
Multiple cancer susceptible genes sequencing in BRCA-negative breast cancer with high hereditary risk
Introduction
Breast cancer susceptibility is demonstrated to be associated with hereditary background, and it is estimated that hereditary and genetic factors contributed to 27% of breast cancer incidences (1,2). BRCA1 and BRCA2 germline mutations are the most common cause of hereditary breast cancer. In our previous study, comprehensive screening in Chinese breast cancer patients with high hereditary risk in our cancer centre showed a low BRCA mutation prevalence (3), which suggesting the majority of Chinese hereditary breast cancer is associated with other susceptible genes. Apart from the first discovery of BRCA1 and BRCA2, other breast cancer associated susceptibility genes have been identified constantly, including high-penetrance susceptible genes (TP53 and PTEN), moderate-penetrance susceptible genes (CDH1, STK11, NF1, PALB2, CHEK2, ATM and NBN), and low-penetrance susceptible genes (BARD1, FANCC, MRE11A, MUTYH heterozygotes, RECQL, RAD50, RET1, SLX4, SMARCA4, XRCC2 and so on) (4-6). Despite the fact that breast cancer susceptible genes have been extensively studied and multiple genes testing have been widely performed in Caucasians, Ashkenazi Jewish and African Americans, insufficient data supports the knowledge of hereditary background in Chinese breast cancer patients.
Many retrospective studies proved that clinicopathologic features and outcomes of breast cancer varied between Chinese and Caucasian population. Chinese patients had a younger age at diagnosis of breast cancer, whose peak age onset was between 45 and 55 years old, compared to an average of between 60 and 70 years old in Caucasian breast cancer patients (7). Besides, Chinese patients had a lower rate of incidence of invasive lobular breast cancer. Genomic profiling studies also demonstrated disparities between breast cancers of different ethics. One study compared gene expression and microRNA profiles between Chinese and Italian breast cancers and found lower prevalence of Luminal A subtype among Chinese breast cancers (8). A more recent study revealed a higher mutational prevalence for TP53 and AKT1 in Chinese patients (9).
The National Comprehensive Cancer Network (NCCN) has set criteria of hereditary risk evaluations for breast cancer patients since 2014 (6,10-12). Main concerns in NCCN guidelines include early-age onset breast cancer, triple negative breast cancer under 60 years old, primary bilateral breast cancer, male breast cancer and breast cancer with certain family history. The NCCN guidelines recommend multigene testing should ideally be offered in the context of professional genetic expertise for pre- and post-test counselling, and warranted’ in those who have tested negative for a single inherited syndrome (6,10,11). However, no consensus or guidelines regarding the identification of hereditary mutation (beyond BRCA1 and BRCA2) carriers and clinical management options has been integrated for Chinese breast cancer patients.
Next-generation sequencing (NGS) is driving growth and possibilities in genomic researching, providing reading lengths as long as the entire genomes, reducing the cost of sequencing, and enabling the application of genetic testing as a clinical tool (13,14). Moreover, NGS allows for the sequencing of multiple genes simultaneously at an unprecedented speed. Multiple gene panel testing could not only include high-penetrance susceptible genes associated with a specific cancer, but also include moderate- and low-penetrance susceptible genes as well (15). Meanwhile, multiple gene panels for inherited cancer risk have proved to be a more time- and cost-efficient approach in hereditary risk management.
In our present study, we are aiming to provide more information about and get better knowledge of mutational spectrum in Chinese population, to identify novel mutations in high hereditary risk breast cancer patients with BRCA1 and BRCA2 testing negative, and to aid in updating the clinical recommendations for genetic testing.
Methods
Pathologic data
A triple-negative breast cancer (TNBC) case was defined as a patient whose tumour sample was negative for oestrogen receptor (ER), progesterone receptor (PR) and human epidermal growth factor receptor 2 (HER2) expression upon immunohistochemical (IHC) staining. ER or PR immunostaining was considered positive when >1% of the tumour cells showed positive nuclear staining. Patients showing HER2 expression (IHC, score equal to 2+) were subjected to florescence in situ hybridization (FISH) to determine HER2 gene amplification. The HER2 over-expression subgroup was defined as those patients who were FISH-positive or presented an IHC staining score equal to 3+.
Cases and samples
We selected the breast cancer patients with high-risk hereditary background who was previously tested negative in BRCA1 and BRCA2 genes. Breast cancer patients with any two of the five following risk criteria were defined to harbour high-risk hereditary background in the present study: (I) pathological diagnosis of TNBC, (II) male breast cancer, (III) primary bilateral breast cancer, (IV) early-age onset breast cancer (less than or equal to 40 years of age at diagnosis), or (V) positive family history of breast and/or ovarian cancer. All the cases were collected from three independent hospitals in China, which were Fudan University Shanghai Cancer Center, the Affiliated Union Hospital of Fujian Medical University, and Shanghai First Maternity and Infant Hospital. Finally, a total of 384 patients were enrolled and peripheral blood samples were collected. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013). The study was approved by the Ethics Committee of Fudan University Shanghai Cancer Center (No. 050432-4-1212B) and informed consent was taken from all the patients.
Multigene testing
The Multigene panel includes 30 breast cancer associated susceptibility genes (Table 1). All coding regions and exon-intron boundaries of the genes were screened. The average intronic sequence length was 70 bp (ranging from 5 to 204 bp).
