
Atractyloside targets cancer-associated fibroblasts and inhibits the metastasis of colon cancer
Introduction
The tumor microenvironment (TME) is a complex tissue environment, which comprises cancer cells, cancer-associated fibroblasts (CAFs), inflammatory cells, macrophages, myeloid-derived suppressor cells, mast cells, and extracellular matrix (ECM) (1). Since CAFs are major constituents of the TME, they play a crucial role in the occurrence, development, and metastasis of tumors. Furthermore, CAFs are activated fibroblasts in tumors and are mostly derived from fibroblasts, but they may be also derived from smooth muscle cells of vasculature, pericytes, and bone marrow-derived mesenchymal cells, or from epithelial or endothelial-mesenchymal transition. Fibroblast activation protein (FAP) and α-smooth muscle actin (α-SMA) are usually considered as the molecular markers of CAFs (2,3). However, the COX-2/PGE2 signal, TGF-β, and exosomes are the three main cross-talks, which facilitate extensive interactions between CAFs and cells of the TME. Moreover, CAFs have positive or negative interactions with all protumoral cells, including cancer cells, regulatory T cells, endothelial cells, M2 macrophages, cancer stem cells, myeloid-derived suppressor cells, tumor-associated dendritic cells, cytotoxic T lymphocyte, natural killer (NK) cells, etc. These cross-talks are usually responsible for the progression of tumor (4-6). Furthermore, CAFs modulate anti-tumor immunity responses at various levels, potentially affecting both innate and adaptive anti-tumor immunity responses. Thus, CAFs play a pivotal role in tumor progression (7).
The initial step of metastasis involves the migration and invasion of cancer cells derived from the primary site. Interestingly, CAFs promote the migration and invasion of cancer cells through paracrine signaling of cytokines. In this process, one of the major cytokines is TGF-β. Furthermore, CAFs alter the ECM environment to promote the migration and invasion of cancer cells (8). Because CAFs are frequently found in colon cancer (CC) tissues, they could be a potential marker in the prognosis of CC. In fact, several studies have reported about the relationship between CAFs and CC (9,10). In CC tissues, CAFs secrete cytokines and chemokines that target endothelial cells, such as VEGF, and induce angiogenesis (11). Most anti-cancer drugs target cancer cells, but the TME supports the growth and invasion of cancer cells. Therefore, there are still some targets that need to be investigated. Because CAFs are linked to tumor drug resistance, extensive research studies must be conducted to identify drugs targeting CAFs. This strategy would be useful in solving the issue of tumor drug resistance (12). However, CAFs create a physical barrier by synthesizing and secreting ECM components and by participating in the constitution of the TME. Therefore, drugs targeting CAFs should reorganize the ECM and enhance the permeability of anti-tumor drugs. By secreting proinflammatory cytokines and chemokines, CAFs induce immunosuppressive cells into the tumor stroma and reject effector T cells. Drugs targeting CAFs profoundly enhance the immune treatment of cancers (13).
In this study, our aim is to find small molecular compounds that target CAFs in CC tissues. Immune cell infiltration score was used to determine the intensity of CAFs in CC tissues. In the high-intensity CAFs group and the low-intensity CAFs group, the differentially expressed genes were screened and converted into small molecular compounds. To confirm the relationship between small molecular compounds and CAFs, we analyzed the potential mechanism of target proteins in small molecular compounds. In this study, we were able to find small molecular compounds that target CAFs in CC tissues. These molecules could be used to inhibit the development and metastasis of CC. We presented the following article in accordance with the MDAR checklist (available at http://dx.doi.org/10.21037/atm-20-1531).
Methods
Screening of small molecular compounds targeting CAFs
Estimate the Proportion of Immune and Cancer Cells (EPIC) is a method based on bulk tumor gene expression profiles. In this method, the proportion of immune cells and cancer cells is estimated in unknown tissues by determining the expression of non-malignant cell types in tumor tissues, such as immune, stromal, and endothelial cells (14). The gene expression profile (GSE2109) was obtained from Gene Expression Omnibus (GEO) (15). In this experiment, we collected data from the primary site of 239 CC cases with pathological grade data. The expression profiles of 239 CC cases were transferred to the proportion of infiltrating cells to obtain the infiltrating proportion of B cells, CAFs, CD4 T cells, CD8 T cells, endothelial cells, macrophages, and NK cells in each CC sample. By considering the proportion of CAFs as the standard, we classified the 239 CC cases into the high-intensity CAFs group (n=119) and the low-intensity CAFs (n=119) group based on the median CAFs. Gene Set Enrichment Analysis (GSEA) was used to perform enrichment analysis of the data in the two groups (16,17). The analysis was based on the following parameters: biological processes, molecular functions, and cell components.
