Infection biomarkers in assisting the judgement of blood stream infection and patient prognosis: a retrospective study incorporating principal components analysis
Introduction
Blood stream infection (BSI) is a severe infectious disease, accounting for an overall mortality of 18.1–41.2% (1-3). Patients with positive blood cultures are associated with a greater severity of illness, longer hospitalizations, and an increased risk of death compared to those with negative blood cultures (4-9). The microbiological culture of the blood sample remains the gold standard to confirm BSI. However, the average time to obtain a blood culture result requires at least 24–48 hours, and an even longer period to identify the causative organisms. Only relying on blood culture results may delay clinical decision making, especially for patients who need escalated antimicrobial treatment. Thus, it is crucial to identify patients who are likely to have positive blood cultures using inexpensive and quickly available laboratory tests.
Among all the laboratory biomarkers of systemic inflammation and infection, procalcitonin (PCT), C-reactive protein (CRP), white blood cell (WBC) counts and percentage of neutrophils (N%) are widely used because they can be assessed promptly and repeatedly at a reasonable cost. Although several studies have evaluated the diagnostic value of these biomarkers (PCT, CRP, WBC, and N%) in the prediction of blood culture results, most of the studies have employed multivariate regression analysis by assuming that these biomarkers are irrelevant. However, CRP, PCT, WBC, and N% are highly relevant in a number of infectious diseases. Incorporating relevant covariates into a multiple regression model simultaneously may raise problems of collinearity, which may undermine the precision of model estimation, thus resulting in erroneous interpretations and misleading conclusions (10,11). To rule out the effect of collinearity on evaluation, we applied principal component analysis (PCA) to transform the 4 sets of primary possibly correlated variables (CRP, PCT, WBC, and N%) into newly derived uncorrelated variables, which are called components. In this study, we mainly explored the relationships between PCA-derived components and blood culture results, as well as patient prognosis. Moreover, the relationships between the primary biomarkers and blood culture results and patient prognosis were also investigated, as appropriate. This study was presented in accordance with the STROBE reporting checklist (available at http://dx.doi.org/10.21037/atm-20-3425).
Methods
Study design and patients
We conducted a retrospective study by searching the Electronic Medical Record System (EMRS) of Changzheng Hospital affiliated to Second Military Medical University, Shanghai, China. Patients who were admitted to the intensive care unit (a 25-bed comprehensive ICU) from July 2015 to June 2017 and had at least one episode of blood culture with matched laboratory results (PCT, CRP, WBC, and N%) were considered potentially eligible for inclusion. Patients were excluded if they (I) were younger than 18 years old; (II) had incomplete laboratory results; (III) had hematological disease; or (IV) received chemotherapy. Data on patient age, gender, Acute Physiology and Chronic Health Evaluation II (APACHE II) scores and Sequential Organ Failure Assessment (SOFA) scores, source of infection, diagnosis, laboratory parameters, length of ICU stay and hospital stay, ICU mortality, and in-hospital mortality were collected. The research was conducted ethically in accordance with the World Medical Association Declaration of Helsinki (as revised in 2013). The anonymous use of data retrieved from EMRS was approved by the ethics committee of Second Military Medical University (approval file number: 2020SL013). The consent was waived since no individual person’s data was shown separately. All data are presented as a whole.
Microbiology and infection biomarkers
Blood cultures were performed when it was deemed necessary by treating physicians. Routinely, two pairs of aerobic and anaerobic bottles of blood samples were collected and incubated for 5 to 7 days or until they became positive. Organisms from positive blood cultures were further identified at the species level by using an automated microbiology identification system (VITEK® 2 COMPACT, bioMerieux Inc. Hazelwood, MO, USA). Contamination, according to previously described criteria, was defined as the isolation of common contaminant species or environmental bacteria (e.g., coagulase-negative Staphylococcus species) in only 1 of a series of blood culture samples (12). A negative blood culture was defined as no growth of any organism within the incubation period. We included all of the eligible blood cultures when analyzing the relationship between infection biomarkers and the blood culture results. However, when analyzing the relationship between infection biomarkers and patient prognosis, only the last episode of positive blood culture was selected because the last episode of positive blood culture had more prognostic implications than prior blood cultures. The WBC counts and N% were measured by Sysmex XN900 (Sysmex Corporation, Kobe, Japan). PCT concentrations were measured by automated immunofluorescent assays with an analytical detection limit of 0.08 ng/mL (Cobas e601, Roche, Switzerland). CRP levels were determined using an immunoturbidimetric by assay with modular analytics (DELTA-I, Radim Diagnosis Corporation, Italy).
