
Types and viral load of human papillomavirus, and vaginal microbiota in vaginal intraepithelial neoplasia: a cross-sectional study
Introduction
Vaginal intraepithelial neoplasia (VAIN) is a preinvasive lesion, which potentially leads to vaginal cancer. The natural history of VAIN is similar to that of cervical intraepithelial neoplasia (CIN), including categorization into three grades, VAIN1, VAIN2 and VAIN3 (1). Previously, only a very low rate of intraepithelial neoplasia of the lower genital tract was reported, with an incidence of 0.2 to 0.3 per 100,000 women (2). Thanks to the widespread application of cytology, human papilloma virus (HPV) testing, and colposcopy cervical cancer screening, the diagnosis of VAIN has been growing steadily over time (3). Nevertheless, owing to a lack of symptoms and doctors’ attention to cervical lesions, VAIN can be easily overlooked.
It is acknowledged that VAIN shares common risk factors with CIN and HPV, while high-risk HPV (hrHPV) in particular plays an important role in its development (4). Studies on distribution of HPV types in VAIN and vaginal cancer have been inconsistent in their conclusions (5,6). Over recent years, an increasing number of research has investigated the relationship between hrHPV viral load and cervical lesions (7,8). These studies discovered that a group of α9 species including HPV16, 31, 33, 35, 52 and 58 were positively associated with the severity of CIN, and the optimal cutoffs of the log10-transformed viral loads of these specific HPV types might help identify high-grade squamous intraepithelial lesions (HSIL). Controversy surrounds the correlation between CIN and α7 species such as HPV18 (7-9). However, few studies have explored the correlation of HPV types, viral load and vaginal lesions, or the potential role of HPV viral load to serve as an indicator for VAIN.
Although HPV infection is prevalent worldwide, most infections are not persistent and do not develop into vaginal neoplasm, suggesting other factors are involved in the process. Emerging evidence shows the stability and composition of vaginal microbiota may play a significant part in the development of persistent HPV infection, the progression of CIN, and cervical cancer (10-16). Increased vaginal microbiome diversity, high levels of potential pathogenic bacteria such as Sneathia and Anaerococcus, and decreased abundance of Lactobacillus have been connected with cervical lesions. However, up to now, there have been no reports on a possible association between vaginal microbiota and the development of vaginal neoplasm.
This study took place at the Obstetrics and Gynecology Hospital of Fudan University, the largest obstetrics and gynecology specialist hospital in China. In 2015, 834 and 5303 women were diagnosed with VAIN and CIN respectively in the hospital, and the diagnosis rate of VAIN has been increasing (3). We aimed to explore the distribution of HPV types in patients with VAIN, and the correlation between viral load and vaginal lesions. We also sought to characterize the composition and alteration of vaginal microbiome of VAIN. We present the following article in accordance with the MDAR checklist (available at http://dx.doi.org/10.21037/atm-20-622).
Methods
Sample collection and study design
Patients were recruited at the Obstetrics and Gynecology Hospital of Fudan University, Shanghai from December 2018 to May 2019. The eligible population consisted of two arms, (I) patients with VAIN and (II) patients without VAIN but with HPV infection. The exclusion criteria were as follows: (I) pregnant or currently menstruating; (II) known immunosuppressive diseases or receiving immunosuppressive therapy; (III) use of antibiotics within 14 days of sampling; (IV) use of vaginal applied medications and vaginal lavage within 3 days of sampling; (V) current vaginitis.
At screening, a sterile, disposable speculum without lubricant was inserted into the patient’s vagiba. Swabs were taken from the vaginal fornix, placed immediately in dry ice, and then stored at −80 °C within 3 hours. Vaginal cells were collected with plastic brushes and stored in specimen preservation solution.
A total of 176 female patients with a definite pathological diagnosis were enrolled in this study. Among them, 109 HPV-positive patients qualified for viral load assay. The vaginal microbiota of 122 HPV-positive patients, matched by severity of cervical lesions and menopause status, were determined using 16S rDNA amplicon sequencing (Figure 1).
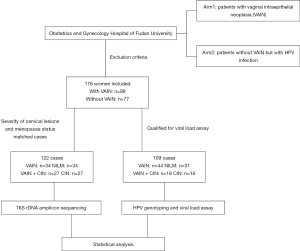
The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013). Our study was performed in accordance with the norm of the Ethical Committee of Obstetrics and Gynecology Hospital of Fudan University (Approval number: 2019-79). All the subjects had signed the informed consent forms and study outcomes will affect the future management of the patients.
