Treatment strategy for papillary renal cell carcinoma type 2: a case series of seven patients treated based on next generation sequencing data
Introduction
Renal cell carcinoma (RCC) is a heterogeneous disease comprising a number of different types of cancer (1,2). The genetic background differs among various forms of RCC, and treatment response and prognosis vary according to subtype (3-6).
Papillary renal cell carcinoma (PRCC), which accounts for 15–20% of RCCs, is divided into type 1 and type 2 (7). In terms of clinicopathologic characteristics, type 1 and 2 diseases are different, and type 1 disease has a relatively good prognosis compared to type 2 (1). Genetic alterations also differ between type 1 and type 2 PRCC (7-9). In particular, type 1 PRCC frequently harbors MET alteration regardless of sporadic or hereditary disease (7).
PRCC type 2 (PRCC2) comprises a number of different types of non-PRCC type 1 cancers, including hereditary leiomyomatosis and renal cell carcinoma (HLRCC) (10). HLRCC is the most aggressive subtype of PRCC and harbors germline pathologic variants of fumarate hydratase (FH), which encodes one of the Krebs cycle enzymes (11). In addition, other genetic alteration, TFE3 translocation was reported in RCC in children and young adults and was associated with a grave prognosis (12).
There has been a steady effort to find genetic alterations of PRCCs. Exome and genome sequencing data suggest that the driver genetic alterations of PRCCs are varied, and somatic copy number alterations might be pathogenic events during the genetic evolution of PRCCs (13). Furthermore, large-scale genomic characterization of PRCC demonstrated that PRCC2 is divided into three subgroups according to genetic alterations. CDKN2A silencing, SETD2 mutation, TFE3 fusion, and NRF2-antioxidant response element (ARE) pathway activation were found in PRCC2 including FH mutation (7). Moreover, these genetic alterations are significantly related to cancer prognosis.
However, current treatment guideline could not reflect the advance of comprehensive genomic analysis in PRCC2 (14). Current treatment guideline suggested sunitinib or clinical trial as the first line treatment for metastatic non-clear RCC, including PRCC2 (8,15). Treatment guideline recommended only bevacizumab plus erlotinib treatment for HLRCC among PRCC2 (14). Moreover, there have been no clinical trials of treatment based on genomic information of PRCC2 because of disease rarity.
In this study, we performed exome and genome sequencing of PRCC2 patient samples to characterize their genomic landscapes. Potentially functional rare germline mutations in a wide range of known cancer genes as well as somatic mutations of a few PRCC-related genes such as ALK, CSF1R, NF2, SETD2, and FH were identified from our sequencing analysis. We also found previously reported recurrent copy gains of chr7, chr16, and chr17 in PRCC from our data as well. Our analysis confirmed the heterogeneous nature of PRCC2 genomic characteristics and identified genetic biomarkers for the prediction of prognosis and drug response in PRCC2. We present the following article in accordance with the MDAR checklist (available at http://dx.doi.org/10.21037/atm-20-3466).
Methods
Patients and sample preparation
We performed genetic analysis of PRCC2 using prospectively collected, surgically removed, fresh frozen samples of RCC and paired normal tissues in the Seoul National University Hospital tissue bank. Two other formalin-fixed paraffin-embedded samples from patients with metastatic PRCC2 were also included. These seven samples were reviewed by qualified pathologists and histologically classified as PRCC2. We obtained informed consent from all subjects, and this study was reviewed and approved by the Institutional Review Board of Seoul National University Hospital (IRB No. 1204-026-403) and have therefore been performed in accordance with the ethical standards laid down in the 1964 Declaration of Helsinki and its later amendments.
Genomic DNA was extracted from FFPE samples using a QIAgen FFPE Tissue DNA kit. DNA and RNA were extracted from the fresh frozen samples using GeneAll exgene cell SV kit and Ambio PureLink RNA mini kit, respectively. Concentrations of DNA and RNA were measured using Qubit.