After comparing the biological processes of the high-intensity CAFs group and the low-intensity CAFs, 20 gene sets were selected with higher enrichment. From each gene set, genes that had enrichment function were collected. Thus, we obtained enrichment genes from the two groups. After comparing the expression data of 239 CC cases in the high-intensity CAFs group and the low-intensity CAFs, we screened the differentially expressed genes. GeneSpring software was used for screening the differentially expressed genes. The parameters were compared with unpaired t-test, the P value was calculated, and multiple testing was corrected with Bonferroni FWER (18). We selected differentially expressed genes (P<0.05) that had a folder change of one time. In the two CAFs groups, intersections were analyzed between the screened differentially expressed genes and enriched genes. The genes obtained by intersection were imported into Connectivity Map. Then, small molecular compounds of higher relativity were screened by considering their enrichment score (Figure 1). Connectivity Map collected the expression profiles of tumor cells treated with 1,309 small molecular compounds (19,20). The data was used to determine the relationship between differentially expressed genes and small molecular compounds. Thus, we could screen small molecular compounds related to tumor cells by determining the changes in tumor tissue expression profiles, which were converted from the Connectivity Map.
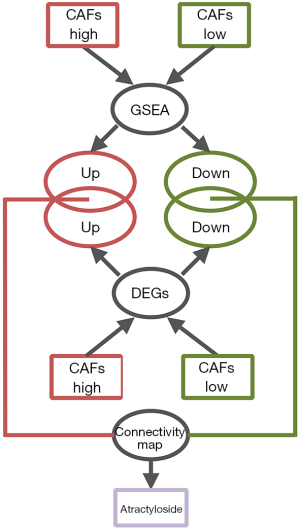
Prediction of target protein of atractyloside and the construction of signal-regulating network
After screening all the small molecular compounds, we considered atractylosides as the target compounds. SwissTargetPrediction (21-23), a tool that can predict the potential target protein of small molecular compounds, was used to predict the binding target protein of atractyloside. It contains a library of 376,342 compounds known to be experimentally active on an extended set of 3,068 macromolecular targets. SwissTargetPrediction compares target compounds with 376,342 small molecules in the database. It uses a combination of 2D and 3D similarity measures for the comparison and predicts the binding target protein. ToppGene Suite database (24) was used to perform gene ontology enrichment analysis on the target protein of atractyloside. Thus, the biological function of the target protein was determined. First intersection was analyzed between the up-regulated genes in the high-intensity CAFs group and target proteins, while second intersection was analyzed with the enriched genes in the high-intensity CAFs group. Hence, the enrichment of up-regulated target proteins was achieved, and a signal regulating network was constructed with Cytoscape software. The network modules were calculated by using the plug-in MCODE in Cytoscope software (25-27). Thus, we determined the relationship between the target protein and the biological processes of the high-intensity CAFs group.
CC cell (CCC) migration was inhibited by atractyloside
To verify the effect of atractyloside on CCCs migration, two CCC lines (RKO and HCT116 purchased from the American Type Culture Collection) were selected to conduct the assay on cell scratch-wound healing. Cell culture medium and fetal bovine serum (FBS) were used for culturing the cells were purchased from Gibco Laboratories (Thermo Fisher Scientific, USA). The cell line was cultured in RPMI-1640 culture medium containing 10% FBS. The cell culture was maintained at 37 °C in a 5% CO2 incubator. Cells in good growing condition were removed and added into a six-well plate at 8×105 cells per well. After the growth of cells reached 90%, the tip of a 100 µL pipette was used to scratch the plate bottom, and the culture medium was replaced by atractyloside serum-free medium at a different concentration in each well to sequentially culture the cells. The concentrations of atractyloside were 0, 5, 25, 50, 75, and 100 µM. The six wells were photographed using an optical microscope at 0 and 24 h to record cell migration (at magnification of 10×).