Outcomes
The primary outcome was defined as the microbiological result of the blood culture. The secondary outcome was the patient prognosis (survivors or non-survivors) during hospitalization.
Statistical analysis
All analyses were performed using SPSS version 19.0 (SPSS, Chicago, IL, USA). For continuous variables, all data were assessed for normality before analysis. Continuous normally distributed variables were described as mean and standard deviation [mean (SD)] and were compared using one-way analysis of variance (ANOVA). If ANOVA revealed a significant result (P<0.05), the Fisher’s Least Significant Difference (LSD) post-hoc test was performed for pairwise comparisons. Continuous non-normally distributed variables were described as median and interquartile range [median (IQR)] and were compared using Mann-Whitney U test (comparison between two groups) or Kruskal-Wallis H test (comparison between three or more groups). Theses analyses were based on non-missing data. No imputation was made for missing data.
To determine the relationship between the possibly correlated biomarkers (PCT, CRP, WBC, and N%) and blood culture results, PCA was performed. This statistical procedure allows the transformation of a set of intercorrelated variables into a smaller number of independent variables called principal components, which can represent most of the information in the original variables. Given the wide range of serum PCT and CRP concentrations, PCT and CRP values were first transformed into 0 to 5 points and 0 to 4 points based on previous findings, respectively (Table S1). Principal components with eigenvalues above 0.9 were retained for subsequent analysis. Then, the relationship between the primary biomarkers and the retained principal components was visualized by loading the biomarkers on the component coordinates using a varimax rotation method. We next calculated the factor scores for all of the blood culture results and scatter-plotted every blood culture as a point on the component map using factor scores as coordinates. We explored the distribution characteristics of (I) positive blood cultures vs. negative blood cultures; (II) positive blood cultures according to different subgroups, including multiple bacterial isolates, gram negative bacterial isolates, gram positive bacterial isolates, and fungal isolates; and (III) positive blood cultures according to different outcomes (survivors and non-survivors). To determine whether the results were biased by demographic characteristics or illness severity, we recalculated the results by forcing the retained components and confounding covariates (age, gender, APACHE II score, and SOFA score) into a multivariate logistic regression model for adjustment. Otherwise, to determine the relationship between the primary biomarkers (PCT, CRP, WBC, and N%) and blood culture results, we explored the proportion of positive blood cultures in different scales of PCT scores and CRP scores, as well as WBC scores and N% scores (Table S1). Furthermore, the levels of the primary biomarkers were also compared between patients with various blood culture results and prognosis. Finally, the diagnostic performance of the components and infection biomarkers in the discrimination of blood culture results and patient prognosis were compared using receiver operating characteristic (ROC) curves. For each ROC curve, Youden’s index was calculated (13). The optimal cut-off value was the point on the ROC curve where the Youden’s index reached the maximum value. The corresponding diagnostic parameters at the optimal cut-off points were also computed, including the sensitivity, specificity, correctly classified ratio, positive likelihood ratio, and negative likelihood ratio.