Exome capture, library construction and sequencing
Up to 3 µg of genomic DNA was sheared with a Covaris SS Ultrasonicator and adaptors were then ligated to both ends of the fragments. Adaptor-ligated templates were purified using Agencourt AMPure SPRI beads, and fragments with an insert size of ~200 bp were isolated. Exons were captured from adaptor-ligated DNAs using SureSelect Human All Exon v4 + UTRs kit (71 Mb) according to the manufacturer’s instructions (Agilent Technologies). PCR amplification of the libraries was carried out for four cycles in the pre-capture step and for ten cycles after capture. Paired-end sequencing, resulting in sequences of 101 base pairs from each end of the fragments, was performed on the HiSeq 2000 platform (Illumina) following the manufacturer’s instructions. Image analysis and base calling were performed using the Illumina pipeline with default settings.
Bioinformatics analysis
Whole-genome sequencing analysis was performed for the five pairs of PRCC2 and matched control samples (four adjacent normal tissues and one blood sample). Whole-exome sequencing analysis was conducted for the two pairs of PRCC2. Reads were mapped to human reference genome (hg19) using Burrows-Wheeler Alignment (BWA) tool 0.7.12 (16). The mapped reads were processed using GATK best practice pipeline (http://fp.amegroups.cn/cms/59e8a8bf8e7cf142d5298efd3f7e76e6/atm-20-3466-1.docx) (17). We used the Mutect2 (version 3.6) (18) and GATK HaplotypeCaller (version 3.6) algorithms in order to identify somatic and germline single nucleotide variants (SNVs)/short insertions and deletions (indels). Germline variants were categorized by the American College of Medical Genomics and Genetics (ACMG) guidelines using InterVar (19,20). Germline variants classified as benign or uncertain significance by InterVar were discarded if their allele frequencies (AFs) are higher than 0.01 based on both 1,000 genomes project (21) and gnomAD (22). Somatic variants were further filtered by visual inspection of panel of normal data using Integrative Genomics Viewer (23). Both germline and somatic variants were annotated using Variant Effect Predictor (version 92) (24), and only the protein-altering variants were used for subsequent analyses (http://fp.amegroups.cn/cms/cf916fe8dd33be9ffb96d81d425f881e/atm-20-3466-2.docx).
Copy-number alterations (CNAs) were identified using Control-FREEC (25). For WES data, exonic regions were used as initial windows, and for WGS data, we used 5 kb for both initial window and step size for CNA detection (http://fp.amegroups.cn/cms/02ef9eaf1b6b4d30901c76d9ec6d4557/atm-20-3466-3.docx). Structural variations (SVs) were detected from WGS data using Meerkat (version 0.189) program (26). We set the standard deviation cutoff to call and cluster discordant repairs as 5 and the number of supporting split reads to call a SV event as 3 (http://fp.amegroups.cn/cms/b7f5aa78e9c0e90bf1e94315b355bc81/atm-20-3466-4.docx).
Results
Clinical characteristics of patients with PRCC type 2
We obtained a total of seven PRCC2 tissues and paired normal samples. The clinical characteristics of the patients are described in Tables 1,2. Of seven patients, only one was female. Median age at cancer diagnosis was 51 (range, 25–82). Four patients underwent curative resection of PRCC2 and these patients were alive without metastasis, however, three patients with metastatic disease received systemic chemotherapy and ultimately died due to disease progression. Two patients had poor prognostic risk groups and one of intermediate risk group according to Memorial Sloan Kettering Cancer Center Prognostic Model (27).

Full table

Full table
The systemic treatments administered to patients with metastatic disease are listed in Table 2. All three patients received temsirolimus as first-line systemic treatment. After disease progression, two patients received axitinib and one patient received high-dose interleukin-2 as second-line treatment. In terms of patients with metastatic disease, we performed genome sequencing at disease progression after first line systemic treatment. In other patients who had stage I–III disease, genome sequencing was performed after curative surgery.
