The role of chemokine receptor 4 and its ligand stromal cell derived factor 1 in breast cancer
Introduction
Stromal cell derived factor1 (SDF1), a chemokine and its receptor C-X-C chemokine receptor type 4 (CXCR4) are responsible for the trafficking and homeostasis of immune cells such as T lymphocytes. Subsequently, it has been determined that the CXCR4/SDF1 axis have prominent role in primary and metastatic breast cancer, as it is involved in tumour progression, angiogenesis, metastasis, and survival. Recently, intensive research has demonstrated that CXCR4/SDF1 interaction also regulates several key events in wide variety of cancers (1). CXCR4 expression is low or absent on normal breast epithelium (2). Thus, CXCR4 expression is generally a characteristic of the malignant epithelial cells and not its normal counterpart. Breast tumours that express CXCR4 preferentially metastasize to specific target organs such as liver, lung, bone marrow and lymph nodes. Müller et al. [2001] hypothesized that chemokines are secreted by metastatic target organs and can function as specific attractants for the tumour cells, analogous to their chemo attractive function for hematogenous cells during the inflammatory process. Primary breast tumours expressed the CXCR4 receptor, whereas target sites of breast cancer metastases expressed SDF1 more than other organs (2).
Up-regulation of cytoplasmic expression of CXCR4/SDF1 might be one of the molecular mechanisms facilitating lymph node metastasis of invasive carcinoma. Also CXCR4/SDF1 is critical in determining the metastatic destination of breast cancer cells, and blocking of CXCR4 in vivo results in significant inhibition of breast cancer metastasis (2). High levels of functional SDF1 have been identified in tumour microenvironments in ovarian cancer (3), breast cancer (4), glioblastoma (5) and prostatic cancer (6). SDF1 induces synthesis of matrix metalloproteinases (7) which can break down extracellular matrix and promote tumour invasion, and modulate the expression and function of cell surface integrin molecules (8). Taken together, these molecular mechanisms triggered by SDF1 may lead to tumour metastasis. Different expression patterns of CXCR4/SDF1 by cells of different tumours indicate differences in the biological behaviour of the respective tumour cells. The steroid hormone, estradiol, plays an important role in the progression of breast cancer and a majority of the human breast cancers start out as estrogen dependent. The chemokine SDF1 was identified as a key mediator of E2-induced breast cancer cell proliferation and survival. CXCR4 and CXCR7 were differentially regulated by E2, which enhanced the expression of both CXCL12 and CXCR4 (9). Kubarek et al. [2007] observed that E2 and 4OHT increased the expression of CXCR4 and SDF1 transcripts and proteins in estrogen receptor positive but not in negative endometrial adenocarcinoma cell lines (10).
The present study hypothesizes that the expression patterns of CXCR4 and SDF1 have prognostic or predictive importance in breast cancer patients. This has been studied by cell proliferation, viability assays, expression and colocalization of CXCR4 and SDF1 in breast cells and tissues in order to verify the correlation of CXCR4 and SDF1 with clinicopathological factors and overall survival of patients. This study also analyzed the modulatory effect of 17-β estradiol (E2), 4-hydroxytamoxifen (4OHT) and its combined effect in the expression and colocalization of CXCR4/SDF1 in breast cancer cells.
Materials and methods
Study subjects
Breast tumour samples were collected prospectively from previously untreated patients who underwent surgery for breast cancer at Regional Cancer Centre, Thiruvananthapuram, India. Normal breast tissues were obtained from patients subjected to reduction mammoplasty and from patients undergoing resection of benign breast lesions. Fresh tissues were collected and stored in RNAlater (Ambion) for RNA extraction, and a tissue bit was transferred into 10% buffered formalin for immunohistochemical analysis. The study group included a total of 152 breast tissue samples, of which 23 were normal tissue samples and 129 were tumour tissue samples. A total of 124 breast tumour and 23 normal breast tissue samples were used for mRNA analysis. For protein analysis, 68 breast tumour and 11 normal breast tissue samples were used. Patient’s details and other clinical parameters were obtained from the patient’s medical records. The study was approved by the Institution Review Board and the Human Ethics Committee of the Regional Cancer Centre, Thiruvananthapuram, India. Informed consent was obtained from all patients included in the study.