The transwell cell-migration assay was conducted to verify the effect of atractylosides on CCC migration. Twenty percent of the culture medium was added to a 24-well plate at 650 µL per well. RKO/HCT116 cells cultured in serum-free medium for 24 h were extracted for 500 µL (with a density of 2×105 cells/mL) and centrifuged to collect sediment. To each of the tubes was added 200 µL atractyloside solution with a final concentration set to 0, 5, 25, 50, 75, and 100 µM, respectively. After mixing evenly, each of these solutions was added into the upper chamber of a transwell membrane. The control group contained an equal volume of PBS and three duplicate wells were set for each concentration. The membrane was immersed in a culture medium in a 24-well plate and incubated in a 5% CO2 incubator for 24 h, after which it was fixed in 4% paraformaldehyde for 30 min and received Giemsa staining for 30 min. Finally, the membrane was rinsed three times with PBS. A cotton swab was used to remove the cells that had not passed through the upper layer of the membrane. The cells were placed on glass slides and three fields of view were randomly selected for counting the cells under an optical microscope and photographed at 20× magnification.
Confirming the effect of atractylosides on the microtubule cytoskeleton of CCCs
Changes in the cell microtubule cytoskeleton could affect the movement and morphology of cancer cells, which is closely related to tumor metastasis. Therefore, we observed the effect of atractylosides on the microtubule cytoskeletons of CCCs. Atractylosides were used to treat CCCs followed by immunofluorescence staining, and the distribution of α-tubulin in the cells was observed under a laser confocal microscope. The details were as follows: RKO/HCT116 cells were placed in a confocal dish (Wuxi NEST Biotechnology Co., Ltd., Jiangsu, China), treated with atractylosides at different concentrations (0.1% dimethyl sulfoxide and 0, 25, and 50 µM atractyloside) for 24 h, fixed with 4% paraformaldehyde, treated with 0.25% Triton-X, blocked with 5% bovine serum albumin, incubated with the first antibody (α-tubulin, dilution 1:1,000; ProteinTech America) in 4 °C overnight, washed three times with PBS for 5 min, incubated with the second antibody (Alexa Fluor 594-labeled goat anti-mouse immunoglobulin, dilution 1:100; Beijing Zhongshan Golden Bridge Biotechnology Co., Ltd., Beijing, China) for 30 min in the dark, washed with PBS and stained with 4',6-diamidino-2-phenylindole (DAPI) for 5 min, washed again with PBS, and observed under a laser confocal microscope (at magnification of 120×).
The study was conducted in accordance with the Declaration of Helsinki (as is revised in 2013).
Statistical analyses
SPSS 19.0 (IBM Corp., Armonk, NY, USA) was used to analyze the data. Data between the two groups were analyzed using the Student’s t-test for independent samples. ANOVA was used to analyze the data for multiple comparisons.
Results
GSEA enrichment analysis of CAFs in CC
For the importance of CAFs in the TME and tumor immunity, we used the EPIC method to perform immune infiltration analysis on the CC expression profiles of 239 cases that were included in the dataset GSE2109. After obtaining the infiltration proportion of CAFs in each tumor sample, the data of 239 cases was classified into the high-intensity CAFs group and the low-intensity CAFs group. By performing GSEA enrichment analysis, we found that CC tissues of the high-intensity CAFs group were involved in various biological processes, such as extracellular structure organization, angiogenesis, ossification, and endothelial cell migration (Figure 2). The CC tissues of the high-intensity CAFs group were also included as cellular components in the composition of ECM, collagen, vesicle, endoplasmic reticulum, lysosome, the granule of platelets, and the lumen of secretory granules. In the high-intensity CAFs group, the CC tissues performed the molecular function of ECM structural constituent, binding with glycosaminoglycan, heparin, integrin, glycoprotein, cell adhesion molecule, cytokine, collagen, fibronectin, laminin, and proteoglycan. The above results indicate that the high-intensity CAFs group was related to the biological function of the TME, thereby confirming the role of CAFs in the development of tumors. Thus, we also verified the accuracy of the results of immune infiltration analysis.