Germline and somatic SNVs/indels
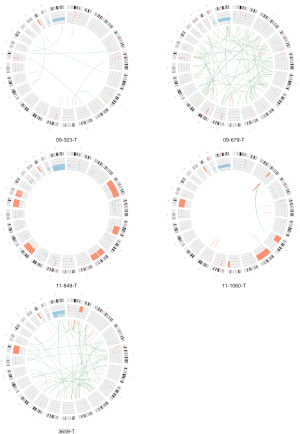
From genome and exome sequencing analysis, we determined germline and somatic SNVs/indels (Figure 1, http://fp.amegroups.cn/cms/cf916fe8dd33be9ffb96d81d425f881e/atm-20-3466-2.docx). In terms of germline mutations, a wide range of known cancer genes involved in various kinds of biological processes such as positive regulation of biosynthetic process, phosphate containing compound metabolic process, tissue development, and others were seemed to be affected by potentially functional rare germline variants reflecting the heterogeneous nature of PRCC2.

Among a few recurrently mutated genes by germline variants, FH germline mutations were detected in two patients (Figure 1). For the patients with metastatic disease, one with familial leiomyoma had a germline missense mutation of FH (p.Lys230Glu) (Figure S1A), and the other patient without family history of HLRCC harbored a germline splicing site mutation (c.1108+1G>A) in FH (Figure S1B).
Other germline mutations were also found in PRCC2 (Figure 1, http://fp.amegroups.cn/cms/cf916fe8dd33be9ffb96d81d425f881e/atm-20-3466-2.docx). Germline MKL1, NACA, PRDM16, TET2, RGPD3, CSMD3 and KMT2C alterations were found in more than one PRCC2 sample. Genetic alterations were present in different loci of coding region. In addition, BRCA1, BRCA2 and PMS2 germline mutations were also detected. In terms of functional alteration according to base change, BRCA1 alteration (p.Lys181Gln), BRCA2 (p.Gly2508Ser) and PMS2 (p.Val196Phe) resulted in amino acid changes be deleterious by SIFT prediction (28). In addition, 16 genes were affected by pathogenic or likely pathogenic germline mutations based on the ACMG criteria in at least one patient. Among them ALDH2, BCR, FH and NBEA genes were known cancer driver genes according to COSMIC Cancer Gene Census database (29) identified (http://fp.amegroups.cn/cms/cf916fe8dd33be9ffb96d81d425f881e/atm-20-3466-2.docx).
A significant number of somatic mutations were detected from tissue development-related genes such as SETD2, COL2A1, FLNA, NF2, NOTCH2, and RAD51B. Especially, SETD2 was altered also by germline mutation in other samples (Figure 1). Interestingly, a somatic FH frameshift deletion (p.Leu132Ter) was detected from the patient with metastatic disease who already harbored the germline splicing site mutation (c.1108+1G>A) (Figure S1B).
Copy number alterations and structure variations
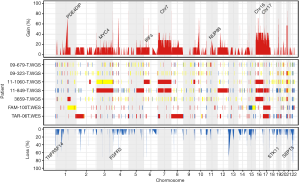
CNAs were analyzed using both whole-exome and whole-genome sequencing data. Recurrent copy number gains of chromosome 7, 16, and 17 were observed in our data (Figure 2), which is well concordant with that reported by The Cancer Genome Atlas (7). Other than chromosomal copy number alterations, recurrent focal amplifications of potential oncogenes such as MYC4, IRF4 and NUP98 were identified. Frequent copy number losses of TNFRSF14, FGFR3, STK11, and SEPT5 that are known to have tumor suppressor properties were also found. For SV analysis, we used whole-genome sequencing data and identified average 34.3 SV events (sd: 29.7) per samples. We identified a number of gene fusions in PRCC2 samples but none affecting known cancer driver genes (Figure 3).
Precision patient treatment strategy according to genetic alterations
We evaluated the effect of genetic alterations on patient prognosis (Tables 1,2). Regardless of genetic alterations, primary carcinoma that received curative surgery did not recur. Three patients with metastatic carcinomas were treated with systemic chemotherapy and eventually died due to disease progression. All patients had received temsirolimus (mTOR inhibitor) as their first line treatment according to treatment guideline (14). After disease progression, sequencing results were reported and two of the three patients with metastatic disease had genetic alteration of FH.