Results
Patient characteristics and outcomes
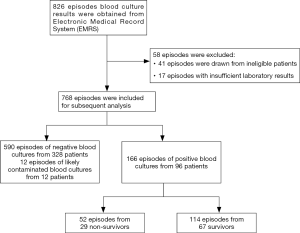
Of 826 episodes of blood culture results retrieved from the EMRS, 768 episodes from 436 patients were eligible for inclusion. There were 590 episodes of negative blood cultures from 328 patients and 166 episodes of positive blood cultures from 96 patients. Among the patients with at least one episode of positive blood culture, 29 died during the hospitalization, accounting for a mortality of 30.2% (Figure 1). The baseline characteristics of patients with positive and negative blood cultures are summarized in Table 1. Patients with positive blood cultures at admission were associated with higher illness severity scores (APACHE II score and SOFA score), and higher levels of infection biomarkers (CRP, PCT, WBC, and N%) than those with negative blood cultures. There were no significant differences between patients with positive blood cultures and those with negative blood cultures at admission in terms of age, gender, presence of systemic inflammatory response syndrome (SIRS), presence of local infection, and disease classification. Patients with positive blood cultures were associated with higher ICU mortality (28.1% vs. 9.8%, P<0.001) and in-hospital mortality (30.2% vs. 10.4%, P<0.001) than patients with negative blood cultures (Table 2). Patients with positive blood cultures also had a longer ICU stay and hospital stay than patients with negative blood cultures (Table 2).

Full table

Full table
Relationship between the primary infection biomarkers and the newly derived components
In the PCA, eigenvalues for each component are shown in scree plot (Figure S1). The first two components returned eigenvalues larger than 0.9 and thus they were retained for subsequent analysis. This meant that 4-dimensional data (PCT scores, CRP scores, WBC, and N%) were compressed into 2-dimensional data (principal components 1 and 2). Figure 2 shows the plot of the loadings of the 4 biomarkers on the components 1 and 2. The loadings of WBC and N% on principal component 1 were 0.833 and 0.793 respectively, indicating that WBC and N% were highly correlated and that the main effects of WBC and N% could be interpreted by component 1.The loadings of PCT scores and CRP scores on principal component 2 were 0.810 and 0.798 respectively, indicating that PCT scores and CRP scores were highly correlated and their main effects could be interpreted by component 2.

Component coordinates as a useful tool in assisting the judgement of blood culture results
The factor scores for each blood culture on component coordinates are plotted in Figure 3A,B. A total of 129 of 166 (77.7%) positive blood cultures were distributed in the first and second quadrants, while only 215 of 590 (36.4%) episodes of negative blood culture were distributed in the same quadrants (OR, 6.28, 95% CI, 4.14–9.64, P<0.001). However, we failed to find a difference in the distribution between positive blood cultures and negative blood cultures within the first and fourth quadrants (OR, 1.27, 95% CI, 0.89–1.83, P=0.17). In subgroup analyses, there were 47, 96, 9, and 14 isolates of Gram-positive bacteria, Gram-negative bacteria, fungi, and multiple bacteria, respectively. Compared with negative blood cultures, multiple bacterial isolates were more likely to be distributed in the first and second quadrants (OR, 23.40, 95% CI, 3.46–996.76, P<0.001), followed by gram negative bacterial isolates (OR, 7.29, 95% CI, 4.22–13.09, P<0.001) and gram positive bacterial isolates (OR, 4.24, 95% CI, 2.15–8.76, P=0.002). Fungal isolates were not associated with a higher frequency of distribution in the first and second quadrants than negative blood cultures (OR, 3.60, 95% CI, 0.76–22.42, P=0.06) (Figures 4,5A). These effects were consistent when the multivariate logistic regression analysis was applied and adjusted for predefined covariates (Figure 5B).



Component coordinates as a useful tool for mortality risk stratification in patients with positive blood cultures
For the 96 patients with positive blood cultures, the distribution map of non-survivors and survivors on the component coordinates is shown in Figure 6. Of 29 non-survivors, 23 (79.3%) were distributed in the first and second quadrants, and 45 of 67 (67.2%) survivors were distributed in the same quadrants. Compared to those with negative blood cultures, non-survivors with positive blood cultures were more likely to be distributed in the first and second quadrants (OR, 6.90, 95% CI, 2.67–20.98, P<0.001), followed by survivors with positive blood cultures (OR, 3.44, 95% CI, 1.97–6.13, P<0.001) (Figure 7A). These effects were consistent after an adjustment for predefined covariates using multivariate logistic regression analysis (Figure 7B).