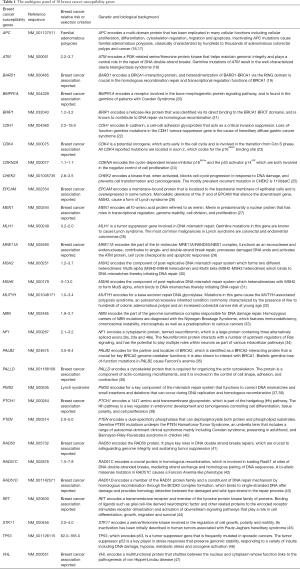
Full table
Multiplex PCR
Genomic DNA was isolated from peripheral lymphocytes using a TGuide M16 automatic extraction machine (Tiangen Biotechnology, Beijing, China). The DNA concentration was quantified using a NanoDrop ND2000 (NanoDrop Technologies, Wilmington, DE, USA) spectrophotometer, and the samples were diluted to 20–50 ng/µL if the DNA concentration was higher than 50 ng/µL. Thirty-microliter aliquots of the DNA samples were transferred to the wells of a 96-well-plate. A total of 384 extracted genomic DNA samples were used for target capture and sequencing.
All DNA samples were amplified in two separate multiplex PCR assays. Each amplification reaction was prepared by mixing 3 µL of the genomic DNA, 8 µL of each primer panel, 12.5 µL of the KAPA2G Robust hot start ready mix (Kapa Biosystems, Wilmington, MA, USA) and 1.5 µL of H2O. The PCR program was 95 °C for 4 min followed by 18 cycles of 98 °C for 15 s and 60 °C for 4 min. The PCR products were cleaned up using AMPure XP Beads (Beckman Coulter, Pasadena, CA, USA). The procedure was performed according to the manufacturer’s protocol and described in the supplementary materials.
Barcoding and Illumina sequencing
Barcoding was performed in a 20-µL reaction mixture that contained 8 µL of the cleaned PCR products, 10 µL of KAPA2G Robust hot start ready mix (Kapa Biosystems, Pasadena, CA, USA), 1 µmol/L barcode F primers and 1 µmol/L barcode R primer. The reaction was performed in a conventional PCR thermal cycler using the following conditions: 95 °C for 30 seconds; 5 cycles of 95 °C for 15 seconds, 55 °C for 15 seconds, and 72 °C for 1 minute; and a completion step at 72 °C for 5 minutes.
The barcoded PCR products from the various samples were cleaned up using AMPure XP Beads (Beckman Coulter, Pasadena, CA, USA). The procedure was performed according to the manufacturer’s protocol and described in the supplementary materials. The purified PCR product library was quantified using a Qubit Fluorometer (Thermo Fisher Scientific, Waltham, MA, USA). Based on library quantitation, the PCR products were pooled together in equal molar ratios. The purified libraries were routinely sequenced on a NextSeq 500 sequencer (Illumina, San Diego, CA, USA) using the 2×150 bp end sequencing protocol.
Analysis of sequencing data
Demultiplexed, compressed FASTQ files were generated from BCL using bcl2fastq Conversion Software v1.8.4 (Illumina, San Diego, CA, USA). For all successful sequencing runs, the read depth was 30× at any given position, with 100× mean coverage across the entire targeted sequence and Q30 at greater than 75% of reads. The variant calling and coverage of each captured region were analysed using an in-house-developed bioinformatics pipeline based on the general analysis algorithm pipeline. Briefly, the reads were mapped to the hg19 version of the human reference genome (GRCh37) and then filtered to remove off-target and poor-quality reads. Variants were identified and annotated. The variants and annotation results were transferred into Excel spreadsheets.
Interpretation of the mutation testing results
The mutations were classified as benign, likely-benign, variants of uncertain significance, likely-pathogenic, and pathogenic. If applicable, detailed information was obtained using the gene-specific databases dbSNP (http://www.ncbi.nlm.nih.gov/projects/SNP), ClinVar (http://www.ncbi.nlm.nih.gov/clinvar/). Subsequently, a manual literature search was performed using a Google search in PubMed, Science-Direct, and BioMed Central to confirm that there had been no previous reports on each specific mutation. Novel mutations were defined when there was no match to the reference single-nucleotide polymorphism (RS) numbers in the dbSNP database. Mutations were classified according to American College of Medical Genetics and Genomics recommendations (48) and interpreted as positive for a oncogenic mutation when (I) frameshift insertions or deletions resulted in the expression of an abnormal or truncated protein product; (II) mutations in noncoding intervening sequence at splicing sites caused abnormal processing of the mRNA transcript; or (III) missense mutations and non-frameshift insertions or deletions were defined as pathogenic in a database and/or published study. The mutations with clear oncogenic impacts reported in previous studies were selected for further analysis.
Variant confirmation
A subset of variants, including known variants that were pathogenic or likely pathogenic and newly identified variants with functional damage, was confirmed by conventional Sanger sequencing using the BigDye Terminator v3.1 Cycle Sequencing Kit (Thermo Fisher Scientific, Waltham, MA, USA). Variants that could not be confirmed were excluded from further analysis.
Statistical analysis
The Chi-square test, t-test and Fisher’s exact test were applied in statistical analysis. The statistical analyses were performed using SPSS software version 20.0 (IBM institute, Chicago, IL, USA). All P values in the study were two-sided, and P<0.05 was considered statistically significant.
Results
Description of the NGS dataset
Our NGS analysis revealed 18,435 candidate variants in the 30 genes’ coding regions and the adjacent splice sites, with a range of 34–78 genetic variants in individual samples. These candidate variants included 27 splicing variants, and 18,408 exonic variants. The exonic variants represented 7,266 missense variants, 11,102 silent variants, 11 stop-gain variants, 3 stop-loss variants, and 26 insertion variants.