One patient with familiar history of leiomyoma had a germline missense mutation of FH. Among his family, his mother and her sisters have been diagnosed with myoma, thyroid cancer, and early gastric cancer (Figure S2). Germline FH mutation testing of his family members revealed germline missense mutation of FH at the same locus in his maternal family members but absent in his father (Figure S1A). In clinical course, this patient did not respond to mTOR inhibitor, VEGFR TKI, cytotoxic chemotherapy, or the anti-PD1 antibody pembrolizumab (Table 2). After we found the FH germline mutation, the patient was treated with a combination of bevacizumab [an anti-VEGF antibody) and erlotinib (an epithelial growth factor receptor (EGFR) TKI) and showed a durable response of 40 weeks (Figure 4A). Disease progression after bevacizumab and erlotinib treatment, there was no effective treatment strategy. In this patient, overall survival (OS) duration between diagnosis to death was 14.5 months.
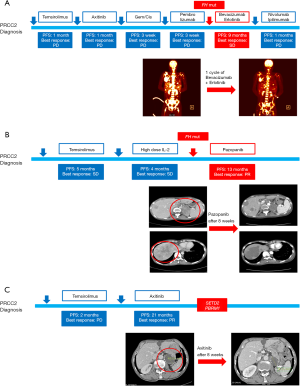
A young patient with germline and somatic FH mutations was treated with pazopanib, a selective multi-targeted receptor TKI, after her genetic alteration was revealed (Table 2). Pazopanib resulted in a decrease in the tumor over one year, but grade 3 lethargy developed and resulted in treatment discontinuation. Time to progression of pazopanib treatment was 52weeks. Her disease also harbored PMS2 alteration (Figure 4B). However, her performance status did not permit further treatment, such as immune check point inhibitor. She had 35.8 months of overall survival.
The patient without genetic alteration of FH showed a durable response to temsirolimus and axitinib [a tyrosine kinase inhibitor (TKI), which targets vascular endothelial growth factor-receptor 1–3 (VEGFR 1–3)]. We observed PBRM1 and SETD2 mutations in his disease and he had relatively long survival duration. He responded axitinib treatment for a long duration (87 weeks) until adverse events forbade him from receiving treatment (Table 2, Figure 4C).
Discussion
In this study, we identified genetic alterations in PRCC2. Mutation in the gene encoding FH, either germline or somatic, was found in patients younger than 30 years and indicated poor prognosis. In terms of treatment, patients harboring FH mutation responded to anti-angiogenesis agents including bevacizumab and pazopanib.
PRCC is defined as RCC with a papillary pattern based on pathologic finding (1). Type 2 PRCC is lined by large cells with abundant eosinophilic cytoplasm compared to the small cells with clear to basophilic cytoplasm present in type 1 PRCC. The best-known genetic alteration of type 1 PRCC is MET mutation, whereas FH nutation is common in type 2, especially in familial cases (30). However, with the exception of familial cases of PRCC, genetic alterations that drive tumorigenesis of PRCC2 have not been revealed. Recent genetic analysis showed that PRCC2 could be classified into three subgroups according to molecular differences (7). One subgroup represented by FH mutation and CDKN2A silencing due to hypermethylation of the CDKN2A promoter had a relatively small number of PRCC2 (13.3%). This subgroup was characterized by relatively young patients and extremely short survival duration compared to other subtypes. In our study, two young patients with FH mutated PRCC2 had similar clinical characteristics to this subtype.
FH, a tricarboxylic acid cycle (TCA) enzyme, catalyzes the hydration of fumarate into malate. Deficiency of FH causes accumulation of fumarate and activation of hypoxia inducible factor (HIF) under normal oxygen levels (31). VEGF and glucose transporter-1 (GLUT-1) are upregulated by an excess of intracellular fumarate through the HIF-dependent pathway. Therefore, inhibition of the VEGF pathway and glucose transport has been suggested as a therapeutic approach in FH mutant cancer (31,32). In our study, we identified two patients with PRCC2 harboring FH mutation characterized by rapid progression. One patient with germline FH mutation did not respond to axitinib, a selective VEGFR inhibitor, but did respond to bevacizumab and erlotinib combination therapy (33). Another patient was treated using pazopanib, a multi-targeted receptor TKI whose targets include VEGFR, and she only responded to this treatment. The mechanism of action of pazopanib does not include glucose metabolism, but hypoglycemia frequently occurs during pazopanib treatment (34). With regard to this adverse event, pazopanib might modify glucose metabolism in FH mutant PRCC2. Therefore, based on our clinical experience, a therapeutic strategy for FH mutant PRCC2 should consider targeting glucose metabolism in addition to inhibition of the VEGF pathway.