Relationship between primary biomarkers and blood culture results and patient prognosis
Based on the nature of components 1 and 2, we formed matrices using PCT scores and CRP scores, and WBC scores and N% scores, respectively. The proportions of positive blood cultures in different domains of the matrices are detailed in Figure 8A and Figure 8B. With the increase of PCT scores and CRP scores, the proportion of positive blood cultures consistently increased. The two domains with the highest PCT scores and CPR scores had the highest proportion of positive blood cultures (94.1% and 94.3%), which was about 35-fold higher than that of the domain with the lowest PCT scores and CPR scores (2.7%) (Figure 8A). Although there was a similar trend that the proportion of positive blood cultures increased with the increase of WBC scores and N% scores, the correlation was weak and incoherent (Figure 8B).

Given that PCT and CRP were more discriminative than WBC and N% in the judgement of blood culture results based on the above analyses, we further explored the relationship between individual biomarkers (serum PCT and CRP level) and blood culture results, as well as patient prognosis. The PCT level was significantly higher in isolates of gram negative bacteria [4.7 (1.1–10.0) ng/mL] than in isolates of gram positive bacteria [vs. 1.6 (0.6–5.7) ng/mL, P=0.003], but not significantly higher than that of fungal isolates [vs. 2.2 (1.5–4.6) ng/mL, P=0.45] or multiple bacterial isolates [vs. 2.5 (2.0–5.9) ng/mL, P=0.83] (Figure S2A). However, the CRP level did not differ between the four subgroups (P=0.12) (Figure S2B). Non-survivors had a significantly higher level of PCT than survivors [3.8 (1.5–12.5) vs. 1.4 (0.4–5.9) ng/mL, P=0.003] (Figure S3A). However, the CRP level was similar between the non-survivors and survivors [82.9 (47.1–148.4) vs. 65.0 (31.6–133.0) mg/L, P=0.29)] (Figure S3B).
Diagnostic performance of the components and infection biomarkers for blood culture results and patient prognosis
The ROC curves of the components and infection biomarkers for discrimination between positive and negative blood cultures and survivors and non-survivors are shown in Figure 9A,B. The AUCs, the optimal cut-off values, and the corresponding diagnostic parameters for the individual components and infection biomarkers are summarized in Table 3. Component 2 yielded the highest AUC (0.81) in the discrimination of blood culture results, followed by PCT (0.77), CRP (0.65), N% (0.63), component 1 (0.54), and WBC (0.54) (P<0.001). In addition, with regard to prognosis prediction, component 2 yielded a larger AUC (0.69) than the other indicators, including PCT (0.67), component 1 (0.63), N% (0.63), WBC (0.57), and CRP (0.56) (P=0.003).


Full table
Discussion
PCA is a multivariate technique that transforms a large set of possibly correlated variables into a smaller set of uncorrelated variables called principal components, which still contain most of the information from the large set (14). In our study, the 4 parameters (PCT, CRP, WBC, and N%) were chosen for PCA in patients with blood cultures because these parameters are routinely monitored, readily accessible, and potentially correlated. The main information extracted from WBC and N% was represented by component 1, which reflects the effect of WBCs and neutrophils in response to pathogen invasion. The main information underlying CRP and PCT was represented by component 2, which reflects the severity of inflammation and infection.
