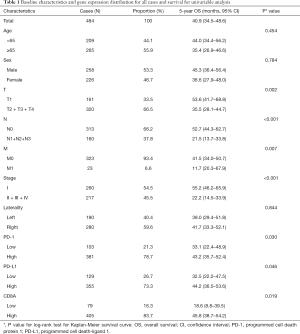
Full table
Definition of cutoff values for CD8A, PD-1, and PD-L1
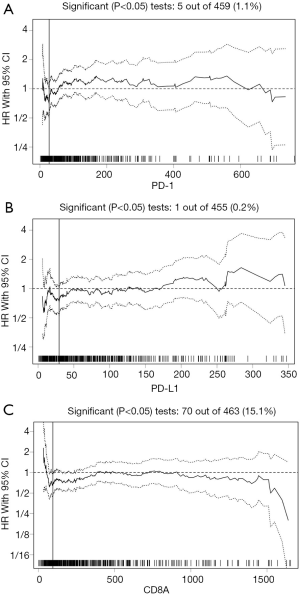
The median mRNA expression values for CD8A, PD-1, and PD-L1 were 245.23, 60.48, and 55.27, respectively. The biostatistical tool Cutoff Finder was used to identify a range of cutoff values for CD8A, PD-1, and PD-L1. The optimal cutoff values for assessing OS were 95.52 for CD8A, 27.4 for PD-1, and 29.41 for PD-L1 (Figure 1A,B,C). The cutoff value for each gene was used to divide cases into high and low expression groups according to whether the gene expression level was higher or lower than the cutoff point. The distribution of cases for each gene in the high and low expression groups is shown in Table 1.
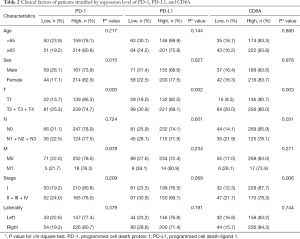
Correlation of PD-1 pathway expression and clinical factors
The clinical factors of cases according to the high and low expression of each gene are shown in Table 2. The results of the chi-square test suggested that women had a significantly higher rate of high expression of PD-1 (P=0.015) and PD-L1 (P=0.027), whereas the high expression of the three genes was significantly associated with early T stage (P=0.003 for PD-1, P=0.002 for PD-L1, and P=0.003 for CD8A). CD8A showed a statistically significantly higher expression in the lymph node–negative (P=0.031) group and in the early clinical stage group (P=0.006).
Full table
The expression of these three genes in patients was positively correlated which shown in Table 3. Cases in the PD-1 high expression group also had high PD-L1 (P<0.001) and CD8A (P<0.001) expression, and similar results were obtained in the PD-L1 and CD8A expression analysis (P<0.001).

Full table
Survival analysis
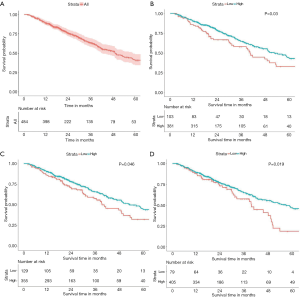
In the 484 cases included in the study, the 5-year OS rate was 40.9% (34.5–48.6%, 95% CI), and the OS curve is shown in Figure 2A. The results of Kaplan-Meier analysis and the log-rank test showed no statistically significant association between survival and age (P=0.454) and sex (P=0.784). However, advanced T stage (P=0.002), lymph-node positivity (P<0.001), distant metastasis (P=0.007), and advanced total stage (P<0.001) were associated with a significantly worse prognosis. In addition, tumor laterality was not a risk factor for LUAC (P=0.844). Assessment of the expression of PD pathway components showed that cases with high expression of PD-1 had a more favorable survival (P=0.030, Figure 2B). High expression of PD-L1 (P=0.046, Figure 2C) or CD8A (P=0.019, Figure 2D) was also associated with better prognosis. All risk factors detected by the Kaplan-Meier method and log-rank test were applied to multivariable analysis. Clinical stage was excluded from multivariable analysis to avoid repetition of stage statistics. The results of the Cox regression hazards model are shown in Table 4, indicating that T stage (P=0.045) and N stage (P<0.001) remained statistically significant, whereas PD-1 (P=0.266), PD-L1 (P=0.740), and CD8A (P=0.193) were not independent risk factors for LUAD.