The Warburg effect (35), a metabolic shift to aerobic glycolysis in normoxia status, is one of the characteristics of FH mutant RCC (33). This ineffective metabolism in tumor cells resulting from the rapid proliferation of cancer cells might induce cachexia of patients (36). In our study, the patient treated with pazopanib suffered cancer-related cachexia even though her disease was well controlled. Consequentially, her general condition gradually deteriorated, and she could not undergo further treatment.
Pathogenic germline alteration of C7, FH, MAK, NBEA, NDUFS1 and TTC37 were also detected in this study. Mutations in NDUFS1, the largest subunit of mitochondrial complex I, are known to reduce the activity of complex I (37). We also identified ten likely pathogenic germline mutations in ALDH1A3, ALDH2, APOE, BCR, CCNL1, COL4A2, EYS, ITGB3, NMNAT1 and PLCZ1. Gao et al. have found that von Hippel-Lindau tumor suppressor (VHL) was directly binding to the promoter of ALDH2 to regulate the transcriptional activity of hepatocyte nuclear factor 4 alpha (HNF4A) in clear-cell RCCs (38). Loss of HNF4A binding activity was often observed in renal cell carcinogenesis (39).
Germline alteration of BRCA1, BRCA2 and PMS2 were also detected in this study. Germline BRCA1 and/or BRCA2 deleterious mutation caused hereditary breast and ovarian cancer as well as other types of familiar cancer syndrome (40). Moreover, cancer caused by BRCA1 mutation was sensitive to PARP1 inhibitors, such as olaparib and talazoparib. Recent clinical trials showed that PARP1 inhibitor prolonged duration of survival in patients with metastatic breast cancer with pathogenic BRCA1 and/or BRCA2 mutations (41,42). We found a deleterious mutation of PMS2, one of mismatch repair (MMR) genes associated with Lynch syndrome (43). With regard to immune checkpoint blockade had more clinical benefit in MMR deficient cancers compared to MMR proficient cancers, anti-PD-1 antibody wound be considered as a therapeutic option in PRCC2 with PMS2 mutation.
Recent genetic study using targeted deep sequencing determined that approximately 20% of advanced RCCs had a germline mutation even though almost all cases were sporadic rather than hereditary cancer. That study found BRCA1, BRCA2, and CHEK2 germline mutations and suggested that half of germline mutations could be potential targets for direct systemic treatment (44). Therefore, these germline mutations are potential therapeutic targets in advanced PRCC2.
Somatic mutations in PRCC2 also indicated disease prognosis. One patient with metastatic disease harboring SETD2 and PBRM1 somatic mutations had overall survival of about 5 years despite having stage IV PRCC2. His disease showed a durable response to axitinib and had a relatively good prognosis. Both SETD2 and PBRM1 were SWI/SNF complex genes associated with chromatin remodeling. In clear cell RCC, SETD2 was associated to poor prognosis while PBRM1 had no impact on patient’s survival or also indicated poor prognosis (45,46). Moreover, in terms of prognostic risk group according to clinical characteristics, this patient was categorized into intermediate risk group, not poor risk group in which other two patients with metastatic PRCC2 harboring FH mutation (27). Therefore, we treated PRCC2 as clear cell RCC with axitinib and he might be having favorable treatment outcome in this case.
The relationship between genetic alteration and clinical characteristics including risk groups have not been revealed neither previous studies nor our study. Further large scaled genetic studies of metastatic PRCC2 would give the information of the relationship between genetic and clinical characteristics to guide treatment of metastatic PRCC2.