Our results showed that positive blood cultures were more likely to be distributed in the first and second quadrants (where the component 2 has a positive value) than negative blood cultures. Whereas, the proportions of positive blood cultures and negative blood cultures in the first and fourth quadrants (where the component 1 has a positive value) were similar. This distribution pattern indicated that component 2 (which represents the severity of inflammation and infection) rather than component 1 (which represents the WBC and neutrophil response) was more discriminative in assisting the judgement of blood culture results. This was further confirmed by the finding that the ROC curves of component 2 yielded the largest AUCs in the discrimination of blood culture results and patient prognosis. We also found that an increased proportion of positive blood cultures was accompanied by a stepwise increase in PCT and CRP levels, whereas more abnormal values of WBC and N% were not necessarily associated with a higher proportion of positive blood cultures. In accordance with our findings, Vargo and colleagues found that the rates of positive blood cultures were not statistically different in patients who had different ranges of WBC [(4.5–9.9) ×103, (10–14.9) ×103, and >14.9×103/µL] (15).
The unreliability of WBC and N% in assisting the judgement of blood culture results can be explained by several reasons. First, the amount of circulating WBC and N% tends to dramatically change because their consumption and supply change over time during the course of infection. The values of WBC and N% measured at a specific moment may not provide a complete picture of a patient’s condition. For instance, normal levels of WBC and N% may occur in patients without infection, patients at the early stage of infection, patients with a severe infection in whom the consumption of WBCs exceeds the capacity of the bone marrow to produce replacements, or patients at the recovery stage of infection. Second, the amount of circulating WBC and N% is influenced by different types of neutrophil cell death such as necrosis, apoptosis, autophagy, and the newly recognized NETosis, all of which actively occur during the process of sepsis (16,17). Therefore, it is difficult to establish a definite relationship between the values of WBC and N% and the probability of obtaining a positive blood culture. Third, WBC and N% are susceptible to various interferences, including genetic polymorphisms on immune response, stress, medication, trauma, cancer, hematological diseases and inflammatory diseases. Thus, WBC and N% often fail to be accurate diagnostic or prognostic indicators of BSI, as reported by a previous study (18).
Our results showed that the distribution map of factor scores was helpful for pathogen categories and risk stratification in patients with positive blood cultures. Multiple bacterial isolates were associated with the highest OR of distribution in the first and second quadrants. This finding suggested that BSI caused by multiple bacteria may more easily provoke a severe inflammatory response, which consequently resulted in high levels of infection biomarkers, including PCT and CRP (17-22). Consistent with our findings, a previous study found that patients with polymicrobial BSI had the highest PCT levels compared with those with any other monomicrobial BSI (23). Interestingly, fungal isolates were not associated a significantly higher OR of distribution in the first and second quadrants when compared to those with negative blood cultures. This may be because patients with fungal infection often have an impaired immune response and therefore elicit lower PCT and CRP levels than patients with bacterial infection (24-27). Our results also showed that non-survivors with positive blood cultures had the highest OR of distribution in the first and second quadrants, which may serve as an early warning signal for more intensive care and escalated therapy.
This study should be interpreted in light of several limitations. First, although blood culture is widely accepted as the gold standard for the diagnosis of BSI, there was a risk that true causative organisms were not identified by conventional blood culture techniques, or that contaminant pathogens were isolated and considered as pathogenic microorganisms, both of which can obscure the relationship between blood culture results and the values of the laboratory parameters. For instance, if a patient actually has a BSI but the causative organism is not detected by blood culture, this patient will be classified as a “patient with negative blood cultures”. However, the laboratory parameters may have been affected by BSI and changed. If this kind of misleading data were present, and were incorporated into the analysis, the effect of the conclusion would be weakened. Second, due to the retrospective nature of this study, the several baseline parameters were imbalanced between patients with negative blood cultures and patients with positive blood cultures. Thus, we performed logistic regression analysis for adjustment by simultaneously incorporating components and important clinical variables (age, gender, APACHE II score, and SOFA score) into the same model. The adjusted ORs (95% CIs) did not change significantly as compared to the crude estimates. Third, several episodes of blood culture results (17 cases) were precluded from the analysis due to incomplete laboratory data, which may have introduced selection bias and undermined the effect of the conclusion. Fourth, the generalizability of our findings may be limited, since it focused only on patients from one center. The results of our study should be interpreted cautiously when applied to patients in other settings. Based on the limitations mentioned above, further prospective studies are necessary to illustrate the relationship between blood culture results and infection biomarkers, and how we can use these biomarkers to predict patient blood culture results and prognosis more precisely.