Immunohistochemistry
Four µm thick sections of the paraffin embedded tissue samples were taken on poly L-lysine coated slides. Histopathologic evaluation was done by Haematoxylin and Eosin staining. Serial sections from representative paraffin blocks containing normal/tumour cells from each case were used for immunohistochemistry. A total of 79 samples were used for immunohistochemical analysis. Briefly, sections were deparaffinized in xylene and passed through graded alcohol. Endogenous peroxidase activity was blocked with 0.3% H2O2 in methanol for 30 min, followed by antigen retrieval by heating sections in 10 mM citrate buffer (pH 6.0). Sections were then incubated with 3% bovine serum albumin (BSA) for 20 min at room temperature followed by incubation at 4 °C overnight with primary antibodies specific for CXCR4 and SDF1 (Santa Cruz Biotechnology, Inc). The reactions were visualized using Super Sensitive Polymer-HRP detection system, (Biogenex, CA.) following manufacturer’s protocol. Sections were counterstained with haematoxylin, dehydrated in graded alcohol, cleared in xylene and mounted. Immunostained slides were scored for CXCR4 and SDF1 using Allred scoring system (11).
Maintenance of breast cancer cell line, MCF-7 and treatment
MCF-7 cells were routinely grown in DMEM (Sigma) supplemented with 10% fetal bovine serum (Sigma), antibiotic antimytotic mix (Sigma) containing penicillin (100 U/mL), streptomycin (100 µg/mL) and amphotericin B (0.25 µg/mL) and the cells were incubated at 37 °C, under 5% CO2. The cells were steroid depleted by growing in phenol-red-free (PRF) DMEM supplemented with 5% dextran activated double charcoal stripped fetal bovine serum (DCC) and the cells were maintained in a humidified atmosphere at 37 °C and 5% CO2.
MTT assay
Cell proliferation and cytotoxicity were assessed by 3-(4, 5-dimethylthiazol-2-yl)-2, 5-diphenyltetrazolium bromide (MTT) (Sigma) assay. Five thousand cells per well were seeded in a 96 well plate, the cells cultured in steroid depleted condition were maintained at 37 °C and 5% CO2. MCF-7 cells were incubated with or without different concentrations of E2, 4OHT and its different combinations for 24, 48 and 72 h. The cells were then washed with 1XPBS and MTT was added to each wells. The formazan crystals were dissolved using dimethysulfoxide (Sigma). The absorbance at 570 nm was read on ELISA reader.
Reverse transcriptase-polymerase chain reaction (RT-PCR)
Gene expression for CXCR4 and SDF1 were assessed using Reverse Transcriptase—conventional PCR. Housekeeping gene glyceraldehyde-3-phosphate dehydrogenase (GAPDH) served as internal control and the expression values for each PCR product were normalised to their GAPDH expression. Total RNA was extracted using TRI reagent (Sigma, USA) following manufacturer’s protocol. Integrity and purity of final RNA extracts were assessed by agarose gel electrophoresis and spectrophotometry. That 2 µg of total RNA was reverse transcribed to cDNA in a 20 µL reaction mix containing 200 U of MMLV Reverse Transcriptase (RT) in 1× reaction buffer with 2 µg of random hexamer, 6 U of RNasein and 100 µM dNTP mix at 42 °C for 1 hour. And 2.5 µL of the cDNA was used for PCR amplification in a 20 µL reaction buffer containing 1 U Taq DNA polymerase in MgCl2 rich 1× reaction buffer, 150 µM dNTP mix and 20 picomoles of oligonucleotide sense and antisense primers (Table 1). The thermal cycling conditions comprised an initial denaturation step at 94 °C for 4 min, then 30 cycles at 94 °C for 30 s, and annealing temperatures (Table 1) for different parameters. PCR products were separated on a 1.2% agarose gel. The presence and absence of corresponding bands on gel were considered as positive and negative respectively. In the case of RT-PCR from MCF-7 cell extracts, The bands obtained were quantified using Image J 1.45S, USA.