Associations between clinical characteristics and mutation status
As it was described above, a total of 384 Chinese breast cancer patients with high hereditary risks were recruited. All the participants were tested to be BRCA-negative who came from our previous study (3). The baseline characteristics of breast cancer patients and its relationship with oncogenic mutations were showed in Table 2.
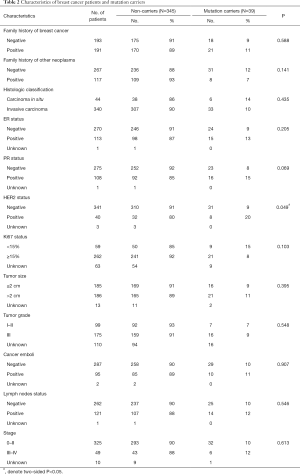
Full table
A total of 39 (39/384, 10.2%) mutation carriers were identified in our multigene screening. Most kinds of clinical characteristics didn’t have statistically significant associations with multigene mutation status, except that breast cancer patients with HER2 positive tended to have a higher mutation prevalence than those with HER2 negative (20% versus 9%, P=0.049).
In our study, the average age at diagnosis of breast cancer was similar between patients with and without germline mutations in these BRCA-negative cases (42 versus 39, P=0.431; Table 3). However, we found the average age at diagnosis of breast cancer was significantly older for patients with deleterious RET mutations than the patients without germline mutations (49 versus 39, P=0.028; Table 3). We further evaluated whether patients with mutations in the 30 predisposition genes were associated with a stronger family history of breast or ovarian cancers than non-mutated patients. In particular, all patients with RET mutations were enriched for a family history of breast cancer (100% versus 49%, P=0.014; Table 3). However, no carriers had a family history of ovarian cancer.
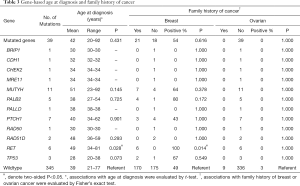
Full table
We also evaluated associations between mutation status of single predisposition gene and clinical stages (Table 4) as well as tumor pathology (Table 5). Overall, carriers and non-carriers had similar tumor stages (Table 4). When each receptor was examined alone, we observed PALB2 mutation carriers were more likely to be ER-positive than non-carriers (80% versus 28%, P=0.027; Table 4). Notably, TP53-mutated breast cancers were significantly more likely to be ER−, PR− and HER2-positive (100% versus 28%, P=0.024 for ER; 100% versus 27%, P=0.020 for PR; 100% versus 9%, P=0.001 for HER2; Table 5).
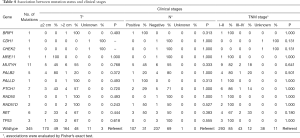
Full table
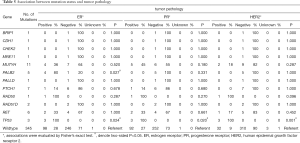
Full table
Associations between hereditary risk factors and mutation status
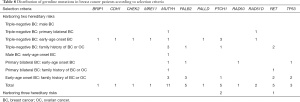
According to the study design, all patients were specifically chosen to harbour at least two known risk factors of hereditary background. Breast cancer patients with two risk factors took the main part of our cohort (354/384, 92%), while breast cancer patients with three risk factors took the rest (30/384, 8%). We didn’t observe any person who harboured four or five risk factors as described in the selection criteria. In the meanwhile, no male patients with primary bilateral breast cancer or a positive family history of breast/ovarian cancer could be enrolled in our cohort. In our study, most of the participants included were early-age onset patients with triple negative (147/384, 38%), followed by early-age onset patients with a positive family history of breast cancer or ovarian cancer (99/384, 26%) (Table 6).

Full table
Though the number of patients is rare, male breast cancer patients under 40 years old were very likely to be tested positive in multigene screening (1/3, 33%). The early-age onset patients with primary bilateral breast cancer showed a high prevalence of germline mutation (4/18, 22%), followed by primary bilateral breast cancer with a positive family history of breast/ovarian cancer (2/13, 15%). Interestingly, multigene mutation frequency was similar between breast cancer patients with two risk factors (36/354, 10%) and those with three factors (3/30, 10%).
Multigene germline mutations
Among the 39 patients (39/384, 10.2%) with pathogenic/likely-pathogenic germline mutations, one participant (patient code, 295860) carried two distinct mutations, which were RET c.341G>A and MUTYH c.C55T (Table 7). The major mutant non-BRCA genes were MUTYH (n=11), PTCH1 (n=7), RET (n=6) and PALB2 (n=5). Other mutant genes included TP53 (n=3), RAD51D (n=2), CHEK2 (n=1), BRIP1 (n=1), CDH1 (n=1), MRE11 (n=1), RAD50 (n=1) and PALLD (n=1). We identified 4 novel mutations which were never reported before, including PALB2 c.2964_2965insAA, PALB2 c.T1352G, RAD50 c.C1966T and RAD51D c.331_332insTA. A splicing germline mutation, MUTYH c.934-2A>G, was demonstrated to be a hotspot (9/384, 2.3%) in Chinese breast cancer. Besides, we observed two recurrent mutations in our cohort, including RET c.341G>A (4/384, 1.0%) and PTCH1 c.2479A>G (6/384, 1.6%) mutations.
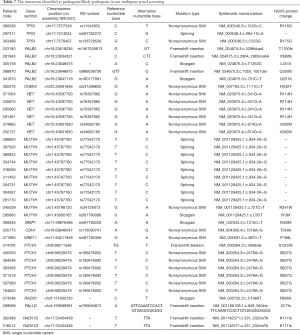
Full table
The association between distribution of multigene germline mutations and hereditary risks was not statistically apparent. We could merely tell PALB2 and RET mutations possibly tend to occur in breast cancer patients with family history of breast or ovarian cancer, for all those mutations were only observed in groups carrying risk factor of a positive family history of breast or ovarian cancer (Table 8). Similarly, TP53 mutations might associate with breast cancer taking place at a young age for they were all falling into groups carrying risk factor of early-age onset.