Conclusions
Our results indicate the role of genetic alterations in precision systemic treatment of PRCC2. FH alterations were found in not only HLRCC, but also in sporadic PRCC2. Sporadic PRCC2 with somatic FH frameshift deletion had similar clinical outcome to HLRCC. In addition, genetic alterations of PBRM1 and SETD2 had a predictive role of PRCC2 prognosis.
In this genetic study, we successfully applied genetic information to patient treatment and prolonged survival. Therefore, genome sequencing should be performed to identify candidates for targeted therapy in PRCC2, a genetically heterogeneous disease.
Acknowledgments
This work partially contained Ji-Yeon Kim’s thesis of the Doctor of Philosophy degree in Seoul National University College of Medicine.
Funding: This research was supported by a grant of the Korean Health Technology R&D Project through the Korea Health Industry Development Institute (KHIDI), funded by the Ministry of Health & Welfare, Republic of Korea (grant number: HI15C1551) and a grant from the National Research Foundation of Korea (NRF-2017R1D1A1B03028446). This work is also supported by the 2019 Research Fund (1.190007.01 and 1.190033.01) of UNIST, the Next-Generation Information Computing Development Program through the National Research Foundation of Korea (NRF) funded by the Ministry of Science and ICT (MSIT) (NRF-2016M3C4A7952635), the Collaborative Genome Program for Fostering New Post- Genome Industry of the NRF funded by the MSIT (NRF-2017M3C9A5031004), and the Basic Science Research Programs through the NRF funded by the MSIT (NRF- 2018R1C1B6008448) and the Ministry of Education (NRF-2018R1A6A1A03025810). And this research was supported by a grant of the Korea Health Technology R&D Project through the Korea Health Industry Development Institute (KHIDI), funded by the Ministry of Health & Welfare, Republic of Korea (grant number: HR20C0025).
Footnote
Reporting Checklist: The authors have completed the MDAR checklist. Available at http://dx.doi.org/10.21037/atm-20-3466
Data Sharing Statement: Available at http://dx.doi.org/10.21037/atm-20-3466
Peer Review File: Available at http://dx.doi.org/10.21037/atm-20-3466
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/atm-20-3466). SHL reports personal fees from AstraZeneca/MedImmune, personal fees from Roche, grants, and personal fees from Merck, outside the submitted work. The authors have no other conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. This study was reviewed and approved by the Institutional Review Board of Seoul National University Hospital (IRB No. 1204-026-403) and have therefore been performed in accordance with the ethical standards laid down in the 1964 Declaration of Helsinki and its later amendments. We obtained informed consent from all subjects.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Rini BI, Campbell SC, Escudier B. Renal cell carcinoma. Lancet 2009;373:1119-32. [Crossref] [PubMed]
- Cohen HT, McGovern FJ. Renal-cell carcinoma. N Engl J Med 2005;353:2477-90. [Crossref] [PubMed]
- Motzer RJ, Bacik J, Mariani T, et al. Treatment outcome and survival associated with metastatic renal cell carcinoma of non-clear-cell histology. J Clin Oncol 2002;20:2376-81. [Crossref] [PubMed]
- Cancer Genome Atlas Research Network. Comprehensive molecular characterization of clear cell renal cell carcinoma. Nature 2013;499:43-9. [Crossref] [PubMed]
- Davis CF, Ricketts CJ, Wang M, et al. The somatic genomic landscape of chromophobe renal cell carcinoma. Cancer Cell 2014;26:319-30. [Crossref] [PubMed]