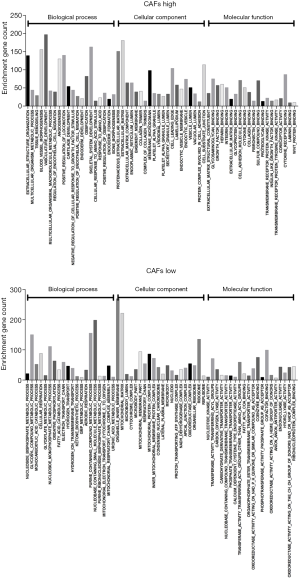
In the low-intensity CAFs group, the CC tissues were involved in the biological processes of nucleoside metabolism, cellular respiration, oxidative phosphorylation, and mitochondrial respiratory process. In the low-intensity CAFs group, the CC tissues were also included as cellular components in the composition of mitochondrial matrix, microbody, cytochrome complex, respiratory chain, and oxidoreductase complex. In the low-intensity CAFs group, the CC tissues performed the molecular function of nucleotide kinase activity, oxidoreductase activity, transferase activity, and coenzyme binding. The above results indicate that the low-intensity CAFs group was involved in cellular respiration and mitochondrial ATP synthesis. The early study (28) reported that in the metastatic stage of CC, up-regulated genes were related to ECM and inflammatory cell infiltration, and down-regulated genes were related to mitochondrial respiratory chain function. This indicated that the intensity of CAFs was closely related to the metastasis of CC. Hence, small molecular compounds that target CAFs in CC tissues can also effectively inhibit the metastasis of CC.
Screening of small molecular compounds targeting CAFs
By performing GSEA enrichment analysis, we obtained enriched genes that were related to various biological processes in the high-intensity CAFs group and the low-intensity CAFs group. After deleting duplicate genes, we collected 545 enriched genes from the high-intensity CAFs group and 325 enriched genes from the low-intensity CAFs group. To further verify the reliability of differentially expressed of enriched genes, we compared the expression data of the high-intensity CAFs group and the low-intensity CAFs group. Then, we screened the differentially expressed genes and obtained 1,572 genes up-regulated expression in the high-intensity CAFs group and 668 genes down-regulated expression in the low-intensity CAFs group. An intersection was analyzed between these differentially expressed genes and enriched genes, and we obtained 350 intersected genes in the high-intensity CAFs group and 78 intersected genes in the low-intensity CAFs group. Then, these genes were imported into the Connectivity Map. According to the hint of Connectivity Map, compounds with negative enrichment score could reverse the expression of differentially expressed genes, which correlated with anti-tumor CAFs in CC tissues. Therefore, compounds with negative enrichment score were selected with a P value of less than 0.01 and experiment was repeated more than 5 times. Finally, 25 small molecular compounds were obtained. After considering the enrichment score, P value, and the results of reported studies, we decided that atractylosides would be the target compounds in this study.
Target protein prediction of atractylosides and network analysis
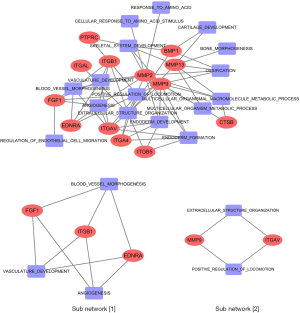
Based on the intensity of CAFs in CC tissues, we obtained the small molecular compounds from the Connectivity Map. Hence, the biological function of the ideal small molecular compounds should be closely related to CAFs. The SwissTargetPrediction tool was used to predict the target protein that would bind with atractyloside. In this process, we screened 97 target proteins, including matrix metalloproteinase family proteins (MMP1, MMP2, MMP3, MMP8, MMP9, MMP10, MMP12, and MMP13), integrin family proteins (ITGB1, ITGA2B, ITGA4, ITGB5, ITGB7, ITGAV, and ITGAL) and caspase family proteins (CASP2, CASP1, CASP3, CASP7 and CASP8). Gene ontology enrichment analysis was performed with ToppGene Suite; the enrichment analysis results indicated that target proteins were mainly involved in cell migration and cell apoptosis. Moreover, included as cellular components in the ECM, cell membrane, and some target proteins showed matrix metalloproteinase activity and fibronectin activity. An intersection was analyzed between 97 target proteins and 350 intersected genes in the high-intensity CAFs group. Thus, we identified 13 target proteins that had an up-regulated expression in the high-intensity CAFs group and were believed to be involved in the biological processes of CAFs. The 13 target proteins were as follows: FGF1, MMP13, MMP2, EDNRA, MMP9, PTPRC, ITGB1, ITGAV, ITGB5, ITGAL, CTSB, BMP1, and ITGA4. Obviously, these 13 target proteins belong to the following families: matrix metalloproteinase, integrin, endothelin receptor, fibroblast growth factor, bone morphogenetic protein, cathepsin, and protein tyrosine phosphatase receptor. Thus, they were closely related to the biological function of CAFs, which confirms the results of GSEA enrichment analysis.