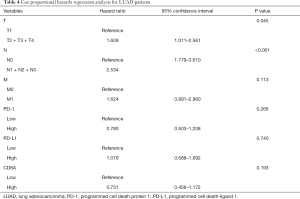
Full table
Discussion
PD-1 is a 288 amino-acid transmembrane protein that was identified in 1992 (20). PD-1 is expressed not only in mature T cells, but also in double-negative T cells, natural killer T cells, and B cells at low levels (21,22). PD-1 is downregulated by T cell antigen receptor and cytokine receptors during activation of naïve T cells (23). PD-1 has two types of ligand, and the main one is PD-L1, a transmembrane glycoprotein (24). PD-L1 is expressed in antigen-presenting cells such as dendritic cells, and in organs including the spleen, lung, heart, and liver (20). The PD-1 pathway plays an important role in the immune system by delivering a negative co-stimulatory signal to the activation of T cells when PD-1 is bound to a ligand such as PD-L1 (25). Under normal physiological conditions, the PD-1 pathway is a critical factor for immune responses, and it induces self-tolerance in peripheral organs such as the placenta, testes, and eyes (26). However, in the tumor microenvironment, the PD-1 pathway contributes to immune tolerance and evasion of host immunity by downregulating antitumor T-cell responses and providing protection from immune destruction (27). Immune checkpoint inhibitors targeting PD-1 and PD-L1 have improved the survival of patients with advanced LUAD both in the second-line and first-line settings (28-30). However, there are few studies focusing on the relationship between PD-1 pathway expression at the genetic level and the clinicopathological factors of LUAD. It would be important to determine the value of the expression of these genes as a prognostic factor in patients with LUAD. Furthermore, the cut-off values used to define the expression of PD-1 pathway genes are inconsistent, which may contribute to the heterogeneity of grouping criteria in PD-1 immunotherapy.
Previous studies have demonstrated that prognosis of LUAD were associated with a number of factors such as TNM stage, historical types or gene mutations (31,32). In this research, we defined the prognostic cutoff values for the expression of PD-1, PD-L1, and CD8A. Based on these cutoff values, we demonstrated that PD-1 high expression was a favorable prognostic factor in patients with LUAD; however, it was not an independent factor for OS. There is limited data on the prognostic and/or predictive value of PD-1 expression, and few studies have investigated the association of PD-1 protein expression with the outcomes of patients with LUAD. A study including 536 patients with completely resected NSCLC showed that PD-1 protein expression assessed using immunohistochemistry was significantly associated with disease-specific survival, which was in line with our results (33). However, no significant differences in disease-specific survival were observed in the adenocarcinoma subgroup. In another study including 321 cases of NSCLC, multivariate analyses in the adenocarcinoma subgroup identified PD-1 expression as an independent favorable prognostic factor (34). A recent study based on 161 patients also showed that PD-1 expression at the protein level was associated with better prognosis in patients with resected LUAD (35). This result could be related to the fact that PD-1 expression is more frequently detected in TILs, and a high positive rate of TILs in the tumor microenvironment leads to better survival rates (36).
Analysis of the prognostic significance of PD-L1 gene expression in LUAD showed that patients with high expression of PD-L1 were more likely to have a longer OS. The prognostic significance of PD-L1 remains controversial, and several studies have reported conflicting results. A large study including 340 NSCLC patients that assessed PD-L1 expression at both the protein and gene levels reported that PD-L1 expression is associated with increased local lymphocytic infiltrates and increased OS (37), which is consistent with the present results. A large study using immunohistochemistry to analyze PD-L1 expression in 678 stage I–III NSCLC cases reported that patients with high PD-L1 expression had significantly longer OS than those with low PD-L1 expression in the univariate analysis, and high PD-L1 expression was confirmed as a significant predictor of improved OS in the multivariable analysis (38). Another study that evaluated 321 NSCLC cases showed that PD-L1 protein expression was a favorable factor for lung squamous cell carcinoma but not for LUAD (34). In addition, high PD-L1 expression was reported as a favorable prognostic factor in small cell lung carcinomas, breast carcinoma, colorectal adenocarcinoma, gastric cancer, and melanoma (39-43). However, several studies suggested a negative prognostic value for high PD-L1 expression in LUAD (44,45), whereas others did not find a prognostic impact (46,47).