HPV genotyping and viral load assay
Type-specific HPV viral load measurement and genotyping were simultaneously conducted by using the Bioperfectus Technologies Human Papillomavirus Genotyping Real Time PCR Kit (Bioperfectus Ltd, Jiangsu, China) to detect 21 different types of HPV in patient specimens, comprising 18 high-risk HPV types (HPV16, 18, 31, 33, 35, 39, 45, 51, 52, 53, 56, 58, 59, 66, 68, 26, 73, and 82) and 3 low-risk HPV types (HPV 6, 11, and 81). Amplification was carried out on the ABI 7500 Real Time PCR System. The distribution of HPV infection within the specimens was uneven, so the calculated viral load was the average viral load of all cells entering the test.
Bacterial DNA extraction and 16S rRNA amplicon sequencing
Total bacterial genomic DNA samples were extracted and stored at −20 °C until analysis. The concentration of the whole genomic DNA samples was tested by fluorescence. Agarose Gel Electrophoresis was used to test the integrity of the samples. The V4 hypervariable regions of 16S rRNA genes were amplified through dual-index paired-end sequencing approach by polymerase chain reaction (PCR). The Hiseq 2500 (MiSeq Reagent Kit) was employed for pair-end sequencing of the qualified sample libraries.
Sequencing analysis
The 16S rRNA sequencing analysis was performed by BGI Co., Ltd. Raw data was filtered to obtain clean reads and then merged to tags. Tags were clustered to operational taxonomic units (OTU) at 97% sequence similarity. Based on the abundance of OTU, most analysis was performed with software R (v3.1.1), including Venn diagram, principal component analysis (PCA), alpha diversity, and heat map analysis. To detect differentially abundant genera and species among groups, linear discriminant analysis effect size (LEfSe) was carried out using LEFSE software. Metastats (http://metastats.cbcb.umd.edu/) was used to determine which taxonomic groups were significantly different and adjusted the obtained P-value by a Benjamini-Hochberg false discovery rate correction.
Statistical analysis
HPV genotype and viral load were analyzed. The mean, standard deviation, median and quartile interval of the log10-transformed virus copy numbers per 10,000 human cells were considered the type-specific HPV viral load. Continuous variables were compared using Student’s t-test or the Mann-Whitney U-test. Categorical variables were compared by Pearson’s chi squared test. A receiver operating characteristic (ROC) curve and Youden’s index were utilized to identify the optimal cutoff value of the type-specific HPV viral load for predicting VAIN. SPSS, version 22.0 (SPSS, Chicago, IL, USA) was used for all analysis. All statistical tests were two-sided, and P values of <0.05 were considered statistically significant.
Results
HPV genotyping characteristics of patients with VAIN
A total of 176 female patients with a definite pathological diagnosis were enrolled in this study, comprising 99 patients with vaginal lesions and 77 patients without vaginal lesions. The HPV detection rate for VAIN was 89.7% (87/99). Patients with VAIN2/3 (n=12) or vaginal cancer (n=2) were all HPV positive, and 57.14% (8/14) of those patients were HPV16 positive.
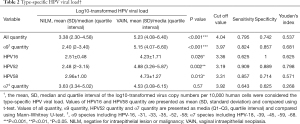
For the 109 qualified cases, which included separate vaginal lesions (n=44), NILM (n=31), separate cervical lesions (n=16) and vaginal lesions concomitant with cervical lesions (n=18), HPV genotyping and viral load were detected. The mean age of the patients with VAIN was 48.68 years (SD, 11.19) (Table 1). The HPV genotyping assay revealed the top 5 HPV types of vaginal lesions to be HPV16 (24.2%), HPV52 (24.2%), HPV53 (16.1%), HPV58 (14.5%) and HPV66 (14.5%) (Table S1). Unlike CIN patients, HPV18 was rare in VAIN patients, especially in VAIN1, with only 2 patients infected by HPV18. Compared with histopathologically normal cases, separate vaginal lesions had a higher rate of HPV multiple infection (47.7% vs. 29.0%), although the difference was not statistically significant (P=0.104). Taken together, these data suggested that HPV16, 52, 53, 58, and 66 were more prevalent in vaginal lesions, and infection with HPV16 might be a higher risk for VAIN2/3 or vaginal cancer.

Full table
Higher level of viral load of HPV16, 52, and 58 might be associated with VAIN
By comparing the viral load of separate vaginal lesions with normal samples, we found that, irrespective of HPV types, the viral load was significantly higher for women with VAIN (P all <0.001) after the log10-transformation. The combined viral load of α9 HPV, as well as the individual viral load of HPV16, 52, and 58 increased as the pathological diagnosis changed from NILM to VAIN (P HPV-α9 <0.001, P HPV16 =0.026, P HPV52 =0.002, P HPV58 =0.013), whereas the combined viral load of α7 HPV (including HPV18, 39, 45, 59, and 68) was not significantly increased (P HPV-α7 =0.57). The ROC curve model was used to estimate an appropriate cut-off value of type-specific HPV for the identification of VAIN. The optimal cutoff value of α9 HPV was 3.97 (Se = 0.824, and SP=0.857) and the area under the curve (AUC) of α9 HPV was 0.899 (95% CI: 0.791–1.000, P<0.001). HPV52 had an optimal cutoff value of 3.19 (Se =0.909, and SP=0.889) and an AUC of 0.884 (95% CI: 0.722–1.000, P=0.004). HPV58 had an optimal cutoff value of 3.31 (Se =0.857, and SP=0.714) and an AUC of 0.878 (95% CI: 0.697–1.000, P=0.018) (Table 2).