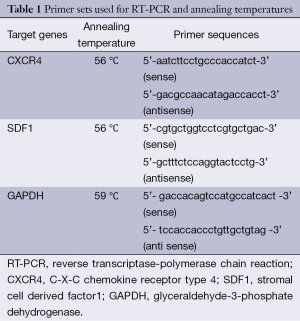
Full table
Immunofluorescence microscopy and colocalization of CXCR4 and SDF1
The fixed cells were incubated in BSA for 30 min. Primary antibodies Rabbit antihuman SDF1 and Mouse antihuman CXCR4 (Santa Cruz Biotechnology, Inc.) were added and incubated for 60 min at 37 °C. Then the sections were incubated with corresponding Alexa fluor conjugated secondary antibodies for 30 min at 37 °C. For colocalization studies, first primary antibody was added and incubated for 60 min for 37 °C then second primary antibody was added and incubated for 60 min at 37 °C with an intermittent PBS wash. Subsequently the sections were incubated with a mixture of Alexa Fluor 405 conjugated anti rabbit antibody and Alexa Fluor 488 conjugated anti mouse antibody for 30 min at 37 °C. Slides were examined under an Axioscope 2 plus fluorescent microscope, Carl Zeiss, Germany. The images were captured by Canon Zoom Browser EX, USA. Colocalization of the proteins and Pearson’s correlation coefficient (Rr) were calculated using the NIS elements software. The mean Rr coefficient was calculated, mean ± SD values for each condition were plotted on the histogram.
Statistical analysis
Statistical analysis was carried out using SPSS statistical software library. To analyze the correlation of expression levels between different genes, proteins and clinicopathologic factors, Spearman’s correlation was used. The survival analysis was performed using Kaplan Meier method with log rank test to establish the status of CXCR4 and SDF1 as a predictor of overall survival. A P value <0.05 was considered as statistically significant. The results of analysis of MCF-7 cells are presented as the mean ± SD of at least three independent experiments were subjected to students ‘t’ test for comparison of the means between two groups wherein P<0.05 were considered as significant.
Results
Expressional and correlation analysis of chemokine receptor CXCR4 and its ligand SDF1 in breast cancer
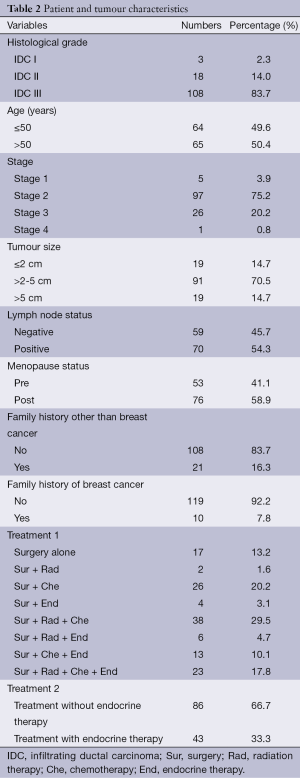
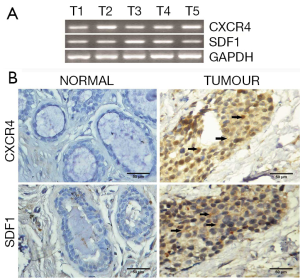
The study group included a total of 152 breast tissue samples of which 23 were normal tissue samples and 129 were tumour tissue samples. Patient and tumour characteristics for the entire study population are shown in Table 2. The mRNA expression levels of chemokine receptors were observed to be higher in tumour samples than in normal samples. Agarose gel electrophoresis of PCR products from breast tissue samples showed clear sharp bands of CXCR4 (367 bp) and SDF1 (237 bp) (Figure 1A). CXCR4 was positive in 83.1% of tumour samples and 69.6% normal samples. SDF1 was positive in 75.8% tumour samples and 65.2% normal samples. CXCR4 and SDF1 proteins were localized in human breast tissue using immunohistochemistry. CXCR4 and SDF1 immunoreactivity were observed in the cytoplasm and to a lesser extent in the nuclei of tumour epithelial cells (Figure 1B). CXCR4 showed cytoplasmic immunoreactivity in 38.2% of tumour sample and 9.1% in normal samples. SDF1 exhibited cytoplasmic positivity in 70.6% of tumour samples and 45.5% of normal samples respectively. Allred score in malignant breast tissues for CXCR4 and its ligand SDF1 is shown in Table 3. Spearman correlation analysis between CXCR4 and SDF1 showed a strong positive correlation in the gene (r=0.498, P=0.000) and protein level (r=0.375, P=0.002).