Conclusions
In this study, we have established a new method called PCA to study the relationships between infection biomarkers and blood culture results and patient prognosis. In the PCA, we found that PCT- and CRP-derived component yielded the largest AUCs and appeared to be more discriminative in the prediction of blood culture results and patient prognosis than WBC- and N%-derived component and other primary biomarkers. Observations distributed in the first and second quadrants were more likely to be positive blood cultures and have a higher risk of death.
Acknowledgments
We thank all the patients involved in this study. We appreciate the work of ANNANE Djillali (General intensive care unit, Raymond Poincaré Hospital (APHP), University of Versailles SQY/INSERM 104 bd Raymond Poincaré, 92380 Garches, France) in language checking and revision.
Funding: None.
Footnote
Reporting Checklist: The authors have completed the STROBE reporting checklist. Available at http://dx.doi.org/10.21037/atm-20-3425
Data Sharing Statement: Available at http://dx.doi.org/10.21037/atm-20-3425
Peer Review File: Available at http://dx.doi.org/10.21037/atm-20-3425
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/atm-20-3425). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. The research was conducted ethically in accordance with the World Medical Association Declaration of Helsinki (as revised in 2013). The anonymous use of data retrieved from EMRS was approved by the ethics committee of Second Military Medical University (approval file number: 2020SL013). The consent was waived since no individual person’s data was shown separately. All data are presented as a whole.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Reddy EA, Shaw AV, Crump JA. Community-acquired bloodstream infections in Africa: a systematic review and meta-analysis. Lancet Infect Dis 2010;10:417-32. [Crossref] [PubMed]
- Prowle JR, Echeverri JE, Ligabo EV, et al. Acquired bloodstream infection in the intensive care unit: incidence and attributable mortality. Crit Care 2011;15:R100. [Crossref] [PubMed]
- Vallés J, Leon C, Alvarez-Lerma F. Nosocomial bacteremia in critically ill patients: a multicenter study evaluating epidemiology and prognosis. Spanish Collaborative Group for Infections in Intensive Care Units of Sociedad Espanola de Medicina Intensiva y Unidades Coronarias (SEMIUC). Clin Infect Dis 1997;24:387-95. [Crossref] [PubMed]
- Phua J, Ngerng W, See K, et al. Characteristics and outcomes of culture-negative versus culture-positive severe sepsis. Crit Care 2013;17:R202. [Crossref] [PubMed]
- Roberts FJ, Geere IW, Coldman A. A three-year study of positive blood cultures, with emphasis on prognosis. Rev Infect Dis 1991;13:34-46. [Crossref] [PubMed]
- Bates DW, Pruess KE, Lee TH. How bad are bacteremia and sepsis? Outcomes in a cohort with suspected bacteremia. Arch Intern Med 1995;155:593-8. [Crossref] [PubMed]
- Søgaard M, Norgaard M, Pedersen L, et al. Blood culture status and mortality among patients with suspected community-acquired bacteremia: a population-based cohort study. BMC Infect Dis 2011;11:139. [Crossref] [PubMed]
- Kayange N, Kamugisha E, Mwizamholya DL, et al. Predictors of positive blood culture and deaths among neonates with suspected neonatal sepsis in a tertiary hospital, Mwanza-Tanzania. BMC Pediatr 2010;10:39. [Crossref] [PubMed]
- Schaffner J, Chochua S, Kourbatova EV, et al. High mortality among patients with positive blood cultures at a children's hospital in Tbilisi, Georgia. J Infect Dev Ctries 2009;3:267-72. [PubMed]