- Choueiri TK, Motzer RJ. Systemic Therapy for Metastatic Renal-Cell Carcinoma. N Engl J Med 2017;376:354-66. [Crossref] [PubMed]
- Cancer Genome Atlas Research Network, Linehan WM, Spellman PT, et al. Comprehensive Molecular Characterization of Papillary Renal-Cell Carcinoma. N Engl J Med 2016;374:135-45. [Crossref] [PubMed]
- Ravaud A, Oudard S, De Fromont M, et al. First-line treatment with sunitinib for type 1 and type 2 locally advanced or metastatic papillary renal cell carcinoma: a phase II study (SUPAP) by the French Genitourinary Group (GETUG)dagger. Ann Oncol 2015;26:1123-8. [Crossref] [PubMed]
- Pignot G, Elie C, Conquy S, et al. Survival analysis of 130 patients with papillary renal cell carcinoma: prognostic utility of type 1 and type 2 subclassification. Urology 2007;69:230-5. [Crossref] [PubMed]
- Lopez-Beltran A, Scarpelli M, Montironi R, et al. 2004 WHO classification of the renal tumors of the adults. Eur Urol 2006;49:798-805. [Crossref] [PubMed]
- Tomlinson IP, Alam NA, Rowan AJ, et al. Germline mutations in FH predispose to dominantly inherited uterine fibroids, skin leiomyomata and papillary renal cell cancer. Nat Genet 2002;30:406-10. [Crossref] [PubMed]
- Macher-Goeppinger S, Roth W, Wagener N, et al. Molecular heterogeneity of TFE3 activation in renal cell carcinomas. Mod Pathol 2012;25:308-15. [Crossref] [PubMed]
- Kovac M, Navas C, Horswell S, et al. Recurrent chromosomal gains and heterogeneous driver mutations characterise papillary renal cancer evolution. Nat Commun 2015;6:6336. [Crossref] [PubMed]
- NCCN Clinical Practice Guidelines on Oncology (NCCN Guidelines) Kidney Cancer. Available online: https://www.nccn.org/professionals/physician_gls/default.aspx. 2019.
- Ronnen EA, Kondagunta GV, Ishill N, et al. Treatment outcome for metastatic papillary renal cell carcinoma patients. Cancer 2006;107:2617-21. [Crossref] [PubMed]
- Li H, Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics 2009;25:1754-60. [Crossref] [PubMed]
- Van der Auwera GA, Carneiro MO, Hartl C, et al. From FastQ data to high confidence variant calls: the Genome Analysis Toolkit best practices pipeline. Curr Protoc Bioinformatics 2013;43:11.10.1-11.10.33.
- Cibulskis K, Lawrence MS, Carter SL, et al. Sensitive detection of somatic point mutations in impure and heterogeneous cancer samples. Nat Biotechnol 2013;31:213-9. [Crossref] [PubMed]
- Richards S, Aziz N, Bale S, et al. Standards and guidelines for the interpretation of sequence variants: a joint consensus recommendation of the American College of Medical Genetics and Genomics and the Association for Molecular Pathology. Genet Med 2015;17:405-24. [Crossref] [PubMed]
- Li Q, Wang K. InterVar: Clinical Interpretation of Genetic Variants by the 2015 ACMG-AMP Guidelines. Am J Hum Genet 2017;100:267-80. [Crossref] [PubMed]
- 1000 Genomes Project Consortium, Auton A, Brooks LD, et al. A global reference for human genetic variation. Nature 2015;526:68-74.
- Lek M, Karczewski KJ, Minikel EV, et al. Analysis of protein-coding genetic variation in 60,706 humans. Nature 2016;536:285-91. [Crossref] [PubMed]
- Robinson JT, Thorvaldsdottir H, Winckler W, et al. Integrative genomics viewer. Nat Biotechnol 2011;29:24-6. [Crossref] [PubMed]
- McLaren W, Gil L, Hunt SE, et al. The Ensembl Variant Effect Predictor. Genome Biol 2016;17:122. [Crossref] [PubMed]
- Boeva V, Popova T, Bleakley K, et al. Control-FREEC: a tool for assessing copy number and allelic content using next-generation sequencing data. Bioinformatics 2012;28:423-5. [Crossref] [PubMed]