Full table
For women with cervical lesions, the viral load of HPV showed little difference whether or not there was coexistence of vaginal lesions (Table S2). These data demonstrated the association between some type-specific HPV viral load and vaginal lesions, and that viral load of HPV16, 52, and 58 might provide indication for vaginal lesions.
Vaginal microbial diversity of menopausal women was increased compared with that of pre-menopausal women
After being matched according to the severity of cervical lesions and menopause status, 122 HPV-positive women were enrolled in 16S rDNA amplicon sequencing for vaginal microbiota. The women were divided into the following groups: the s-VAIN group (separate vaginal lesions, n=34); the NILM group (negative for intraepithelial lesion or malignancy, n=34); the s-CIN group (separate cervical lesions, n=27), and the VAIN+CIN group (vaginal lesions concomitant with cervical lesions, n=27). We combined s-VAIN group with VAIN+CIN group to form the with-VAIN group (patients with vaginal lesions), and the NILM and s-CIN groups to form the without-VAIN group (patients without vaginal lesions). Among the women with vaginal lesions, there were 47 with VAIN1 and 14 with VAIN2/3.
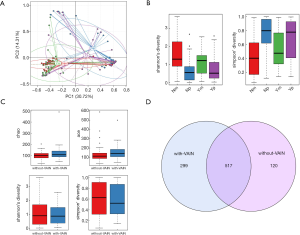
PCA was applied to confirm whether menopausal status is an important factor in the composition of vaginal microbiota. Menopausal patients, whether with or without VAIN, shared a different construction of microbiome compared with pre-menopausal patients (Figure 2A). Alpha diversity was applied for analyzing complexity of species diversity. For women without VAIN, a significant difference was found between menopausal and pre-menopausal group (Shannon index, P<0.001; Simpson’s index, P<0.001), indicating that there was a higher level of microbial diversity in the menopausal group. Menopausal women with VAIN also showed a higher diversity, but no statistical significance was observed (Shannon index, P=0.11; Simpson’s index, P=0.18) (Figure 2B). Also, heat map and LEfSe analysis suggested that the abundance of Lactobacillus was decreased, while some anaerobes such as Streptococcus, Prevotella, and Anaerococcus were increased in menopausal women versus pre-menopausal women [absolute value of linear discriminant analysis (LDA) score >3.6] (Figure S1). These data indicated that the microbial diversity of menopausal women was increased, and that the microbiota became less stable after menopause with a decreased abundance of Lactobacillus, which also suggested that it is necessary to take menopausal status into consideration when matching patients.
The characteristics and composition of vaginal microbiota in patients with VAIN
High microbial diversity is commonly recognized to be associated with cervical lesions. However, no significant differences in the microbial diversity of vaginal lesions were found between the with-VAIN and without-VAIN groups, the s-VAIN and NILM groups, or the VAIN+CIN and s-CIN groups. The with-VAIN group showed increased abundance of OTU and species richness (chao index, P=0.008; ace index, P=0.004) (Figure 2C,D). Data indicated that HPV-positive patients with VAIN had a tendency for increased microbial species richness but did not exhibit a large alteration in diversity.
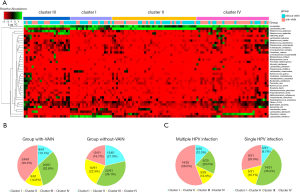
Species heat map analysis was utilized to further study the structure of vaginal microbiota. The vaginal microbiota was grouped into four major clusters based on the differences in species composition and their relative abundances. Clusters I, II, III were dominated by Lactobacillus helveticus, Lactobacillus iners, and Streptococcus anginosus, respectively; cluster IV was dominated by Atopobium vaginae and other species, such as Escherichia coli, Peptostreptococcus anaerobius, and Burkholderia bryophila (Figure 3A). Cluster IV was the most abundant cluster in the with-VAIN group, while cluster II was predominant in the without-VAIN group (Figure 3B). The number with cluster IV was twice as large in the with-VAIN group as in the without-VAIN group. Among the women with VAIN, HPV genotype was available for 56 women. Cluster IV was most frequently observed in women with multiple HPV infections (14/25, 56.0%), while single HPV-positive women were most likely to have cluster II (14/31, 45.2%) (Figure 3C). These data suggested that Lactobacillus iners was a dominant member in the microbiota of patients without VAIN. In patients with VAIN, cluster IV was predominant, particularly in women with multiple HPV infections.