Full table

Full table
Correlation of chemokine receptor CXCR4 and its ligand SDF1 with clinicopathological variables
Association between the expression of Chemokine receptor CXCR4 and its ligand SDF1 with clinicopathological parameters was examined. Histological grade showed significant positive correlation with CXCR4 positivity in the mRNA level (r=0.204, P=0.023). Increased tumour size showed positive correlation with SDF1 (r=0.210, P=0.019). mRNA level expression of the Chemokine receptor CXCR4 and its ligand SDF1 when compare with patient age, menopause status, family history, tumour stage and lymph node metastasis showed no correlation. No significant correlation was observed between any of the protein expression status and clinicopathological variables.
Survival analysis according to status of chemokine receptor CXCR4 and its ligand SDF1
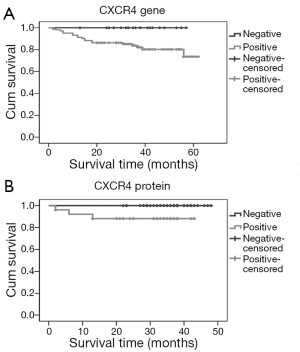
Survival analysis showed that negative immunostaining for CXCR4 in breast cancer patients was associated with a better overall survival (P=0.023), that translated to overexpression of CXCR4 associated with decreased survival in breast cancer patients. The association of CXCR4 negativity in gene level with a better overall survival is on the bordeline of statistical significance (P=0.049) (Figure 2). The median follow up period is 37 months; during the follow up period three patients succumbed to the disease. Immunostaining for SDF1 in breast cancer patients were not significantly associated with overall survival (data not shown) in our study. Hence,the positive expression of CXCR4 can be considered as a predictor of decreased overall survival in breast cancer patients.
Effect of 17β-estradiol, 4-hydroxytamoxifen and its combinations on MCF-7 cells
Different concentrations of estradiol (E2) ranging from 0.5 to 1,000 (nM) were used to study the proliferative effect of E2 after incubation for 24, 48 and 72 h in MCF-7 cells. The growth stimulatory effects of E2 on MCF-7 cells were determined by MTT assay and direct cell counting. After 24 h incubation, no obvious survival stimulation was observed. It was not until 48 h incubation that E2 induced 10% more increase in percentage survival in 100 nM E2 treated cells. Increasing the incubation time to 72 h further increased the percentage survival. At this time period, 50 nM E2 attained 40% increase in survival with respect to controls. On the other hand, 1,000 nM E2 reduced cell survival from 14% to 48% after 24 and 72 h treatment respectively when compared to the maximum survival stimulation of 110% in 100 nM E2 after 24 h and 140% in 50 nM after 72 h. The maximum proliferative effect of E2 was found to be 100, 50, and 50 nM for 24, 48 and 72 h incubation respectively (Figure 3A). From these experiments, E2 with a concentration of 50 nM was used for further studies.
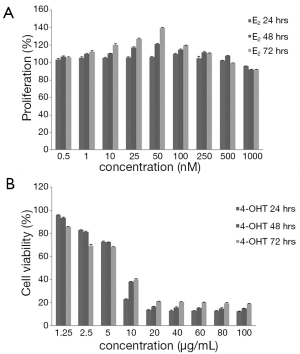
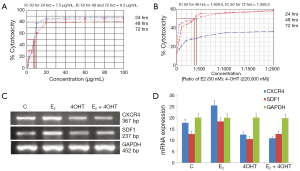
Different concentrations of 4OHT 1.25 to 100 (µg/mL) were used to study the effect of 4OHT after incubation for 24, 48 and 72 h in MCF-7 cells. The cell growth inhibitory effect of 4OHT was studied in terms of cell viability and cytotoxicity of MCF-7 cells which are shown in Figure 3B. IC50 values of 4OHT were found to be 7.5 µg/mL (19.35 µM), 8.3 µg/mL (21.42 µM) and 8.3 µg/mL (21.42 µM) for 24, 48 and 72 h incubations respectively (Figure 4A). From these experiments, 4OHT with a concentration of 22,000 nM (22 µM) was used for further studies. MCF-7 cells were seeded to study the combined effect of E2 and 4OHT in MCF-7 cells. Combination of E2 and 4OHT with different concentrations from in a ratio of 1:1, 1:50, 1:100, 1:200, 1:300, 1:400, 1:500, 1:1,000 and 1:2,000 were used to treat MCF-7 cells and cytotoxicity was assessed after 24, 48 and 72 h. When cells were incubated with E2 and 4OHT in a ratio of 1:400 and above, the cytotoxicity was increased by more than 50% and the IC50 values for 48 and 72 h were found to be 1:409.5 and 1:358.3 respectively (Figure 4B). From these experiments, E2 (50 nM) and 4OHT (22,000 nM) was used in a ratio of 1:400 for further studies.