- Yang L, Luquette LJ, Gehlenborg N, et al. Diverse mechanisms of somatic structural variations in human cancer genomes. Cell 2013;153:919-29. [Crossref] [PubMed]
- Motzer RJ, Bacik J, Murphy BA, et al. Interferon-alfa as a comparative treatment for clinical trials of new therapies against advanced renal cell carcinoma. J Clin Oncol 2002;20:289-96. [Crossref] [PubMed]
- Ng PC, Henikoff S. SIFT: Predicting amino acid changes that affect protein function. Nucleic Acids Res 2003;31:3812-4. [Crossref] [PubMed]
- Forbes SA, Beare D, Boutselakis H, et al. COSMIC: somatic cancer genetics at high-resolution. Nucleic Acids Res 2017;45:D777-83. [Crossref] [PubMed]
- Linehan WM, Srinivasan R, Schmidt LS. The genetic basis of kidney cancer: a metabolic disease. Nat Rev Urol 2010;7:277-85. [Crossref] [PubMed]
- Frezza C, Zheng L, Folger O, et al. Haem oxygenase is synthetically lethal with the tumour suppressor fumarate hydratase. Nature 2011;477:225-8. [Crossref] [PubMed]
- Isaacs JS, Jung YJ, Mole DR, et al. HIF overexpression correlates with biallelic loss of fumarate hydratase in renal cancer: novel role of fumarate in regulation of HIF stability. Cancer Cell 2005;8:143-53. [Crossref] [PubMed]
- Linehan WM, Rouault TA. Molecular pathways: Fumarate hydratase-deficient kidney cancer--targeting the Warburg effect in cancer. Clin Cancer Res 2013;19:3345-52. [Crossref] [PubMed]
- Motzer RJ, Hutson TE, Cella D, et al. Pazopanib versus sunitinib in metastatic renal-cell carcinoma. N Engl J Med 2013;369:722-31. [Crossref] [PubMed]
- Warburg O. On the origin of cancer cells. Science 1956;123:309-14. [Crossref] [PubMed]
- Fearon KC, Glass DJ, Guttridge DC. Cancer cachexia: mediators, signaling, and metabolic pathways. Cell Metab 2012;16:153-66. [Crossref] [PubMed]
- Hoefs SJ, Skjeldal OH, Rodenburg RJ, et al. Novel mutations in the NDUFS1 gene cause low residual activities in human complex I deficiencies. Mol Genet Metab 2010;100:251-6. [Crossref] [PubMed]
- Gao YH, Wu ZX, Xie LQ, et al. VHL deficiency augments anthracycline sensitivity of clear cell renal cell carcinomas by down-regulating ALDH2. Nat Commun 2017;8:15337. [Crossref] [PubMed]
- Sel S, Ebert T, Ryffel GU, et al. Human renal cell carcinogenesis is accompanied by a coordinate loss of the tissue specific transcription factors HNF4 alpha and HNF1 alpha. Cancer Lett 1996;101:205-10. [Crossref] [PubMed]
- Levy-Lahad E, Friedman E. Cancer risks among BRCA1 and BRCA2 mutation carriers. Br J Cancer 2007;96:11-5. [Crossref] [PubMed]
- Litton JK, Rugo HS, Ettl J, et al. Talazoparib in Patients with Advanced Breast Cancer and a Germline BRCA Mutation. N Engl J Med 2018;379:753-63. [Crossref] [PubMed]
- Robson M, Im SA, Senkus E, et al. Olaparib for Metastatic Breast Cancer in Patients with a Germline BRCA Mutation. N Engl J Med 2017;377:523-33. [Crossref] [PubMed]
- Senter L, Clendenning M, Sotamaa K, et al. The clinical phenotype of Lynch syndrome due to germ-line PMS2 mutations. Gastroenterology 2008;135:419-28. [Crossref] [PubMed]
- Carlo MI, Mukherjee S, Mandelker D, et al. Prevalence of Germline Mutations in Cancer Susceptibility Genes in Patients With Advanced Renal Cell Carcinoma. JAMA Oncol 2018;4:1228-35. [Crossref] [PubMed]
- Hakimi AA, Ostrovnaya I, Reva B, et al. Adverse outcomes in clear cell renal cell carcinoma with mutations of 3p21 epigenetic regulators BAP1 and SETD2: a report by MSKCC and the KIRC TCGA research network. Clin Cancer Res 2013;19:3259-67. [Crossref] [PubMed]
- Pawłowski R, Muhl SM, Sulser T, et al. Loss of PBRM1 expression is associated with renal cell carcinoma progression. Int J Cancer 2013;132:E11-7. [Crossref] [PubMed]