Finally, we identified the difference in microbiota composition. In the with-VAIN group, there were 10 significant different genera (P<0.05). In the without-VAIN group, there were 15 enriched genera (P<0.05) (Figure 4). Gardnerlla, Atopbium, Allobaculum and Clostridium were far more abundant in the with-VAIN group as well as in the s-VAIN and VAIN + CIN groups, while Finegoldia, Actinobaculum and Blautia were significantly over-represented in the without-VAIN as well as in the NILM and s-CIN groups (Figure S2). At species level, Clostridium celatum (P<0.001, FDR =0.018) and Atopobium vaginae (P=0.001, FDR =0.049) were significantly enriched in the with-VAIN group. However, despite a decrease of Lactobacillus and Lactobacillus iners in the microbiota of VAIN patients, the difference was not statistically significant. Taken together, cluster IV was predominant in patients with VAIN, and an increased abundance of Atopobium, Gardnerella, Allobaculum, and Clostridium, as well as a decreased abundance of Finegoldia, Actinobaculum, and Blautia, might be associated with vaginal lesions.
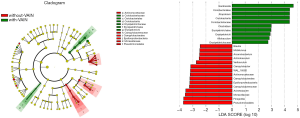
Enterococcus, Allobaculum and some Clostridium apps. increased in VAIN2/3
When comparing the vaginal microbiota of VAIN1 and VAIN2/3, there was a trend of VAIN2/3 exhibiting increasing species diversity and abundance, however, the difference was not significant (Figure S3). LEfSe modeling and differences analysis both showed that VAIN2/3 was characterized by a higher level of Enterococcus (absolute value of LDA score >4, FDR =0.015). The genus Allobaculum, which was much more enriched in VAIN versus non-VAIN, was also observed to be more abundant in VAIN2/3 versus VAIN1 (absolute value of LDA score >3, P=0.015, FDR =0.2) (Figure S4). At species level, we discovered that Prevotella copri was significantly increased in VAIN2/3, and Clostridium ramosun and Clostridium symbiosum were only detected in VAIN2/3. Moreover, Lactobacillus and Lactobacillus iners were decreased in VAIN2/3, without statistical significance, which is similar to the result of comparison between VAIN and non-VAIN. These data suggested that Enterococcus, Allobaculum, and some Clostridium apps. might indicate more severe vaginal lesions.
Discussion
The incidence rate of VAIN is extremely low. Available data on the distribution of HPV is limited, and no data have been reported about the potential influence of vaginal microbiota on VAIN.
In our study, the HPV detection rate in VAIN samples (89.7%) was much higher than the 69.3% (273/394) reported in one large series (n=490) using PCR (5). However, Ki-67 staining revealed 98.8% of VAIN specimens to be HPV positive. Another meta-analysis reported high HPV detection rates of 100% and in 90.1% in VAIN1 and VAIN2/3 patients, respectively (6). These differences might derive from the HPV detection methods chosen and diverse detection cutoff values. Ki-67 staining sites were determined to have vaginal lesions while samples collected by swabs were not always from lesion sites. De Vuyst et al. (6) detected all types of HPV, while we only identified 18 high-risk HPV types and 3 low-risk HPV types. HPV16 was present in 58.3% (7/12) of VAIN2/3 patients and in 50% (1/2) of vaginal cancer patients in our study, De Vuyst et al. (6) reported similar HPV16 infection rates of 57.6% in VAIN2/3 and 53.7% in vaginal cancer. Our results suggested that the five leading HPV types of vaginal lesions were HPV16 (24.2%), HPV52 (24.2%), HPV53 (16.1%), HPV58 (14.5%) and HPV66 (14.5%). In Shanghai, China, the predominant types of HPV in cervical screening are HPV16, 18, 58, and 52 (17), which, except HPV18, are consistent with the most prevalent HPV types in vaginal lesions according to our study. Some other studies also supported the rare detection rate of HPV18 in VAIN (5,18). This may be because HPV18 and other α7 species tended to develop in columnar epithelial cells instead of squamous epithelial cells (7).