mRNA level expressions of SDF1 and CXCR4 in MCF-7 cells
The mRNA expression levels of SDF1 and CXCR4 were assessed in MCF-7 cells treated with media supplemented with E2 (50 nM), 4OHT (22,000 nM), combinations of E2 and 4OHT in a ratio of 1:400 and without E2 or 4OHT as control. Different expression pattern was observed among control, E2, 4OHT and combination of E2 and 4OHT. In the case of SDF1 and CXCR4 the band intensity for E2 treated cells were found to be increased than C, 4OHT, and E2 + 4OHT. The relative expressions of genes in 4OHT treated cells were found to fall between the expression levels of genes in control MCF-7 cells with no treatment and combination of E2 and 4OHT. But the cells treated with 4OHT showed more intense band than in control cells. Clear sharp bands of CXCR4 (367 bp) and SDF1 (237 bp) were observed on 1.2% agarose gel electrophoresis (Figure 4C). The bands obtained were quantified using Image J 1.45S, USA and is represented in Figure 4D.
Immunofluorescence for co localization of CXCR4 and SDF1 in MCF-7 cells
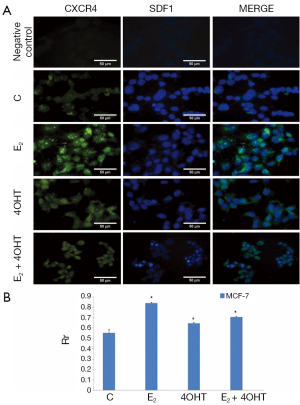
The colocalization of CXCR4 and SDF1 proteins were assessed in MCF-7 cells treated with E2, 4OHT, combination of E2 and 4OHT. Immunoflurescence and colocalization of proteins were observed differently for control, E2, 4OHT, and combination of E2 and 4OHT treated cells. The fluorescence and colocalization of proteins in E2 treated cells were found to be higher than control, 4OHT, and E2 treated cells. The co-associations of CXCR4 and SDF1 with different treatment groups were compared with control in MCF-7 cells. Representative immunofluorescent images showing co association of CXCR4 and SDF1 in MCF-7 cells treated with E2, 4OHT, combination of E2 and 4OHT, control, negative control for antibodies are shown in Figure 5A. Mean ± SD values of Pearson’s correlation coefficient (Rr) for each condition and significant correlation were plotted on the histogram (Figure 5B).
Discussion
This study suggests that the expression of CXCR4 and SDF1 in breast cancer tissue samples has prognostic and/or predictive importance in breast cancer patients. In the present study, there is significant correlation of histological grade with gene level expression of CXCR4. Positive correlation of increased breast tumour size >5 cm with SDF1and reduced overall survival of cancer patients with gene and protein level expression of CXCR4 is also noteworthy. The modulatory effect of 17-β estradiol, 4OHT and its combinations in the expression and colocalization of CXCR4/SDF1 in breast cancer cells has been validated. In our study, we found the co-expression of CXCR4 and SDF1 in the cytoplasm of breast tumour cells. Sacanna et al. [2011] showed the role of CXCR4 in the prediction of bone metastases from breast cancer, Cytoplasmic CXCR4 expression was high in bone metastasis patients, much lower in no evidence of disease patients and negative in the visceral metastasis group. CXCR4 coexpression in the nucleus and cytoplasm was observed in about half of the bone metastasis tumours but in patients with no evidence of disease or visceral metastasis (12). In a study by Lee et al. [2004] expression of SDF1, was about 2-fold higher in microdissected human breast cancer cells as compared against normal epithelial cells. Immunohistochemical analysis indicated that SDF1 expression is consistently higher in primary breast tumour cells than in normal breast epithelial cells (13).