In the present study, correlation analysis showed that the expression of the PD-1 and PD-L1 genes was positively associated with CD8A expression, which may explain why high expression of PD-1 or PD-L1 was a favorable prognostic factor. CD8A expression can be detected in natural killer cells and dendritic cells, although CD8A is predominantly expressed on the surface of cytotoxic T-cells (48). CD8-positive cytolytic T cells (CD8+ CTLs) play a crucial role in the cellular immune system and cell-mediated immune responses (49). Strong lymphocytic infiltration is associated with a better clinical outcome in many types of malignant tumors (50). Our results showed that high CD8A expression was associated with longer OS, which is in line with previous studies. Current evidence suggests that the relationship between PD-1/PD-L1 expression and CD8+ CTLs is complex. A mechanistic study of melanoma revealed that PD-L1 upregulation in the tumor microenvironment may depend on the presence of CD8+ CTLs (51). Another recent study in lung cancer showed a significant correlation between PD-L1 positivity and CD8+ CTLs, and cases with PD-L1 overexpression showed consistently dense CD8+ TILs, even in LUAD subgroup analyses (52). However, most studies suggest that the high expression of PD-1 and PD-L1 impairs the function of CD8+ CTLs in the tumor microenvironment, and a longer OS is more likely in the PD-L1 low and CD8 high expression groups of NSCLC (53,54).
The present study had several limitations. First, this was a retrospective study and is likely to have some selection bias. The cutoff values for PD-1, PD-L1, and CD8A expression require external validation, as the ability to predict high and low expression should be clinically validated. Finally, we did not assess the relationship between PD-1, PD-L1, and CD8A expression and the therapeutic strategies for patients with LUAD, as treatment data are not provided by TCGA.
In conclusion, to differentiate high from low mRNA expression, we analyzed TCGA data of LUAD patients and defined the cutoff values of PD-1, PD-L1, and CD8A expression as 27.4, 29.41, and 95.52 respectively. A high expression of PD-1 and PD-L1 was more frequently observed in women. A high expression of PD-1, PD-L1, and CD8A was associated with early T status, and CD8A showed a significantly higher expression in both early stage and early N stage groups. PD-1, PD-L1, and CD8A were significantly positively correlated in all pairs. Although the high expression of each of these three genes was associated with favorable OS, they were not independent prognostic factors.
Acknowledgments
Funding: None.
Footnote
Reporting Checklist: The authors have completed the REMARK reporting checklist. Available at http://dx.doi.org/10.21037/atm-20-6486
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/atm-20-6486). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013).