Full table
Discussion
The present study demonstrated about 10% of Chinese breast cancer patients with high hereditary risk who were previously tested BRCA-negative could benefit from multigene testing. Our study contributed to the knowledge of germline variations in multiple cancer susceptible genes in Chinese population. In previous studies, beyond BRCA1 and BRCA2, the prevalence of germline mutations varied from 4.3% to 34.3% according to different recruiting criteria, gene panels or sequencing methods (49-55). Li et al. conducted a multi-centre study to investigate mutational frequency in Chinese patients with high hereditary risk breast cancer patients, and the study showed 23.8% of participants contained germline mutations, including 6.8% in 38 other non-BRCA genes (52). Similarly, the study also defined multiple hereditary risks as selection criteria. A more recent study carried out by Wang et al. found 8.5% of patients harboured non-BRCA oncogenic mutations through a 22-gene panel screening, which were mainly found in ATM, CHEK2, PALB2, and BRIP1 genes (55). In a much larger study with more susceptibility genes testing, the data from 8,085 cases demonstrated a mutation frequency of 2.9% in non-BRCA susceptibility genes (54). In spite of the fact that a more general gene panel was applied, the mutation frequency didn’t go up as the rising number of sequenced genes. However, it seemed quite different when genes number crossing over one hundred. There is another study using a panel of 152 genes associated with hereditary cancer, and the study identified 16.1% of hereditary breast cancer patients as non-BRCA germline mutation carriers. Taken together, these collective evidences suggested criteria should be carefully chosen when using a small gene panel to detect genetic variations in hereditary breast cancer patients.
In our previous study, we observed BRCA mutation frequency raised up with hereditary risk factors added up (3). However, the theory didn’t work well in non-BRCA mutations. It was noted that multigene mutation frequency was similar between breast cancer patients with two risk factors (36/354, 10%) and those with three factors (3/30, 10%) in our present cohort. Due the limited sample size and the lack of comparable study, it is hard to tell a difference for now, so more data and larger studies await to demonstrate such phenomenon.
PALB2 germline mutation frequency was demonstrated to be 1.3% in our study, and the results varied from 0.7–1.2% in other Chinese studies (52,56,57). We further observed a potential association between PALB2 mutation carriers and breast cancer with a positive history of breast/ovarian cancer, and other studies also proved the conclusion (56,57). Wu et al. performed PALB2 mutation screening a large Chinese breast cancer cohort, and demonstrated that compared with non-carriers, PALB2 mutation carriers were significantly more likely to have a familial aggregation of breast cancer and/or ovarian cancer (27.8% vs. 8.4%, P<0.001) (57). In the meanwhile, we also RET mutations tended to occur in breast cancer patients with family history of breast or ovarian cancer, but no further studies support the conclusion for RET mutations were less studied in breast cancer. A previous study only found one RET mutation carriers out of 8,085 consecutive unselected Chinese breast cancer patients (54). It seemed RET mutations could be more prevalent in breast cancer with high hereditary risk which needed to be confirmed by further investigation.
As mentioned before, we identified a hotspot germline mutation, MUTYH c.934-2A>G, in Chinese breast cancer. MUTYH is a human base excision repair gene involved in preventing 8-oxo-dG-induced mutagenesis (58). Bi-allelic germline mutations of the MUTYH gene lead to autosomal recessive colorectal adenomatous polyposis and very high colorectal cancer risk in Caucasian population (59,60). MUTYH c.934-2A>G was first found in Japanese familial gastric cancer patients and also demonstrated to cause a splicing abnormality that led to the production of an aberrant mRNA transcript encoding a truncated MYH protein and lead to an impaired ability of excision repair (61). Interestingly, experts hold converse opinion about the MUTYH mutation, saying that some support its pathogenicity (62-65), while some do not (52,66,67). Notably, a Chinese study reported a relatively high variant rate (4.2%, 5/120) of MUTYH c.892-2A>G in their high-risk group, but lower rate (0.8%, 1/120) in their breast cancer group (66). According to the 5-tier rating system in American College of Medical Genetics and Genomics recommendations, MUTYH c.934-2A>G is likely pathogenic (48). Besides, another Chinese study also noticed 8 MUTYH mutation carriers out of 937 patients with high hereditary risk breast cancer (52). Moreover, a more recent study identified a MUTYH germline pathogenic variant and somatic loss of the wild-type allele which contributed to tumorigenesis (65). Considering all above, with currently available evidence suggesting that the variant is pathogenic, but the available data is insufficient to prove that conclusively. Therefore, this variant was classified as likely pathogenic in our study.