In our study, the expression of both CXCR4 and SDF1 were cytoplasmic in infiltrating ductal carcinoma epithelial cells and their expression were found to be significantly associated to each other. This coexistence and correlation of CXCR4 and SDF1 favors the stimulation of CXCR4 by SDF1 and plays an important role in enhancing motility as well as regulating adhesive and invasive changes during breast cancer metastasis. CXCR4 is expressed at a low level in normal breast epithelium but becomes more strongly expressed in the early stages of carcinogenesis as evidenced by a more intense immunohistochemical staining pattern and an altered cellular localization in studies of human ductal carcinoma in situ (DCIS) (14,15). In our study, tumour samples showed CXCR4 positivity in 83.1% cases at the gene level and 38.2% at the protein level. Prominent CXCR4 expression is a feature of all major histological forms of invasive breast cancer, including ductal, lobular, mucinous (14), and the distinctive and highly aggressive inflammatory form of the disease (16). In our study significant positive correlation of CXCR4 and SDF1 was observed with histological grade and tumour size.
The CXCR4 has been shown to play an important role in lymph node metastasis. Various studies reported that CXCR4 expression detected by immunohistochemistry or reverse transcription-polymerase chain reaction was a prognosis factor for node involvement or survival in primary breast cancer patients (17-20). Different opinion about CXCR4 expression also exist dependent up on difference in cellular location, with membrane CXCR4 expression predicting good survival whilst cytoplasmic expression predicts the contrary. Blot et al. [2008] showed that membrane-localized CXCR4 staining was a strong prognostic factor for survival in node negative patients, whereas cytoplasmic staining was not (21). This finding is in contrast with previous reports of the prognostic influence of CXCR4 expression showing a weak link between cytoplasmic staining and cancer progression (19,20,22). The CXCR4 expression in our study is highly correlated with decreased breast carcinoma patient survival. Similar correlation between CXCR4 expression and decreased breast carcinoma patient survival had been reported (14,19,20,23). When compared to CXCR4, SDF1 does not show correlation to patient survival in the study. This may be explained by ubiquitin, the recently identified natural ligand for CXCR4. CXCR4 activation with SDF1 and ubiquitin results in partially synergistic effects on cellular signaling events and in differential effects on receptor desensitization (24). This might be an explanation for the significant role of CXCR4 in cancer metastasis and chemotaxis.
A study submitted by Boudot et al. reported a differential E2 regulation of SDF1 chemokine receptors CXCR4 and CXCR7 that contributes to the growth effect of estrogens in breast cancer cells. The inhibition of the expression or activity of either SDF1 or CXCR4 significantly blunted the E2-mediated stimulation of cellular growth (9). In this study, SDF1 was identified as a key mediator of E2-induced breast cancer cell proliferation and survival. These results also showed a positive regulation of E2 in both SDF1 and its receptor CXCR4. Anti-estrogens, such as 4OHT, elicited an E2 withdrawal effect mainly by competitive binding to the hormone binding domain of ERα and subsequent alteration of the conformation necessary for recruitment of transcription co-activators to transcription activation function 2 (AF2). In our study the mRNA level expression of SDF1 and CXCR4 was found to be higher in 4OHT treated cells but less than the E2 treated cells when compared to that of untreated MCF-7 cells. The increase in MCF-7 cells treated with a combination of E2 and 4OHT shows that the mitogenic effect of E2 is ahead of the competitive and cytotoxic effect of 4OHT. This is more evident in the case of SDF1 expression.