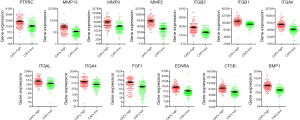
A network was constructed from the 13 target proteins and their corresponding biological processes related with CAFs. Out of the 20 biological processes related to CAFs, we found that 16 were closely linked with the 13 target proteins (Figure 3). This indicated that the 13 target proteins were mainly involved in the biological processes related to CAFs. Gene expression analysis showed that the expression of 13 genes in the high-intensity CAFs group was higher than that in the low-intensity CAFs group (Figure 4). Using the plug-in MCODE in Cytoscape software, we analyzed the network modules and obtained two subnetworks. In the subnetwork [1], we found that the target proteins FGF1, ITGB1, and EDNRA were closely related to angiogenesis. This indicated that atractylosides could inhibit the angiogenesis of CC tissues by binding with the three target proteins. The other subnetwork [2] revealed that the target proteins MMP9 and ITGAV were associated with the ECM and cell motility, which indicated that atractylosides could inhibit cell motility by binding with these two target proteins.
The atractyloside inhibits CCC migration
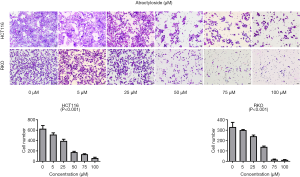
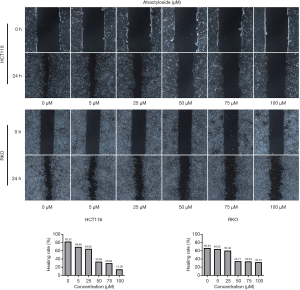
Because there is a close relationship between CAFs and cancer metastasis, our analyses suggested a relationship between the target protein of atractylosides and CCC migration. To examine the effect of atractylosides on CCC migration, transwell cell migration and scratch-wound healing assays were conducted on RKO and HCT116 cells using different atractyloside concentrations (0, 5, 25, 50, 75, and 100 µM). After the cells were cultured in different concentrations of atractyloside for 24 h, the number of CCCs that passed through the transwell membrane significantly decreased with increasing atractyloside concentration (Figure 5). We observed that inhibition of CCCs migration first appeared at an atractyloside concentration of 5 µM, while significant inhibition was observed at a concentration of 50 µM. One-way ANOVA tests were conducted to compare the differences among the concentrations of RKO/HCT116 cells, and the results were statistically significant (P<0.001). Similarly, we observed using the scratch wound-healing assay that the healing speed distinctly decreased with an increase in concentration, which was significant at an atractyloside concentration of 50 µM too (Figure 6). By calculating the healing rate after 24 h [healing rate = (1 − area after healing)/area before healing], the healing rate obviously decreased with an increase in concentration. All of these results suggested that the atractyloside could effectively inhibit the migration of CCCs.
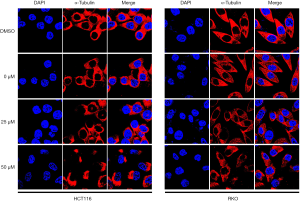
Effects of atractylosides on the CC microtubule cytoskeleton
Immunofluorescence staining and observation under the laser confocal microscope exhibited the effects of atractylosides at concentrations of 0, 25, and 50 µM on the microtubule cytoskeletons of RKO and HCT116 cells. We found that with an increasing concentration of atractyloside, the microtubule cytoskeletons located in the cytoplasm of the CCCs significantly changed. Compared with that in the cells of the control group, the microtubule cytoskeletons in the CCCs became sparse and disordered. In particular, at a concentration of 50 µM, obvious cell damage was observed and many nuclei were exposed (Figure 7). The above results suggested that atractylosides affected the microtubule cytoskeleton of CCCs and was closely related to the CCC migration.