We also explored whether the mutation status could impact the survival in these BRCA-negative breast cancer (data not showed), but no significant results were observed in comparing disease-free survival (DFS) or overall survival between the germline mutation carriers and non-carriers. Previous studies came to inconsistent conclusions about BRCA mutation status as a prognostic factor in breast cancer (68-73). Among other predisposition genes, CHEK2 1100delC was demonstrated to be associated with increased risk of second breast cancer and a worse long-term recurrence-free survival rate (74). Another study indicated CHEK2 H371Y mutation carriers were more likely to respond to neoadjuvant chemotherapy than non-carriers (75). However, we failed to identify these two mutations in our cohort. Moreover, breast cancer patients with PALB2 mutations were considered to be at a higher risk of death from breast cancer compared with non-carriers (76). A more recent study involved 16 DNA-repair genes including ATM, BLM, CHEK2, FANCC, MER11A, MLH1, MSH2, MSH6, MUTHY, NBN, PALB2, PMS2, RAD50, RAD51C, RAD51D and TP53 (77), where most genes were also comprised in our study. The study concluded that 3.4% of BRCA-negative breast cancer patients carried germline mutations in the 16 DNA-repair genes, and the DNA-repair gene mutation carriers exhibited an aggressive phenotype and had poor survival compared with non-carriers. By virtue of the germline mutations, breast cancers harboring these mutations had unique mechanisms that could be rationally targeted for therapeutic opportunities. Increasing evidences demonstrated mutations in non-BRCA1/2 DNA-repair genes contributed to sensitivity to PARP inhibitors, which suggested carriers of mutated DNA-repair genes could undergo treatment with PARP inhibitors (78). Besides PARP, there were other key components, like PTEN (79-81), ATM (82), MSH2 (83,84) and APC (85), showing potentials for targeted therapy.
In conclusion, appropriately selected patients may gain benefit from multigene sequencing, and comprehensive gene panels could help understand hereditary mutations in genetic counselling, for hereditary breast cancer could be associated with more than breast cancer specific susceptibility genes especially when it was tested BRCA-negative. As the costs of genomic testing decline and the benefits of sequencing appearing, it is inevitable that the use of gene-panel testing, even whole-exome and whole-genome sequencing, will become widespread and come into daily clinical practice in China.
Conclusions
Our study demonstrated 10% of Chinese breast cancer patients with high hereditary risk who were previously tested BRCA-negative could benefit from multigene testing. Comprehensive gene panels could help understand hereditary mutations in genetic counselling, for hereditary breast cancer could be associated with more than breast cancer specific susceptibility genes when it was tested BRCA-negative.
Acknowledgments
Funding: This work was supported by grants from the National Natural Science Foundation of China (81572583, 81502278, 81672601, 81872137, 81502289 and 81602311), the Municipal Project for Developing Emerging and Frontier Technology in Shanghai Hospitals (SHDC12010116), the Cooperation Project of Conquering Major Diseases in Shanghai Municipality Health System (2013ZYJB0302), the Innovation Team of Ministry of Education (IRT1223), the Shanghai Key Laboratory of Breast Cancer (12DZ2260100), the Ministry of Science and Technology of China (2018YFE020160), the Shanghai Natural Science Foundation (19ZR1411100) and Shanghai Health Commission Foundation (201940167). The funders had no role in the study design, collection and analysis of the data, decision to publish, or manuscript preparation.
Footnote
Data Sharing Statement: Available at http://dx.doi.org/10.21037/atm-20-2999
Peer Review File: Available at http://dx.doi.org/10.21037/atm-20-2999
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/atm-20-2999). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013). The study was approved by the Ethics Committee of Fudan University Shanghai Cancer Center (No. 050432-4-1212B) and informed consent was taken from all the patients.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Paradiso A, Formenti S. Hereditary breast cancer: clinical features and risk reduction strategies. Ann Oncol 2011;22 Suppl 1:i31-6. [Crossref] [PubMed]
- Peto J, Mack TM. High constant incidence in twins and other relatives of women with breast cancer. Nat Genet 2000;26:411-4. [Crossref] [PubMed]
- Lang GT, Shi JX, Hu X, et al. The spectrum of BRCA mutations and characteristics of BRCA-associated breast cancers in China: Screening of 2,991 patients and 1,043 controls by next-generation sequencing. Int J Cancer 2017;141:129-42. [Crossref] [PubMed]
- Nielsen FC, van Overeem Hansen T, Sorensen CS. Hereditary breast and ovarian cancer: new genes in confined pathways. Nat Rev Cancer 2016;16:599-612. [Crossref] [PubMed]
- Zhang B, Beeghly-Fadiel A, Long J, et al. Genetic variants associated with breast-cancer risk: comprehensive research synopsis, meta-analysis, and epidemiological evidence. Lancet Oncol 2011;12:477-88. [Crossref] [PubMed]
- Daly MB, Pilarski R, Berry M, et al. NCCN Guidelines Insights: Genetic/Familial High-Risk Assessment: Breast and Ovarian, Version 2.2017. J Natl Compr Canc Netw 2017;15:9-20. [Crossref] [PubMed]