Studies have explored whether some types of HPV with a higher viral load might indicate an increased risk of CIN (7,8). With respect to VAIN, few data were accessible. Our study demonstrated that the viral loads of α9 species, especially HPV16, 52, and 58 were related to vaginal lesions, and were also positively correlated with the severity of cervical lesions (7). When no obvious cervical lesions are found by colposcopy, a higher viral load may remind the clinician that careful detection for vaginal lesions may be necessary. However, the viral loads of α7 species did not bear a similar connection. Studies have demonstrated that different HPV genotypes share distinct biological properties. As mentioned above, α7 species tend to affect columnar epithelial cells, indicating that diverse HPV types enjoy different tropism for host cells. Furthermore, one of the α7 species, HPV45, is more often in the state of integration instead of duplication (19). Tropism and behavior of different types of HPV are suggested to partly account for this type-dependent association with VAIN. A possible mechanism underlying the positive correlation between viral loads and incidence of CIN and VAIN is that the number of cells infected and integrated by HPV rises as increased virus copies in individual cells (20). Several studies have also suggested that higher viral load is connected with persistent HPV infection, and persistence of HPV usually indicates high risk of vaginal lesions, which may be another possible underlying pathogenic mechanism (21,22). A higher viral load is also likely to be associated with immunological factors affecting VAIN progression, such as genetic susceptibility. Further studies are needed to verify these mechanisms (9). Furthermore, we discovered that, for women with cervical lesions, the viral load of HPV showed little difference whether accompanied with vaginal lesions or not. The threshold of viral load resulting in VAIN is likely to be similar to that leading to CIN.
HPV infection is a significant risk factor for vaginal lesions, however, only a small number of women have a persistent infection and subsequently develop VAIN. Vaginal microbiota may play an important role. Some influencing factors of vaginal microbiota composition include ethnicity, HPV infection status, and female hormones (10). In a twin cohort study, HPV-positive women showed higher diversity and less Lactobacillus compared to their uninfected sisters (11). After menopause, reduced oestrogen and progestin might result in increased diversity and Lactobacillus depletion (23), which was confirmed in our study. For women without VAIN, the difference in diversity was significant, while for those with VAIN, the gap in the difference was smaller, indicating that the microbiome becomes less stable in VAIN. We also found that some anaerobes such as Streptococcus, Prevotella, and Anaerococcus were increased in menopausal women versus pre-menopausal women. The condition, characterized by Lactobacillus depletion and anaerobic species overgrowth, is associated with bacterial vaginosis (BV), which has been related to delayed clearance of the virus (24), meaning that menopausal women probably have a higher risk of VAIN.
An increasing number of studies suggest that the structure of vaginal microbiota differs at various stages of CIN and cervical cancer. To control the variables influencing vaginal composition, we matched the species according to the patients’ severity of cervical lesions and menopause status, and classified them into four groups before analysis. Ravel et al. (25) studied vaginal microbiome of asymptomatic and reproductive-age women, grouped into five “community-state types” (CSTs). CSTs I, II, III, and V were dominated by Lactobacillus crispatus, L. gasseri, L. iners and L. jensenii, respectively. CST IV was devoid of Lactobacillus spp. and instead enriched with Atopobium vaginae and other anaerobic species such as Gardnerella, Sneathia, and Prevotella. In our data, the microbiota of species was classified into four clusters. Clusters I, and III were dominated by L. helveticus and Streptococcus anginosus, respectively. Cluster II resembled CST III and cluster IV resembled CST V. Higher rates of CST IV were observed to be associated with increasing disease severity of cervical lesions (12). Similarly, we found that cluster IV was the most abundant cluster in VAIN, while cluster I was the least. L. helveticus and L. crispatus are affiliated to the L. delbrueckii group, and are known as probiotics (26). L. helveticus has been shown to produce hydrogen peroxide, hinder pathogens adhesion, and reduce the viability of BV associated bacteria (27). Taken together, vaginal microbiota of VAIN was characterized by increased cluster IV, which might interplay with HPV and cause vaginal lesions. Incremental microbiome diversity is commonly thought to be associated with an increased severity of CIN. Our data did not show significant increased diversity in VAIN versus non-VAIN nor in VAIN2/3 versus VAIN1, although more abundant species richness was observed (12,28). No significant increase of diversity could be attributed to various factors, including sample collection methods (14) and the relatively small number of VAIN2/3 patients in our study. Another possibility is that some bacteria, the abundance of which is too low to obviously alter microbiome diversity, may be important during the process of VAIN.