Discussion
An atractyloside is a diterpenoid glycoside that occurs naturally in some plants, which are found in Europe, Africa, South America, Asia, and the Far East. The atractyloside was first isolated from the rhizomes of Atractylis gummifera. The atractyloside significantly inhibits mitochondrial oxidative phosphorylation and affects the metabolism of sugars and lipids (29). Massive necrosis can occur with high doses of atractylosides, while apoptosis can occur with low doses of atractylosides (30). Therefore, the atractyloside is used as an apoptosis inducer in animal model of tumor (31). The acquired drug resistance hampers the efficacy of chemotherapy and immunotherapy, which are appeared in the treatment of osteosarcoma (32,33) and malignant melanoma (34). The atractylosides can open mitochondrial permeability transition pore (MPTP), and it can enhance the cytotoxicity of TNF-related apoptosis-inducing ligand (TRAIL) in more than two apoptosis-resistant cells. Thus, it is beneficial to the treatment of cancer (35). In liver cancer patients, multidrug resistance (36) develops due to defects in apoptosis and it hampers the efficacy of chemotherapy. According to previous studies (37), the atractyloside acts as an MPTP opener and incubates with drug-resistance cells. Moreover, it reduces the IC50 in cis-diamminedichloroplatinum (CDDP) and enhances the anti-tumor effect. For the treatment of CC tissues, a combination of 5-fluorouracil and atractylosides may be used to promote the effect of chemotherapeutic agent and to inhibit the proliferation of tumor cells (38).
In this study, we found that most target proteins of atractyloside were related to CAFs, especially in the two subnetworks. These target proteins included FGF1, ITGB1, EDNRA, MMP9, and ITGAV. The migration and invasion of CC was closely linked to CAFs. In a previous study (39), it was reported that fibroblast growth factor 1/fibroblast growth factor receptor 3 (FGF1/FGFR-3) signaling were mediators that caused an increased migration and invasion of CC. In an in vitro study, it was reported that FGF1 was released by CAFs, and it acted on tumor epithelial cells to promote the migration of tumor cells. Hence, the inhibitor of FGF1 or a neutralizing antibody may inhibit the metastasis of CC. The target protein ITGB1 is a member of the integrin family. This protein is used for cell adhesion to ECM proteins. In large samples, the immunohistochemistry and the clinical pathology of CC showed that a high expression of ITGB1 was associated with lymph node metastasis and liver metastasis. Moreover, an increased expression of ITGB1 was related to poor prognosis of CC (40). The target protein EDNRA is an endothelin receptor. A previous study (41) analyzed the data of lymph node metastasis in bladder cancer patients. It also included immunohistochemistry results of large samples. In addition, it was found that EDNRA was highly expressed in bladder cancer patients with lymph node metastasis and distant metastasis. Meanwhile, a high expression of EDNRA was associated with decreased cancer-specific survival. MMPs have been extensively investigated to establish its relationship with cancer. Type IV and V collagen can be degraded by enzymes, which are encoded with MMP-9 gene. In CC, the target proteins MMP-2, MMP-7, and MMP-9 were up-regulated and co-expressed together with trypsin. Thus, they promoted the progression and invasion of CC (42). The target protein ITGAV is a member of the integrin family, and its expression is associated with tumor angiogenesis and metastasis. In CC, a high expression of ITGAV protein is correlated with neural invasion (43), indicating that target proteins of the atractyloside are closely related to the development and metastasis of tumor. This finding further confirms that the screening of small molecular compounds can be based on the intensity of CAFs.
In this study, by performing immune cell infiltration analysis, we determined the intensity of CAFs in the expression profile of CC. After converting the differentially expressed genes, we obtained the small molecular compounds of anti-CAFs, atractylosides. By analyzing the target proteins of the atractyloside, we further verified its association with CAFs. In addition, the transwell cell migration and scratch wound-healing assays proved that atractylosides could significantly inhibit CCC migration. Immunofluorescence staining and observation under a laser confocal microscope confirmed that atractylosides can affect the distribution of the microtubule cytoskeleton in CCCs. Thus, the atractyloside might be a small molecular compound that can potentially target CAFs and inhibit the development and metastasis of CC by changing the TME.
Acknowledgments
Funding: This work was supported by the National Key Basic Research Program of China (973 program, 2015CB554002), the National Natural Science Foundation of China (81972754,81773101), the Scientific Research Starting Foundation of Southern Medical University (PY2018N003).
Footnote
Reporting Checklist: The authors have completed the MDAR checklist (available at http://dx.doi.org/10.21037/atm-20-1531).
Data Sharing Statement: Available at http://dx.doi.org/10.21037/atm-20-1531
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/atm-20-1531). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. This study was conducted in accordance with the Declaration of Helsinki (as is revised in 2013).