- Chen DN, Song CG, Ouyang QW, et al. Differences in breast cancer characteristics and outcomes between Caucasian and Chinese women in the US. Oncotarget 2015;6:12774-82. [Crossref] [PubMed]
- Huang X, Dugo M, Callari M, et al. Molecular portrait of breast cancer in China reveals comprehensive transcriptomic likeness to Caucasian breast cancer and low prevalence of luminal A subtype. Cancer Med 2015;4:1016-30. [Crossref] [PubMed]
- Zhang G, Wang Y, Chen B, et al. Characterization of frequently mutated cancer genes in Chinese breast tumors: a comparison of Chinese and TCGA cohorts. Ann Transl Med 2019;7:179. [Crossref] [PubMed]
- Daly MB, Pilarski R, Axilbund JE, et al. Genetic/familial high-risk assessment: breast and ovarian, version 1.2014. J Natl Compr Canc Netw 2014;12:1326-38. [Crossref] [PubMed]
- Daly MB, Pilarski R, Axilbund JE, et al. Genetic/Familial High-Risk Assessment: Breast and Ovarian, Version 2.2015. J Natl Compr Canc Netw 2016;14:153-62. [Crossref] [PubMed]
- Daly MB, Pilarski R, Yurgelun MB, et al. NCCN Guidelines Insights: Genetic/Familial High-Risk Assessment: Breast, Ovarian, and Pancreatic, Version 1.2020. J Natl Compr Canc Netw 2020;18:380-91. [Crossref] [PubMed]
- Koboldt DC, Steinberg KM, Larson DE, et al. The next-generation sequencing revolution and its impact on genomics. Cell 2013;155:27-38. [Crossref] [PubMed]
- Metzker ML. Sequencing technologies - the next generation. Nat Rev Genet 2010;11:31-46. [Crossref] [PubMed]
- Grissom AA, Friend PJ. Multigene Panel Testing for Hereditary Cancer Risk. J Adv Pract Oncol 2016;7:394-407. [PubMed]
- Kerr SE, Thomas CB, Thibodeau SN, et al. APC germline mutations in individuals being evaluated for familial adenomatous polyposis: a review of the Mayo Clinic experience with 1591 consecutive tests. J Mol Diagn 2013;15:31-43. [Crossref] [PubMed]
- Zeineldin M, Neufeld KL. Understanding phenotypic variation in rodent models with germline Apc mutations. Cancer Res 2013;73:2389-99. [Crossref] [PubMed]
- Choi M, Kipps T, Kurzrock R. ATM Mutations in Cancer: Therapeutic Implications. Mol Cancer Ther 2016;15:1781-91. [Crossref] [PubMed]
- Weber-Lassalle N, Borde J, Weber-Lassalle K, et al. Germline loss-of-function variants in the BARD1 gene are associated with early-onset familial breast cancer but not ovarian cancer. Breast Cancer Res 2019;21:55. [Crossref] [PubMed]
- Dahdaleh FS, Carr JC, Calva D, et al. Juvenile polyposis and other intestinal polyposis syndromes with microdeletions of chromosome 10q22-23. Clin Genet 2012;81:110-6. [Crossref] [PubMed]
- Easton DF, Lesueur F, Decker B, et al. No evidence that protein truncating variants in BRIP1 are associated with breast cancer risk: implications for gene panel testing. J Med Genet 2016;53:298-309. [Crossref] [PubMed]
- Figueiredo J, Melo S, Carneiro P, et al. Clinical spectrum and pleiotropic nature of CDH1 germline mutations. J Med Genet 2019;56:199-208. [Crossref] [PubMed]
- Sabir M, Baig RM, Mahjabeen I, et al. Novel germline CDK4 mutations in patients with head and neck cancer. Hered Cancer Clin Pract 2012;10:11. [Crossref] [PubMed]
- Bartsch DK, Sina-Frey M, Lang S, et al. CDKN2A germline mutations in familial pancreatic cancer. Ann Surg 2002;236:730-7. [Crossref] [PubMed]
- Fan Z, Ouyang T, Li J, et al. Identification and analysis of CHEK2 germline mutations in Chinese BRCA1/2-negative breast cancer patients. Breast Cancer Res Treat 2018;169:59-67. [Crossref] [PubMed]
- Pathak SJ, Mueller JL, Okamoto K, et al. EPCAM mutation update: Variants associated with congenital tufting enteropathy and Lynch syndrome. Hum Mutat 2019;40:142-61. [Crossref] [PubMed]
- Lemos MC, Thakker RV. Multiple endocrine neoplasia type 1 (MEN1): analysis of 1336 mutations reported in the first decade following identification of the gene. Hum Mutat 2008;29:22-32. [Crossref] [PubMed]
- Bronner CE, Baker SM, Morrison PT, et al. Mutation in the DNA mismatch repair gene homologue hMLH 1 is associated with hereditary non-polyposis colon cancer. Nature 1994;368:258-61. [Crossref] [PubMed]
- Li Y, Shen Y, Jin K, et al. The DNA Repair Nuclease MRE11A Functions as a Mitochondrial Protector and Prevents T Cell Pyroptosis and Tissue Inflammation. Cell Metabolism 2019;30:477-92.e6. [Crossref] [PubMed]
- Chan TL, Yuen ST, Kong CK, et al. Heritable germline epimutation of MSH2 in a family with hereditary nonpolyposis colorectal cancer. Nat Genet 2006;38:1178-83. [PubMed]
- Edelmann W, Yang K, Umar A, et al. Mutation in the mismatch repair gene Msh6 causes cancer susceptibility. Cell 1997;91:467-77. [PubMed]
- Jasperson KW, Tuohy TM, Neklason DW, et al. Hereditary and familial colon cancer. Gastroenterology 2010;138:2044-58. [Crossref] [PubMed]
- Varon R, Vissinga C, Platzer M, et al. Nibrin, a novel DNA double-strand break repair protein, is mutated in Nijmegen breakage syndrome. Cell 1998;93:467-76. [Crossref] [PubMed]
- Shilyansky C, Lee YS, Silva AJ. Molecular and cellular mechanisms of learning disabilities: a focus on NF1. Annu Rev Neurosci 2010;33:221-43. [Crossref] [PubMed]
- Antoniou AC, Casadei S, Heikkinen T, et al. Breast-cancer risk in families with mutations in PALB2. N Engl J Med 2014;371:497-506. [Crossref] [PubMed]
- Pogue-Geile KL, Chen R, Bronner MP, et al. Palladin mutation causes familial pancreatic cancer and suggests a new cancer mechanism. PLoS Med 2006;3:e516. [Crossref] [PubMed]
- Stelzer G, Rosen N, Plaschkes I, et al. The GeneCards Suite: From Gene Data Mining to Disease Genome Sequence Analyses. Curr Protoc Bioinformatics 2016;54:1.30.1-1.30.33.