We further identified distinctions in microbiota composition. At the genus level, Atopobium, Gardnerella, Allobaculum and Clostridium were detected more abundantly in VAIN. At the species level, Clostridium celatum and Atopobium vaginae were significantly enriched in VAIN. Recent reports have shown that predominance of Atopobium vaginae, Gardnerella vaginalis and Lactobacillus iners has a higher CIN risk (15,16). Atopobium vaginae can facilitate inflammation including interfering with T-cell expression and activating the NF-kB signaling pathway in vaginal epithelial cells (29). All of these may generate damage to the mucosal and epithelial barriers of the vagina (30), resulting in a high risk of persistent HPV infection and virus integration. Clostridium celatum is a potential pathogenic bacteria that can result in severe infection (31), although it is rarely reported in vaginal diseases. Whether an increase in C. celatum is a cause or consequence of VAIN development needs further exploration. Two further Clostridium spp., Clostridium ramosun, and Clostridium symbiosum, were detected only in VAIN2/3. In the studies of colorectal microbiome, C. ramosun is considered as an inducer of Tregs in vivo (32-34). Clostridium symbiosum is identified as a potential microbial marker for the early detection of colorectal cancer and may potentially support the development of carcinogenesis (35). An increase in Allobaculum, associated with BV (36), was observed not only in VAIN versus non-VAIN, but also in VAIN2/3 versus VAIN1. A higher level of Allobaculum is thought to be a potential risk indicator for female hepatocellular carcinoma (37), and may be potential marker for the progress of VAIN. Enterococcus and Prevotella copri were also relatively more enriched in VAIN2/3. Enterococcus, normal commensal bacteria in human vaginal vault, can act as pathogens by adhering to host tissues and secreting potentially toxic products (38). Prevotella is commonly thought to be associated with BV. The level of Prevotella copri was found to be markedly elevated in the mucosa and stools of patients with HIV infection (39,40). Increased levels of P. copri might similarly contribute to driving chronic inflammation in HPV infected individuals.
Moreover, Blautia was detected at significantly reduced levels in VAIN. It is mostly mentioned in the gut microbiome and usually act as beneficial bacteria, owing to its potential anti-inflammatory effect (41). No significant decrease of Lactobacillus was observed in VAIN against non-VAIN, nor in VAIN2/3 against VAIN1, which was detected in CIN. In our study, the most abundant Lactobacillus was L. iners, which did not decline sharply. L. iners is a dominant species when the vaginal flora changes from a normal to an abnormal state (42). It fails to produce effective D-isomer of lactic acid or H2O2 to play a protective role (43). L. iners appears to be more capable of surviving in stress conditions (10).
There are several limitations in our study. First, the sample size was relatively small, especially for the VAIN2/3. Consequently, the differential analysis of viral load between VAIN2/3 and VAIN1 is missing. Second, we did not record data of patients’ behavior, including sexual behavior and contraception, which would influence the composition of vaginal microbiota. Finally, we conducted a cross-sectional study, which means findings need to be further determined in longitudinal studies Our focused was on associations rather than causation. Large-scale prospective design and better controlled trials are required to extend the clinical meaning of viral load tests and to determine specific differential bacteria which are causal factors, causes or consequences of VAIN.
In conclusion, we explored the association between VAIN, HPV, and vaginal microbiota. The study showed that HPV-16, 52, 53, 58, and 66 are more prevalent in vaginal lesions, and viral loads of HPV16, 52, and 58 were significantly increased in VAIN. Data on genotype distribution and viral loads of HPV types in VAIN might help to provide guidance on screening for vaginal neoplasms. Also, we revealed that the proportion of cluster IV rose from non-VAIN to VAIN and the vaginal microbiome of VAIN was characterized by an increased abundance of Atopobium, Gardnerella, Allobaculum, and Clostridium, and a decreased abundance of Finegoldia, Actinobaculum, and Blautia. Furthermore, a higher level of Enterococcus and some specific Clostridium spp. might be correlated with an elevated risk of VAIN2/3. Future studies are required to better understand the roles that vaginal microbiota structure or individual bacterial species may play in the development of VAIN.
Acknowledgments
Funding: This work was supported by Shanghai Medical Center of Key Programs for Female Reproductive Diseases (grant number 2017ZZ01016).