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Spano D, Zollo M. Tumor microenvironment: a main actor in the metastasis process. Clin Exp Metastasis 2012;29:381-95. [Crossref] [PubMed]
- Bremnes RM, Donnem T, Al-Saad S, et al. The role of tumor stroma in cancer progression and prognosis: emphasis on carcinoma-associated fibroblasts and non-small cell lung cancer. J Thorac Oncol 2011;6:209-17. [Crossref] [PubMed]
- Koliaraki V, Pallangyo CK, Greten FR, et al. Mesenchymal Cells in Colon Cancer. Gastroenterology 2017;152:964-79. [Crossref] [PubMed]
- Farhood B, Najafi M, Mortezaee K. Cancer-associated fibroblasts: Secretions, interactions, and therapy. J Cell Biochem 2019;120:2791-800. [Crossref] [PubMed]
- Wang M, Zhang J, Huang Y, et al. Cancer-Associated Fibroblasts Autophagy Enhances Progression of Triple-Negative Breast Cancer Cells. Med Sci Monit 2017;23:3904-12. [Crossref] [PubMed]
- Jia CC, Wang TT, Liu W, et al. Cancer-associated fibroblasts from hepatocellular carcinoma promote malignant cell proliferation by HGF secretion. PLoS One 2013;8:e63243. [Crossref] [PubMed]
- Ziani L, Chouaib S, Thiery J. Alteration of the Antitumor Immune Response by Cancer-Associated Fibroblasts. Front Immunol 2018;9:414. [Crossref] [PubMed]
- Erdogan B, Webb DJ. Cancer-associated fibroblasts modulate growth factor signaling and extracellular matrix remodeling to regulate tumor metastasis. Biochem Soc Trans 2017;45:229-36. [Crossref] [PubMed]
- Zhu HF, Zhang XH, Gu CS, et al. Cancer-associated fibroblasts promote colorectal cancer progression by secreting CLEC3B. Cancer Biol Ther 2019;20:967-78. [Crossref] [PubMed]
- Li T, Yi S, Liu W, et al. Colorectal carcinoma-derived fibroblasts modulate natural killer cell phenotype and antitumor cytotoxicity. Med Oncol 2013;30:663. [Crossref] [PubMed]
- Tommelein J, Verset L, Boterberg T, et al. Cancer-associated fibroblasts connect metastasis-promoting communication in colorectal cancer. Front Oncol 2015;5:63. [Crossref] [PubMed]
- Kadel D, Zhang Y, Sun HR, et al. Current perspectives of cancer-associated fibroblast in therapeutic resistance: potential mechanism and future strategy. Cell Biol Toxicol 2019;35:407-21. [Crossref] [PubMed]
- Truffi M, Mazzucchelli S, Bonizzi A, et al. Nano-Strategies to Target Breast Cancer-Associated Fibroblasts: Rearranging the Tumor Microenvironment to Achieve Antitumor Efficacy. Int J Mol Sci 2019;20:1263. [Crossref] [PubMed]
- Racle J, de Jonge K, Baumgaertner P, et al. Simultaneous enumeration of cancer and immune cell types from bulk tumor gene expression data. Elife 2017;6:e26476. [Crossref] [PubMed]
- Clough E, Barrett T. The Gene Expression Omnibus Database. Methods Mol Biol 2016;1418:93-110. [Crossref] [PubMed]
- Subramanian A, Kuehn H, Gould J, et al. GSEA-P: a desktop application for Gene Set Enrichment Analysis. Bioinformatics 2007;23:3251-3. [Crossref] [PubMed]
- Damian D, Gorfine M. Statistical concerns about the GSEA procedure. Nat Genet 2004;36:663-author reply 663. [Crossref] [PubMed]
- Bland JM, Altman DG. Multiple significance tests: the Bonferroni method. BMJ 1995;310:170. [Crossref] [PubMed]
- Lamb J. The Connectivity Map: a new tool for biomedical research. Nat Rev Cancer 2007;7:54-60. [Crossref] [PubMed]
- Lamb J, Crawford ED, Peck D, et al. The Connectivity Map: using gene-expression signatures to connect small molecules, genes, and disease. Science 2006;313:1929-35. [Crossref] [PubMed]
- Daina A, Michielin O, Zoete V. SwissTargetPrediction: updated data and new features for efficient prediction of protein targets of small molecules. Nucleic Acids Res 2019;47:W357-W364. [Crossref] [PubMed]
- Gfeller D, Michielin O, Zoete V. Shaping the interaction landscape of bioactive molecules. Bioinformatics 2013;29:3073-9. [Crossref] [PubMed]