The colocalization was also found to be reduced than E2 treated group, the treatment of MCF-7 cells with a combination of E2 and 4OHT resulted in the colocalization of CXCR4 and SDF1 between E2 alone and 4OHT alone treated groups. Binding of SDF1 to CXCR4 stimulates phosphatidylinositol-3-kinase, activating protein kinase AKT in distinct carcinomas. Active AKT provides anti-apoptotic and proliferative effect in malignant cells, since this pathway is also important for the progression of breast and other carcinomas (7). The mitogen-activated protein (MAP) kinase pathway is another signal transduction pathway regulated by the liganded CXCR4 receptor. The MAP kinase pathway also up-regulates expression of genes encoding proteins involved in proliferation and survival of cancer cells (2). Moreover, liganded CXCR4 promotes polymerization of actin, which leads to migration of normal and malignant cells (25). Kubarek et al. suggest that 4OHT induces an increase in DNMT 3B expression that is associated with the increase of CpG dinucleotide methylation in the CXCR4 promoter and significant reduction of CXCR4 gene expression in MCF-7 cells (10). Furthermore, Gil et al. showed that CXCR4 antagonists have a significant therapeutic impact on primary and metastatic breast cancer by disrupting tumour vasculature in the microenvironment (26).
In conclusion, the expression of both CXCR4 and SDF1 were found to be associated in breast epithelial cells of infiltrating ductal carcinoma cells and their expression were found to be significantly correlated with each other. This coexistence and correlation of CXCR4 and SDF1 favors the stimulation of CXCR4 by SDF1 and plays an important role in enhancing motility as well as regulating adhesive and invasive changes during breast cancer metastasis. SDF1 overexpression implies that it can lead to invasion, migration, angiogenesis, chemotaxis of circulating lymphocytes and most importantly metastasis of breast cancer cells. On the other hand CXCR4 overexpression is responsible for the enhanced cell proliferation and poor survival in breast carcinoma through CXCR4/SDF1 signalling pathways. The response of SDF1and CXCR4 with E2 and 4OHT shows that CXCR4/SDF1axis mediate an E2 dependent cancer cell proliferation, as indicated by the association of SDF1 with E2 and anti-estradiol in breast cancer. Thus based on these observations it can be concluded that SDF1 overexpression, with significant association with CXCR4 expression in the same cell, itself contribute to the development of mammary cancer and metastatic progression and for aggressive stages of the disease. Thus, understanding SDF1 signaling in breast cancer cells might lead to greater insights into the molecular mechanisms of breast cancer metastasis and design of therapies based on the blocking of the CXCR4/SDF1signaling pathway in breast cancer. The delineation of this pathway is certain to create a new turning point in the field of breast cancer treatment.
Acknowledgements
Funding: This study was supported by grants from Indian Council of Medical Research (No. 5/13/77/06-NCD-III), Government of India.
Disclosure: The authors declare no conflict of interest.
References
- Teicher BA, Fricker SP. CXCL12 (SDF-1)/CXCR4 pathway in cancer. Clin Cancer Res 2010;16:2927-31. [PubMed]
- Müller A, Homey B, Soto H, et al. Involvement of chemokine receptors in breast cancer metastasis. Nature 2001;410:50-6. [PubMed]
- Zou W, Machelon V, Coulomb-L'Hermin A, et al. Stromal-derived factor-1 in human tumors recruits and alters the function of plasmacytoid precursor dendritic cells. Nat Med 2001;7:1339-46. [PubMed]
- Kang H, Watkins G, Parr C, et al. Stromal cell derived factor-1: its influence on invasiveness and migration of breast cancer cells in vitro, and its association with prognosis and survival in human breast cancer. Breast Cancer Res 2005;7:R402-10. [PubMed]
- Barbero S, Bonavia R, Bajetto A, et al. Stromal cell-derived factor 1alpha stimulates human glioblastoma cell growth through the activation of both extracellular signal-regulated kinases 1/2 and Akt. Cancer Res 2003;63:1969-74. [PubMed]