Footnote
Reporting Checklist: The authors have completed the MDAR checklist. Available at http://dx.doi.org/10.21037/atm-20-622
Data Sharing Statement: Available at http://dx.doi.org/10.21037/atm-20-622
Peer Review File: Available at http://dx.doi.org/10.21037/atm-20-622
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/atm-20-622). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013). Our study was performed in accordance with the norm of the Ethical Committee of Obstetrics and Gynecology Hospital of Fudan University (Approval number: 2019-79). All the subjects had signed the informed consent forms and study outcomes will affect the future management of the patients.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Aho M, Vesterinen E, Meyer B, et al. Natural history of vaginal intraepithelial neoplasia. Cancer 1991;68:195-7. [Crossref] [PubMed]
- de Witte CJ, Van de Sande AJ, Van Beekhuizen HJ, et al. Imiquimod in cervical, vaginal and vulvar intraepithelial neoplasia: a review. Gynecol Oncol 2015;139:377-84. [Crossref] [PubMed]
- Cong Q, Wang Q, Gao SJ, et al. Detection trend of vaginal intraepithelial neoplasia diagnosed by colposcopy guided biopsy from 2013 to 2015. Zhonghua Fu Chan Ke Za Zhi 2017;52:239-43. [PubMed]
- Daling JR, Madeleine MM, Schwartz SM, et al. A population-based study of squamous cell vaginal cancer: HPV and cofactors. Gynecol Oncol 2002;84:263-70. [Crossref] [PubMed]
- Chao A, Chen TC, Hsueh C, et al. Human papillomavirus in vaginal intraepithelial neoplasia. International Journal of Cancer 2012;131:E259-68. [Crossref] [PubMed]
- De Vuyst H, Clifford GM, Nascimento MC, et al. Prevalence and type distribution of human papillomavirus in carcinoma and intraepithelial neoplasia of the vulva, vagina and anus: a meta-analysis. Int J Cancer 2009;124:1626-36. [Crossref] [PubMed]
- Fu Xi L, Schiffman M, Ke Y, et al. Type-dependent association between risk of cervical intraepithelial neoplasia and viral load of oncogenic human papillomavirus types other than types 16 and 18. Int J Cancer 2017;140:1747-56. [Crossref] [PubMed]
- Dong B, Sun P, Ruan G, et al. Type-specific high-risk human papillomavirus viral load as a viable triage indicator for high-grade squamous intraepithelial lesion: a nested case- control study. Cancer Manag Res 2018;10:4839-51. [Crossref] [PubMed]
- Malagón T, Louvanto K, Ramanakumar AV, et al. Viral load of human papillomavirus types 16/18/31/33/45 as a predictor of cervical intraepithelial neoplasia and cancer by age. Gynecol Oncol 2019;155:245-53. [Crossref] [PubMed]
- Mitra A, MacIntyre DA, Marchesi JR, et al. The vaginal microbiota, human papillomavirus infection and cervical intraepithelial neoplasia: what do we know and where are we going next? Microbiome 2016;4:58. [Crossref] [PubMed]
- Lee JE, Lee S, Lee H, et al. Association of the vaginal microbiota with human papillomavirus infection in a Korean twin cohort. PLoS One 2013;8:e63514. [Crossref] [PubMed]
- Mitra A, MacIntyre DA, Lee YS, et al. Cervical intraepithelial neoplasia disease progression is associated with increased vaginal microbiome diversity. Sci Rep 2015;5:16865. [Crossref] [PubMed]
- Łaniewski P, Barnes D, Goulder A, et al. Linking cervicovaginal immune signatures, HPV and microbiota composition in cervical carcinogenesis in non-Hispanic and Hispanic women. Sci Rep 2018;8:7593. [Crossref] [PubMed]
- Huang X, Li C, Li F, et al. Cervicovaginal microbiota composition correlates with the acquisition of high-risk human papillomavirus types. Int J Cancer 2018;143:621-34. [Crossref] [PubMed]
- Seo SS, Oh HY, Lee JK, et al. Combined effect of diet and cervical microbiome on the risk of cervical intraepithelial neoplasia. Clin Nutr 2016;35:1434-41. [Crossref] [PubMed]
- Oh HY, Kim BS, Seo SS, et al. The association of uterine cervical microbiota with an increased risk for cervical intraepithelial neoplasia in Korea. Clin Microbiol Infect 2015;21:674.e1-9. [Crossref] [PubMed]
- Zhang C, Zhang C, Huang J, et al. Prevalence and genotype distribution of human papillomavirus among females in the suburb of Shanghai, China. J Med Virol 2018;90:157-64. [Crossref] [PubMed]
- Srodon M, Stoler MH, Baber GB, et al. The distribution of low and high-risk HPV types in vulvar and vaginal intraepithelial neoplasia (VIN and VaIN). Am J Surg Pathol 2006;30:1513-8. [Crossref] [PubMed]
- Vinokurova S, Wentzensen N, Kraus I, et al. Type-dependent integration frequency of human papillomavirus genomes in cervical lesions. Cancer Res 2008;68:307-13. [Crossref] [PubMed]