- Tu YK, Kellett M, Clerehugh V, et al. Problems of correlations between explanatory variables in multiple regression analyses in the dental literature. Br Dent J 2005;199:457-61. [Crossref] [PubMed]
- Tu YK, Clerehugh V, Gilthorpe MS. Collinearity in linear regression is a serious problem in oral health research. Eur J Oral Sci 2004;112:389-97. [Crossref] [PubMed]
- Bekeris LG, Tworek JA, Walsh MK, et al. Trends in blood culture contamination: a College of American Pathologists Q-Tracks study of 356 institutions. Arch Pathol Lab Med 2005;129:1222-5. [PubMed]
- Fluss R, Faraggi D, Reiser B. Estimation of the Youden Index and its associated cutoff point. Biom J 2005;47:458-72. [Crossref] [PubMed]
- Jolliffe IT, Cadima J. Principal component analysis: a review and recent developments. Philos Trans A Math Phys Eng Sci 2016;374:20150202. [Crossref] [PubMed]
- Vargo JD, Grow JN, Yang S, et al. Parameters for Ordering Blood Cultures in Major Burn Injury Patients: Improving Clinical Assessment. J Burn Care Res 2018;39:445-9. [PubMed]
- Shen XF, Cao K, Jiang JP, et al. Neutrophil dysregulation during sepsis: an overview and update. J Cell Mol Med 2017;21:1687-97. [Crossref] [PubMed]
- Lerman YV, Kim M. Neutrophil migration under normal and sepsis conditions. Cardiovasc Hematol Disord Drug Targets 2015;15:19-28. [Crossref] [PubMed]
- Murray Clinton K. Evaluation of White Blood Cell Count, Neutrophil Percentage, and Elevated Temperature as Predictors of Bloodstream Infection in Burn Patients. Arch Surg 2007;142:639-42. [Crossref] [PubMed]
- Tanrıverdi H, Tor MM, Kart L, et al. Prognostic value of serum procalcitonin and C-reactive protein levels in critically ill patients who developed ventilator-associated pneumonia. Ann Thorac Med 2015;10:137-42. [Crossref] [PubMed]
- Rey C, Garcia-Hernandez I, Concha A, et al. Pro-adrenomedullin, pro-endothelin-1, procalcitonin, C-reactive protein and mortality risk in critically ill children: a prospective study. Crit Care 2013;17:R240. [Crossref] [PubMed]
- Ryu JA, Yang JH, Lee D, et al. Clinical Usefulness of Procalcitonin and C-Reactive Protein as Outcome Predictors in Critically Ill Patients with Severe Sepsis and Septic Shock. PLoS One 2015;10:e0138150. [Crossref] [PubMed]
- Ito A, Ishida T, Tachibana H, et al. Serial procalcitonin levels for predicting prognosis in community-acquired pneumonia. Respirology 2016;21:1459-64. [Crossref] [PubMed]
- Lin JN, Lai CH, Chen YH, et al. Characteristics and outcomes of polymicrobial bloodstream infections in the emergency department: A matched case-control study. Acad Emerg Med 2010;17:1072-9. [Crossref] [PubMed]
- Leli C, Ferranti M, Moretti A, et al. Procalcitonin levels in gram-positive, gram-negative, and fungal bloodstream infections. Dis Markers 2015;2015:701480. [Crossref] [PubMed]
- Charles PE, Dalle F, Aho S, et al. Serum procalcitonin measurement contribution to the early diagnosis of candidemia in critically ill patients. Intensive Care Med 2006;32:1577-83. [Crossref] [PubMed]
- Pieralli F, Corbo L, Torrigiani A, et al. Usefulness of procalcitonin in differentiating Candida and bacterial BSIs in critically ill septic patients outside the intensive care unit. Intern Emerg Med 2017;12:629-35. [Crossref] [PubMed]
- Marková M, Brodska H, Malickova K, et al. Substantially elevated C-reactive protein (CRP), together with low levels of procalcitonin (PCT), contributes to diagnosis of fungal infection in immunocompromised patients. Support Care Cancer 2013;21:2733-42. [Crossref] [PubMed]