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Torre LA, Siegel RL, Jemal A. Lung Cancer Statistics. Adv Exp Med Biol 2016;893:1-19. [Crossref] [PubMed]
- Bray F, Ferlay J, Soerjomataram I, et al. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin 2018;68:394-424. [Crossref] [PubMed]
- Walters S, Maringe C, Coleman MP, et al. Lung cancer survival and stage at diagnosis in Australia, Canada, Denmark, Norway, Sweden and the UK: a population-based study, 2004-2007. Thorax 2013;68:551-64. [Crossref] [PubMed]
- Xia L, Zhu Y, Zhang C, et al. Decreased expression of EFCC1 and its prognostic value in lung adenocarcinoma. Ann Transl Med 2019;7:672. [Crossref] [PubMed]
- Planchard D, Popat S, Kerr K, et al. Metastatic non-small cell lung cancer: ESMO Clinical Practice Guidelines for diagnosis, treatment and follow-up. Ann Oncol 2018;29:iv192-237. [Crossref]
- Pardoll DM. The blockade of immune checkpoints in cancer immunotherapy. Nat Rev Cancer 2012;12:252-64. [Crossref] [PubMed]
- Kakavand H, Vilain RE, Wilmott JS, et al. Tumor PD-L1 expression, immune cell correlates and PD-1+ lymphocytes in sentinel lymph node melanoma metastases. Mod Pathol 2015;28:1535-44. [Crossref] [PubMed]
- Thompson RH, Gillett MD, Cheville JC, et al. Costimulatory B7-H1 in renal cell carcinoma patients: Indicator of tumor aggressiveness and potential therapeutic target. Proc Natl Acad Sci U S A 2004;101:17174-9. [Crossref] [PubMed]
- Mittendorf EA, Philips AV, Meric-Bernstam F, et al. PD-L1 expression in triple-negative breast cancer. Cancer Immunol Res 2014;2:361-70. [Crossref] [PubMed]
- Xia L, Huang H, Xiao H, et al. Utilization of combined PD-L1 expression and neutrophil-to-lymphocyte ratio prior to surgery as a prognostic factor in non-small cell lung cancer with brain metastasis. Transl Cancer Res 2019;8:2864-77. [Crossref]
- Ahmadzadeh M, Johnson LA, Heemskerk B, et al. Tumor antigen-specific CD8 T cells infiltrating the tumor express high levels of PD-1 and are functionally impaired. Blood 2009;114:1537-44. [Crossref] [PubMed]
- Duraiswamy J, Freeman GJ, Coukos G. Therapeutic PD-1 pathway blockade augments with other modalities of immunotherapy T-cell function to prevent immune decline in ovarian cancer. Cancer Res 2013;73:6900-12. [Crossref] [PubMed]
- Kazandjian D, Suzman DL, Blumenthal G, et al. FDA Approval Summary: Nivolumab for the Treatment of Metastatic Non-Small Cell Lung Cancer With Progression On or After Platinum-Based Chemotherapy. Oncologist 2016;21:634-42. [Crossref] [PubMed]
- Sul J, Blumenthal GM, Jiang X, et al. FDA Approval Summary: Pembrolizumab for the Treatment of Patients With Metastatic Non-Small Cell Lung Cancer Whose Tumors Express Programmed Death-Ligand 1. Oncologist 2016;21:643-50. [Crossref] [PubMed]
- Muller M, Schouten RD, De Gooijer CJ, et al. Pembrolizumab for the treatment of non-small cell lung cancer. Expert Rev Anticancer Ther 2017;17:399-409. [Crossref] [PubMed]
- Tumeh PC, Harview CL, Yearley JH, et al. PD-1 blockade induces responses by inhibiting adaptive immune resistance. Nature 2014;515:568-71. [Crossref] [PubMed]
- Ock CY, Keam B, Kim S, et al. Pan-Cancer Immunogenomic Perspective on the Tumor Microenvironment Based on PD-L1 and CD8 T-Cell Infiltration. Clin Cancer Res 2016;22:2261-70. [Crossref] [PubMed]