- Brotherton JML, Tabrizi SN, Phillips S, et al. Looking beyond human papillomavirus (HPV) genotype 16 and 18: Defining HPV genotype distribution in cervical cancers in Australia prior to vaccination. Int J Cancer 2017;141:1576-84. [Crossref] [PubMed]
- Mittal S, Basu P, Muwonge R, et al. Risk of high-grade precancerous lesions and invasive cancers in high-risk HPV-positive women with normal cervix or CIN 1 at baseline-A population-based cohort study. Int J Cancer 2017;140:1850-9. [Crossref] [PubMed]
- van der Weele P, van Logchem E, Wolffs P, et al. Correlation between viral load, multiplicity of infection, and persistence of HPV16 and HPV18 infection in a Dutch cohort of young women. J Clin Virol 2016;83:6-11. [Crossref] [PubMed]
- Muhleisen AL, Herbst-Kralovetz MM. Menopause and the vaginal microbiome. Maturitas 2016;91:42-50. [Crossref] [PubMed]
- Gillet E, Meys JF, Verstraelen H, et al. Bacterial vaginosis is associated with uterine cervical human papillomavirus infection: a meta-analysis. BMC Infect Dis 2011;11:10. [Crossref] [PubMed]
- Ravel J, Gajera P, Abdob Z, et al. Vaginal microbiome of reproductive-age women. PNAS 2011;108:4680-7. [Crossref] [PubMed]
- Taverniti V, Guglielmetti S. Health-Promoting Properties of Lactobacillus helveticus. Front Microbiol 2012;3:392. [Crossref] [PubMed]
- Atassi F, Brassart D, Grob P, et al. In vitro antibacterial activity of Lactobacillus helveticus strain KS300 against diarrhoeagenic, uropathogenic and vaginosis-associated bacteria. J Appl Microbiol 2006;101:647-54. [Crossref] [PubMed]
- Audirac-Chalifour A, Torres-Poveda K, Bahena-Roman M, et al. Cervical Microbiome and Cytokine Profile at Various Stages of Cervical Cancer: A Pilot Study. PLoS One 2016;11:e0153274. [Crossref] [PubMed]
- Libby EK, Pascal KE, Mordechai E, et al. Atopobium vaginae triggers an innate immune response in an in vitro model of bacterial vaginosis. Microbes Infect 2008;10:439-46. [Crossref] [PubMed]
- Borgdorff H, Gautam R, Armstrong SD, et al. Cervicovaginal microbiome dysbiosis is associated with proteome changes related to alterations of the cervicovaginal mucosal barrier. Mucosal Immunol 2016;9:621-33. [Crossref] [PubMed]
- Agergaard CN, Hoegh SV, Holt HM, et al. Two Serious Cases of Infection with Clostridium celatum after 40 Years in Hiding? J Clin Microbiol 2016;54:236-8. [Crossref] [PubMed]
- Yissachar N, Zhou Y, Ung L, et al. An Intestinal Organ Culture System Uncovers a Role for the Nervous System in Microbe-Immune Crosstalk. Cell 2017;168:1135-48.e12. [Crossref] [PubMed]
- Narushima S, Sugiura Y, Oshima K, et al. Characterization of the 17 strains of regulatory T cell-inducing human-derived Clostridia. Gut Microbes 2014;5:333-9. [Crossref] [PubMed]
- Sefik E, Geva-Zatorsky N, Oh S, et al. Individual intestinal symbionts induce a distinct population of ROR + regulatory T cells. Science 2015;349:993-7. [Crossref] [PubMed]
- Xie YH, Gao QY, Cai GX, et al. Fecal Clostridium symbiosum for Noninvasive Detection of Early and Advanced Colorectal Cancer: Test and Validation Studies. EBioMedicine 2017;25:32-40. [Crossref] [PubMed]
- Piyathilake CJ, Ollberding NJ, Kumar R, et al. Cervical Microbiota Associated with Higher Grade Cervical Intraepithelial Neoplasia in Women Infected with High-Risk Human Papillomaviruses. Cancer Prev Res (Phila) 2016;9:357-66. [Crossref] [PubMed]
- Huang R, Li T, Ni J, et al. Different Sex-Based Responses of Gut Microbiota During the Development of Hepatocellular Carcinoma in Liver-Specific Tsc1-Knockout Mice. Front Microbiol 2018;9:1008. [Crossref] [PubMed]
- Jett BD, Huycke MM, Gilmore MS. Virulence of enterococci. Clinical Microbiology reviews 1994;7:462-78. [Crossref] [PubMed]
- Ley RE. Gut microbiota in 2015: Prevotella in the gut: choose carefully. Nat Rev Gastroenterol Hepatol 2016;13:69-70. [Crossref] [PubMed]
- Dillon SM, Lee EJ, Kotter CV, et al. Gut dendritic cell activation links an altered colonic microbiome to mucosal and systemic T-cell activation in untreated HIV-1 infection. Mucosal Immunol 2016;9:24-37. [Crossref] [PubMed]
- Jenq RR, Taur Y, Devlin SM, et al. Intestinal Blautia Is Associated with Reduced Death from Graft-versus-Host Disease. Biol Blood Marrow Transplant 2015;21:1373-83. [Crossref] [PubMed]
- Jakobsson T, Forsum U. Lactobacillus iners: a marker of changes in the vaginal flora? J Clin Microbiol 2007;45:3145. [Crossref] [PubMed]
- Witkin SS, Mendes-Soares H, Linhares IM, et al. Influence of vaginal bacteria and D- and L-lactic acid isomers on vaginal extracellular matrix metalloproteinase inducer: implications for protection against upper genital tract infections. MBio 2013;4:e00460-13. [Crossref] [PubMed]