- Prolla TA, Baker SM, Harris AC, et al. Tumour susceptibility and spontaneous mutation in mice deficient in Mlh1, Pms1 and Pms2 DNA mismatch repair. Nat Genet 1998;18:276-9. [Crossref] [PubMed]
- Reinders MG, van Hout AF, Cosgun B, et al. New mutations and an updated database for the patched-1 (PTCH1) gene. Mol Genet Genomic Med 2018;6:409-15. [Crossref] [PubMed]
- Tan MH, Mester JL, Ngeow J, et al. Lifetime cancer risks in individuals with germline PTEN mutations. Clin Cancer Res 2012;18:400-7. [Crossref] [PubMed]
- Bhaskara V, Dupré A, Lengsfeld B, et al. Rad50 adenylate kinase activity regulates DNA tethering by Mre11/Rad50 complexes. Mol Cell 2007;25:647-61. [Crossref] [PubMed]
- Sopik V, Akbari MR, Narod SA. Genetic testing for RAD51C mutations: in the clinic and community. Clin Genet 2015;88:303-12. [Crossref] [PubMed]
- Masson JY, Tarsounas MC, Stasiak AZ, et al. Identification and purification of two distinct complexes containing the five RAD51 paralogs. Genes Dev 2001;15:3296-307. [Crossref] [PubMed]
- Durbec P, Marcos-Gutierrez CV, Kilkenny C, et al. GDNF signalling through the Ret receptor tyrosine kinase. Nature 1996;381:789-93. [Crossref] [PubMed]
- Hemminki A, Markie D, Tomlinson I, et al. A serine/threonine kinase gene defective in Peutz-Jeghers syndrome. Nature 1998;391:184-7. [Crossref] [PubMed]
- Whibley C, Pharoah PDP, Hollstein M. p53 polymorphisms: cancer implications. Nature Reviews Cancer 2009;9:95-107. [Crossref] [PubMed]
- Kaelin WG Jr. Molecular basis of the VHL hereditary cancer syndrome. Nat Rev Cancer 2002;2:673-82. [Crossref] [PubMed]
- Richards S, Aziz N, Bale S, et al. Standards and guidelines for the interpretation of sequence variants: a joint consensus recommendation of the American College of Medical Genetics and Genomics and the Association for Molecular Pathology. Genet Med 2015;17:405-24. [Crossref] [PubMed]
- Kurian AW, Hare EE, Mills MA, et al. Clinical evaluation of a multiple-gene sequencing panel for hereditary cancer risk assessment. J Clin Oncol 2014;32:2001-9. [Crossref] [PubMed]
- Tung N, Battelli C, Allen B, et al. Frequency of mutations in individuals with breast cancer referred for BRCA1 and BRCA2 testing using next-generation sequencing with a 25-gene panel. Cancer 2015;121:25-33. [Crossref] [PubMed]
- Castera L, Krieger S, Rousselin A, et al. Next-generation sequencing for the diagnosis of hereditary breast and ovarian cancer using genomic capture targeting multiple candidate genes. Eur J Hum Genet 2014;22:1305-13. [Crossref] [PubMed]
- Li JY, Jing R, Wei H, et al. Germline mutations in 40 cancer susceptibility genes among Chinese patients with high hereditary risk breast cancer. Int J Cancer 2019;144:281-9. [Crossref] [PubMed]
- Yang X, Wu J, Lu J, et al. Identification of a comprehensive spectrum of genetic factors for hereditary breast cancer in a Chinese population by next-generation sequencing. PLoS One 2015;10:e0125571. [Crossref] [PubMed]
- Sun J, Meng H, Yao L, et al. Germline Mutations in Cancer Susceptibility Genes in a Large Series of Unselected Breast Cancer Patients. Clin Cancer Res 2017;23:6113-9. [Crossref] [PubMed]
- Wang J, Li W, Shi Y, et al. Germline mutation landscape of Chinese patients with familial breast/ovarian cancer in a panel of 22 susceptibility genes. Cancer Med 2019;8:2074-84. [Crossref] [PubMed]
- Deng M, Chen HH, Zhu X, et al. Prevalence and clinical outcomes of germline mutations in BRCA1/2 and PALB2 genes in 2769 unselected breast cancer patients in China. Int J Cancer 2019;145:1517-28. [Crossref] [PubMed]
- Wu Y, Ouyang T, Li J, et al. Spectrum and clinical relevance of PALB2 germline mutations in 7657 Chinese BRCA1/2-negative breast cancer patients. Breast Cancer Res Treat 2020;179:605-14. [Crossref] [PubMed]
- Hegde M, Ferber M, Mao R, et al. ACMG technical standards and guidelines for genetic testing for inherited colorectal cancer (Lynch syndrome, familial adenomatous polyposis, and MYH-associated polyposis). Genet Med 2014;16:101-16. [Crossref] [PubMed]
- Al-Tassan N, Chmiel NH, Maynard J, et al. Inherited variants of MYH associated with somatic G:C-->T:A mutations in colorectal tumors. Nat Genet 2002;30:227-32. [Crossref] [PubMed]
- Sieber OM, Lipton L, Crabtree M, et al. Multiple colorectal adenomas, classic adenomatous polyposis, and germ-line mutations in MYH. N Engl J Med 2003;348:791-9. [Crossref] [PubMed]
- Tao H, Shinmura K, Hanaoka T, et al. A novel splice-site variant of the base excision repair gene MYH is associated with production of an aberrant mRNA transcript encoding a truncated MYH protein not localized in the nucleus. Carcinogenesis 2004;25:1859-66. [Crossref] [PubMed]