- Singh S, Singh UP, Grizzle WE, et al. CXCL12-CXCR4 interactions modulate prostate cancer cell migration, metalloproteinase expression and invasion. Lab Invest 2004;84:1666-76. [PubMed]
- Marchesi F, Monti P, Leone BE, et al. Increased survival, proliferation, and migration in metastatic human pancreatic tumor cells expressing functional CXCR4. Cancer Res 2004;64:8420-7. [PubMed]
- Burger M, Glodek A, Hartmann T, et al. Functional expression of CXCR4 (CD184) on small-cell lung cancer cells mediates migration, integrin activation, and adhesion to stromal cells. Oncogene 2003;22:8093-101. [PubMed]
- Boudot A, Kerdivel G, Habauzit D, et al. Differential estrogen-regulation of CXCL12 chemokine receptors, CXCR4 and CXCR7, contributes to the growth effect of estrogens in breast cancer cells. PLoS One 2011;6:e20898. [PubMed]
- Kubarek Ł, Jagodzinski PP. Epigenetic up-regulation of CXCR4 and CXCL12 expression by 17 beta-estradiol and tamoxifen is associated with formation of DNA methyltransferase 3B4 splice variant in Ishikawa endometrial adenocarcinoma cells. FEBS Lett 2007;581:1441-8. [PubMed]
- Harvey JM, Clark GM, Osborne CK, et al. Estrogen receptor status by immunohistochemistry is superior to the ligand-binding assay for predicting response to adjuvant endocrine therapy in breast cancer. J Clin Oncol 1999;17:1474-81. [PubMed]
- Sacanna E, Ibrahim T, Gaudio M, et al. The role of CXCR4 in the prediction of bone metastases from breast cancer: a pilot study. Oncology 2011;80:225-31. [PubMed]
- Lee BC, Lee TH, Avraham S, et al. Involvement of the chemokine receptor CXCR4 and its ligand stromal cell-derived factor 1alpha in breast cancer cell migration through human brain microvascular endothelial cells. Mol Cancer Res 2004;2:327-38. [PubMed]
- Salvucci O, Bouchard A, Baccarelli A, et al. The role of CXCR4 receptor expression in breast cancer: a large tissue microarray study. Breast Cancer Res Treat 2006;97:275-83. [PubMed]
- Schmid BC, Rudas M, Rezniczek GA, et al. CXCR4 is expressed in ductal carcinoma in situ of the breast and in atypical ductal hyperplasia. Breast Cancer Res Treat 2004;84:247-50. [PubMed]
- Cabioglu N, Gong Y, Islam R, et al. Expression of growth factor and chemokine receptors: new insights in the biology of inflammatory breast cancer. Ann Oncol 2007;18:1021-9. [PubMed]
- Kang H, Watkins G, Douglas-Jones A, et al. The elevated level of CXCR4 is correlated with nodal metastasis of human breast cancer. Breast 2005;14:360-7. [PubMed]
- Cabioglu N, Yazici MS, Arun B, et al. CCR7 and CXCR4 as novel biomarkers predicting axillary lymph node metastasis in T1 breast cancer. Clin Cancer Res 2005;11:5686-93. [PubMed]
- Kato M, Kitayama J, Kazama S, et al. Expression pattern of CXC chemokine receptor-4 is correlated with lymph node metastasis in human invasive ductal carcinoma. Breast Cancer Res 2003;5:R144-50. [PubMed]
- Li YM, Pan Y, Wei Y, et al. Upregulation of CXCR4 is essential for HER2-mediated tumor metastasis. Cancer Cell 2004;6:459-69. [PubMed]
- Blot E, Laberge-Le Couteulx S, Jamali H, et al. CXCR4 membrane expression in node-negative breast cancer. Breast J 2008;14:268-74. [PubMed]
- Andre F, Cabioglu N, Assi H, et al. Expression of chemokine receptors predicts the site of metastatic relapse in patients with axillary node positive primary breast cancer. Ann Oncol 2006;17:945-51. [PubMed]
- Holm NT, Abreo F, Johnson LW, et al. Elevated chemokine receptor CXCR4 expression in primary tumors following neoadjuvant chemotherapy predicts poor outcomes for patients with locally advanced breast cancer (LABC). Breast Cancer Res Treat 2009;113:293-9. [PubMed]
- Tripathi A, Davis JD, Staren DM, et al. CXC chemokine receptor 4 signaling upon co-activation with stromal cell-derived factor-1α and ubiquitin. Cytokine 2014;65:121-5. [PubMed]
- Balkwill F. Chemokine biology in cancer. Semin Immunol 2003;15:49-55. [PubMed]
- Gil M, Seshadri M, Komorowski MP, et al. Targeting CXCL12/CXCR4 signaling with oncolytic virotherapy disrupts tumor vasculature and inhibits breast cancer metastases. Proc Natl Acad Sci U S A 2013;110:E1291-300. [PubMed]