- Tomczak K, Czerwińska P, Wiznerowicz M. The Cancer Genome Atlas (TCGA): an immeasurable source of knowledge. Contemp Oncol (Pozn) 2015;19:A68-77. [Crossref] [PubMed]
- Budczies J, Klauschen F, Sinn BV, et al. Cutoff Finder: a comprehensive and straightforward Web application enabling rapid biomarker cutoff optimization. PLoS One 2012;7:e51862. [Crossref] [PubMed]
- Okazaki T, Honjo T. PD-1 and PD-1 ligands: from discovery to clinical application. Int Immunol 2007;19:813-24. [Crossref] [PubMed]
- Agata Y, Kawasaki A, Nishimura H, et al. Expression of the PD-1 antigen on the surface of stimulated mouse T and B lymphocytes. Int Immunol 1996;8:765-72. [Crossref] [PubMed]
- Velez MA, Burns TF. Is the game over for PD-1 inhibitors in EGFR mutant non-small cell lung cancer? Transl Lung Cancer Res 2019;8:S339-S342. [Crossref] [PubMed]
- Okazaki T, Maeda A, Nishimura H, Kurosaki T, Honjo T. PD-1 immunoreceptor inhibits B cell receptor-mediated signaling by recruiting src homology 2-domain-containing tyrosine phosphatase 2 to phosphotyrosine. Proc Natl Acad Sci U S A 2001;98:13866-71. [Crossref] [PubMed]
- He YF, Zhang GM, Wang XH, et al. Blocking programmed death-1 ligand-PD-1 interactions by local gene therapy results in enhancement of antitumor effect of secondary lymphoid tissue chemokine. J Immunol 2004;173:4919-28. [Crossref] [PubMed]
- Francisco LM, Sage PT, Sharpe AH. The PD-1 pathway in tolerance and autoimmunity. Immunol Rev 2010;236:219-42. [Crossref] [PubMed]
- Fife BT, Pauken KE. The role of the PD-1 pathway in autoimmunity and peripheral tolerance. Ann N Y Acad Sci 2011;1217:45-59. [Crossref] [PubMed]
- Afreen S., Dermime S. The immunoinhibitory B7-H1 molecule as a potential target in cancer: killing many birds with one stone. Hematol Oncol Stem Cell Ther 2014;7:1-17. [Crossref] [PubMed]
- Borghaei H, Paz-Ares L, Horn L, et al. Nivolumab versus Docetaxel in Advanced Nonsquamous Non-Small-Cell Lung Cancer. N Engl J Med 2015;373:1627-39. [Crossref] [PubMed]
- Herbst RS, Baas P, Kim DW, et al. Pembrolizumab versus docetaxel for previously treated, PD-L1-positive, advanced non-small-cell lung cancer (KEYNOTE-010): a randomised controlled trial. Lancet 2016;387:1540-50. [Crossref] [PubMed]
- Rittmeyer A, Barlesi F, Waterkamp D, et al. Atezolizumab versus docetaxel in patients with previously treated non-small-cell lung cancer (OAK): a phase 3, open-label, multicentre randomised controlled trial. Lancet 2017;389:255-65. [Crossref] [PubMed]
- Goldstraw P, Chansky K, Crowley J, et al. The IASLC Lung Cancer Staging Project: Proposals for Revision of the TNM Stage Groupings in the Forthcoming (Eighth) Edition of the TNM Classification for Lung Cancer. J Thorac Oncol 2016;11:39-51. [Crossref] [PubMed]
- Travis WD, Brambilla E, Nicholson AG, et al. The 2015 World Health Organization Classification of Lung Tumors: Impact of Genetic, Clinical and Radiologic Advances Since the 2004 Classification. J Thorac Oncol 2015;10:1243-60. [Crossref] [PubMed]
- Paulsen EE, Kilvaer TK, Khanehkenari MR, et al. Assessing PDL-1 and PD-1 in Non-Small Cell Lung Cancer: A Novel Immunoscore Approach. 2017;18:220-33.e8.