- Wasielewski M, Out AA, Vermeulen J, et al. Increased MUTYH mutation frequency among Dutch families with breast cancer and colorectal cancer. Breast Cancer Res Treat 2010;124:635-41. [Crossref] [PubMed]
- Rennert G, Lejbkowicz F, Cohen I, et al. MutYH mutation carriers have increased breast cancer risk. Cancer 2012;118:1989-93. [Crossref] [PubMed]
- Win AK, Reece JC, Dowty JG, et al. Risk of extracolonic cancers for people with biallelic and monoallelic mutations in MUTYH. Int J Cancer 2016;139:1557-63. [Crossref] [PubMed]
- Nones K, Johnson J, Newell F, et al. Whole-genome sequencing reveals clinically relevant insights into the aetiology of familial breast cancers. Ann Oncol 2019;30:1071-9. [Crossref] [PubMed]
- Jian W, Shao K, Qin Q, et al. Clinical and genetic characterization of hereditary breast cancer in a Chinese population. Hered Cancer Clin Pract 2017;15:19. [Crossref] [PubMed]
- Fulk K, LaDuca H, Black MH, et al. Monoallelic MUTYH carrier status is not associated with increased breast cancer risk in a multigene panel cohort. Fam Cancer 2019;18:197-201. [Crossref] [PubMed]
- Zhong Q, Peng HL, Zhao X, et al. Effects of BRCA1- and BRCA2-related mutations on ovarian and breast cancer survival: a meta-analysis. Clin Cancer Res 2015;21:211-20. [Crossref] [PubMed]
- van den Broek AJ, Schmidt MK, van 't Veer LJ, et al. Worse breast cancer prognosis of BRCA1/BRCA2 mutation carriers: what's the evidence? A systematic review with meta-analysis. PLoS One 2015;10:e0120189. [Crossref] [PubMed]
- Goodwin PJ, Phillips KA, West DW, et al. Breast cancer prognosis in BRCA1 and BRCA2 mutation carriers: an International Prospective Breast Cancer Family Registry population-based cohort study. J Clin Oncol 2012;30:19-26. [Crossref] [PubMed]
- Cortesi L, Masini C, Cirilli C, et al. Favourable ten-year overall survival in a Caucasian population with high probability of hereditary breast cancer. BMC Cancer 2010;10:90. [Crossref] [PubMed]
- Rennert G, Bisland-Naggan S, Barnett-Griness O, et al. Clinical outcomes of breast cancer in carriers of BRCA1 and BRCA2 mutations. N Engl J Med 2007;357:115-23. [Crossref] [PubMed]
- Lee LJ, Alexander B, Schnitt SJ, et al. Clinical outcome of triple negative breast cancer in BRCA1 mutation carriers and noncarriers. Cancer 2011;117:3093-100. [Crossref] [PubMed]
- Schmidt MK, Tollenaar RA, de Kemp SR, et al. Breast cancer survival and tumor characteristics in premenopausal women carrying the CHEK2*1100delC germline mutation. J Clin Oncol 2007;25:64-9. [Crossref] [PubMed]
- Liu Y, Xu Y, Ouyang T, et al. Association between CHEK2 H371Y mutation and response to neoadjuvant chemotherapy in women with breast cancer. BMC Cancer 2015;15:194. [Crossref] [PubMed]
- Cybulski C, Kluźniak W, Huzarski T, et al. Clinical outcomes in women with breast cancer and a PALB2 mutation: a prospective cohort analysis. Lancet Oncol 2015;16:638-44. [Crossref] [PubMed]
- Fan Z, Hu L, Ouyang T, et al. Germline mutation in DNA-repair genes is associated with poor survival in BRCA1/2-negative breast cancer patients. Cancer Sci 2019;110:3368-74. [Crossref] [PubMed]
- Brown JS, Carrigan B, Jackson SP, et al. Targeting DNA Repair in Cancer: Beyond PARP Inhibitors. Cancer Discov 2017;7:20. [Crossref] [PubMed]
- Fan QW, Cheng CK, Nicolaides TP, et al. A dual phosphoinositide-3-kinase alpha/mTOR inhibitor cooperates with blockade of epidermal growth factor receptor in PTEN-mutant glioma. Cancer Res 2007;67:7960-5. [Crossref] [PubMed]
- Yap TA, Yan L, Patnaik A, et al. First-in-man clinical trial of the oral pan-AKT inhibitor MK-2206 in patients with advanced solid tumors. J Clin Oncol 2011;29:4688-95. [Crossref] [PubMed]
- Lin J, Sampath D, Nannini MA, et al. Targeting activated Akt with GDC-0068, a novel selective Akt inhibitor that is efficacious in multiple tumor models. Clin Cancer Res 2013;19:1760-72. [Crossref] [PubMed]
- Kwok M, Davies N, Agathanggelou A, et al. Synthetic lethality in chronic lymphocytic leukaemia with DNA damage response defects by targeting the ATR pathway. Lancet 2015;385 Suppl 1:S58. [Crossref] [PubMed]
- Le DT, Uram JN, Wang H, et al. PD-1 Blockade in Tumors with Mismatch-Repair Deficiency. N Engl J Med 2015;372:2509-20. [Crossref] [PubMed]
- Rizvi NA, Hellmann MD, Snyder A, et al. Cancer immunology. Mutational landscape determines sensitivity to PD-1 blockade in non-small cell lung cancer. Science 2015;348:124-8. [Crossref] [PubMed]
- Lau T, Chan E, Callow M, et al. A novel tankyrase small-molecule inhibitor suppresses APC mutation-driven colorectal tumor growth. Cancer Res 2013;73:3132-44. [Crossref] [PubMed]