- Gfeller D, Grosdidier A, Wirth M, et al. SwissTargetPrediction: a web server for target prediction of bioactive small molecules. Nucleic Acids Res 2014;42:W32-8. [Crossref] [PubMed]
- Chen J, Bardes EE, Aronow BJ, et al. ToppGene Suite for gene list enrichment analysis and candidate gene prioritization. Nucleic Acids Res 2009;37:W305-11. [Crossref] [PubMed]
- Shannon P, Markiel A, Ozier O, et al. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res 2003;13:2498-504. [Crossref] [PubMed]
- Kohl M, Wiese S, Warscheid B. Cytoscape: software for visualization and analysis of biological networks. Methods Mol Biol 2011;696:291-303. [Crossref] [PubMed]
- Bader GD, Hogue CW. An automated method for finding molecular complexes in large protein interaction networks. BMC Bioinformatics 2003;4:2. [Crossref] [PubMed]
- Qi L, Ding Y. Construction of key signal regulatory network in metastatic colorectal cancer. Oncotarget 2018;9:6086-94. [Crossref] [PubMed]
- Obatomi DK, Bach PH. Biochemistry and toxicology of the diterpenoid glycoside atractyloside. Food Chem Toxicol 1998;36:335-46. [Crossref] [PubMed]
- Stewart MJ, Steenkamp V. The biochemistry and toxicity of atractyloside: a review. Ther Drug Monit 2000;22:641-9. [Crossref] [PubMed]
- Tcherkassova J, Tsurkan S, Smirnova G, et al. Binding characterization of the targeting drug AIMPILA to AFP receptors in human tumor xenografts. Tumour Biol 2017;39:1010428317734815. [Crossref] [PubMed]
- Lin BC, Huang D, Yu CQ, et al. MicroRNA-184 Modulates Doxorubicin Resistance in Osteosarcoma Cells by Targeting BCL2L1. Med Sci Monit 2016;22:1761-5. [Crossref] [PubMed]
- Liu ZL, Wang G, Shu Y, et al. Enhanced antitumor activity of epirubicin combined with cerulenin in osteosarcoma. Mol Med Rep 2012;5:326-30. [PubMed]
- Yu Y, Wang X, Li Q, et al. Bioinformatics analysis of gene expression alterations conferring drug resistance in tumor samples from melanoma patients with EGFR-activating BRAF mutations. Oncol Lett 2018;15:635-41. [PubMed]
- Takata N, Ohshima Y, Suzuki-Karasaki M, et al. Mitochondrial Ca2+ removal amplifies TRAIL cytotoxicity toward apoptosis-resistant tumor cells via promotion of multiple cell death modalities. Int J Oncol 2017;51:193-203. [Crossref] [PubMed]
- Hou J, Hong Z, Feng F, et al. A novel chemotherapeutic sensitivity-testing system based on collagen gel droplet embedded 3D-culture methods for hepatocellular carcinoma. BMC Cancer 2017;17:729. [Crossref] [PubMed]
- Ling X, Zhou Y, Li SW, et al. Modulation of mitochondrial permeability transition pore affects multidrug resistance in human hepatocellular carcinoma cells. Int J Biol Sci 2010;6:773-83. [Crossref] [PubMed]
- Fang M, Song Y, Ren J, et al. Atractyloside mimics BORIS knockdown to induce DNA damage in colorectal cancer cells. Int J Clin Exp Pathol 2018;11:3286-93. [PubMed]
- Henriksson ML, Edin S, Dahlin AM, et al. Colorectal cancer cells activate adjacent fibroblasts resulting in FGF1/FGFR3 signaling and increased invasion. Am J Pathol 2011;178:1387-94. [Crossref] [PubMed]
- Liu QZ, Gao XH, Chang WJ, et al. Expression of ITGB1 predicts prognosis in colorectal cancer: a large prospective study based on tissue microarray. Int J Clin Exp Pathol 2015;8:12802-10. [PubMed]
- Laurberg JR, Jensen JB, Schepeler T, et al. High expression of GEM and EDNRA is associated with metastasis and poor outcome in patients with advanced bladder cancer. BMC Cancer 2014;14:638. [Crossref] [PubMed]
- Soreide K, Janssen EA, Korner H, et al. Trypsin in colorectal cancer: molecular biological mechanisms of proliferation, invasion, and metastasis. J Pathol 2006;209:147-56. [Crossref] [PubMed]
- Waisberg J, De Souza VL, Affonso JR, et al. Overexpression of the ITGAV gene is associated with progression and spread of colorectal cancer. Anticancer Res 2014;34:5599-607. [PubMed]