- Schmidt LH, Kümmel A, Görlich D, et al. PD-1 and PD-L1 Expression in NSCLC Indicate a Favorable Prognosis in Defined Subgroups. PLoS One 2015;10:e0136023. [Crossref] [PubMed]
- Zaric B, Brcic L, Buder A, et al. PD-1 and PD-L1 Protein Expression Predict Survival in Completely Resected Lung Adenocarcinoma. Clin Lung Cancer 2018;19:e957-63. [Crossref] [PubMed]
- Mahoney KM, Rennert PD, Freeman GJ. Combination cancer immunotherapy and new immunomodulatory targets. Nat Rev Drug Discov 2015;14:561-84. [Crossref] [PubMed]
- Velcheti V, Schalper KA, Carvajal DE, et al. Programmed death ligand-1 expression in non-small cell lung cancer. Lab Invest 2014;94:107-16. [Crossref] [PubMed]
- Cooper WA, Tran T, Vilain RE, et al. PD-L1 expression is a favorable prognostic factor in early stage non-small cell carcinoma. Lung Cancer 2015;89:181-8. [Crossref] [PubMed]
- Ishii H, Azuma K, Kawahara A, et al. Significance of programmed cell death-ligand 1 expression and its association with survival in patients with small cell lung cancer. J Thorac Oncol 2015;10:426-30. [Crossref] [PubMed]
- Droeser RA, Hirt C, Viehl CT, et al. Clinical impact of programmed cell death ligand 1 expression in colorectal cancer. Eur J Cancer 2013;49:2233-42. [Crossref] [PubMed]
- Schalper KA, Velcheti V, Carvajal D, et al. In situ tumor PD-L1 mRNA expression is associated with increased TILs and better outcome in breast carcinomas. Clin Cancer Res 2014;20:2773-82. [Crossref] [PubMed]
- Taube JM, Anders RA, Young GD, et al. Colocalization of inflammatory response with B7-h1 expression in human melanocytic lesions supports an adaptive resistance mechanism of immune escape. Sci Transl Med 2012;4:127ra37. [Crossref] [PubMed]
- Zheng Z, Bu Z, Liu X, et al. Level of circulating PD-L1 expression in patients with advanced gastric cancer and its clinical implications. Chin J Cancer Res 2014;26:104-11. [PubMed]
- Azuma K, Ota K, Kawahara A, et al. Association of PD-L1 overexpression with activating EGFR mutations in surgically resected nonsmall-cell lung cancer. Ann Oncol 2014;25:1935-40. [Crossref] [PubMed]
- Zhang Y, Wang L, Li Y, et al. Protein expression of programmed death 1 ligand 1 and ligand 2 independently predict poor prognosis in surgically resected lung adenocarcinoma. Onco Targets Ther 2014;7:567-73. [Crossref] [PubMed]
- Yang CY, Lin MW, Chang YL, Wu CT, Yang PC. Programmed cell death-ligand 1 expression in surgically resected stage I pulmonary adenocarcinoma and its correlation with driver mutations and clinical outcomes. Eur J Cancer 2014;50:1361-9. [Crossref] [PubMed]
- Uruga H, Bozkurtlar E, Huynh TG, et al. Programmed Cell Death Ligand (PD-L1) Expression in Stage II and III Lung Adenocarcinomas and Nodal Metastases. J Thorac Oncol 2017;12:458-66. [Crossref] [PubMed]
- Shien K, Papadimitrakopoulou VA, Wistuba II. Predictive biomarkers of response to PD-1/PD-L1 immune checkpoint inhibitors in non-small cell lung cancer. Lung Cancer 2016;99:79-87. [Crossref] [PubMed]
- Fridman WH, Dieu-Nosjean MC, Pagès F, et al. The immune microenvironment of human tumors: general significance and clinical impact. Cancer Microenviron 2013;6:117-22. [Crossref] [PubMed]
- Hao J, Li M, Zhang T, et al. Prognostic Value of Tumor-Infiltrating Lymphocytes Differs Depending on Lymphocyte Subsets in Esophageal Squamous Cell Carcinoma: An Updated Meta-Analysis. Front Oncol 2020;10:614. [Crossref] [PubMed]
- Spranger S, Spaapen RM, Zha Y, et al. Up-regulation of PD-L1, IDO, and T(regs) in the melanoma tumor microenvironment is driven by CD8(+) T cells. Sci Transl Med 2013;5:200ra116. [Crossref] [PubMed]
- Al-Shibli KI, Donnem T, Al-Saad S, et al. Prognostic effect of epithelial and stromal lymphocyte infiltration in non-small cell lung cancer. Clin Cancer Res 2008;14:5220-7. [Crossref] [PubMed]
- Tokito T, Azuma K, Kawahara A, et al. Predictive relevance of PD-L1 expression combined with CD8+ TIL density in stage III non-small cell lung cancer patients receiving concurrent chemoradiotherapy. Eur J Cancer 2016;55:7-14. [Crossref] [PubMed]
- Yang H, Shi J, Lin D, et al. Prognostic value of PD-L1 expression in combination with CD8(+) TILs density in patients with surgically resected non-small cell lung cancer. Cancer Med 2018;7:32-45. [Crossref] [PubMed]
(English Language Editor: J. Gray)


