
Differential expression of genes related to calcineurin and mTOR signaling and regulatory miRNAs in peripheral blood from kidney recipients under tacrolimus-based therapy
Introduction
Chronic kidney disease (CKD) is a major global health problem characterized by the gradual and irreversible loss of kidney function (1). The terminal stage, also known as end-stage renal disease (ESRD), requires renal replacement therapy—dialysis or transplantation—for the maintenance of life (2).
Kidney transplant provides better quality of life and longer survival, however to achieve good long-term outcomes, a life-long immunosuppression is required to prevent the allograft rejection (3). The immunosuppressive protocols usually include calcineurin inhibitors (e.g., tacrolimus) and/or mechanistic target of rapamycin (mTOR) inhibitors (e.g., everolimus), anti-proliferative drug (e.g., mycophenolate sodium) and corticosteroids (4,5).
Calcineurin, a key enzyme in T-cell activation, is a serine/threonine phosphatase composed of catalytic A (Isoforms encoded by PPP3CA, PPP3CB and PPP3CC) and regulatory B (Isoforms encoded by PPP3R1 and PPP3R2) subunits (6). Tacrolimus or FK506 is able to block calcineurin signaling pathway and the production of pro-inflammatory cytokines, e.g., interleukin-2 (IL-2), and thereby attenuates cytokine receptor-dependent mTOR activation and lymphocyte proliferation (7). Tacrolimus immunosuppressive activity is mediated by the complex formed with FK506-binding proteins (FKBPs), best described for FKBP12 (encoded by FKBP1A) (8). It is known that the tacrolimus also binds with different affinities to FKBP12.6 (FKBP1B), the closest homolog of FKBP12, FKBP13 (FKBP2) and FKBP51 (FKBP5) (8,9).
Tacrolimus has high inter-individual variability of blood concentration, which requires therapeutic drug monitoring to maintain drug efficacy and safety. Genetic and non-genetic factors, such as age, gender, kidney and liver function, comorbidities, drug interactions, and others, may contribute to the variability of tacrolimus response (10,11).
Variants in pharmacokinetic-related genes, such as CYP3A4, CYP3A5, ABCB1 and others, have been shown to influence the response to tacrolimus in kidney recipients from Brazil and other sample populations (12-17). However, polymorphisms in pharmacodynamics-related genes have been less explored in patients taking tacrolimus-based treatment (18,19). Our group investigated variants in genes related to calcineurin and mTOR signaling pathways and observed that MTOR, FKBP1A, FKBP2 and FOXP3 are associated with long-term impaired renal function, increased risk of acute rejection, and adverse events in kidney recipients treated with tacrolimus and everolimus-based immunosuppression (20,21).
Epigenetics factors, such as DNA methylation and histone acetylation, modulate the expression of genes and they have been suggested to influence allograft dysfunction and response to immunosuppressive therapies (22,23).
MicroRNAs (miRNAs) are short noncoding RNAs that bind the 3'UTR region of target mRNAs silencing gene expression. miRNAs regulate many biological processes including cell proliferation and differentiation (24). Differential expression of miRNAs has been implicated in CKD, and impaired graft function and rejection in kidney transplantation (25,26). Therefore, expression of miRNAs and target mRNAs in genes involved in pharmacodynamics may also contribute to the variability to immunosuppressive drugs response in kidney transplantation.
This study explored the expression of genes related to the calcineurin and mTOR signaling pathway (PPP3CA, PPP3CB, MTOR, FKBP1A, FKBP1B and FKBP5) and the miRNAs targeting PPP3CA (miR-30a, miR-145), PPP3CB (miR-10b), MTOR (miR-99a, miR-100), and FKBP1A (miR-103a) in the peripheral blood of kidney recipients in the initial phase of tacrolimus-based therapy. The influence of PPP3CA and MTOR polymorphisms on mRNA expression was also investigated.
We present the following article in accordance with the STROBE reporting checklist (available at http://dx.doi.org/10.21037/atm-20-1757).
Methods
Patients and study protocol
Thirty-six recipients of first kidney transplantation were selected at the at the Hospital do Rim, UNIFESP, Sao Paulo, Brazil. The allograft was ABO-compatible with a CDC-negative cross-match and a peak panel reactive antibody lower than 30%. The main exclusion criteria were significant hematologic or severe metabolic abnormalities; focal and segmental glomerulosclerosis; membrane proliferative glomerulonephritis; active infection or positivity for hepatitis B or C or human immunodeficiency viruses; and previous history of malignancy.
This investigation is part of a prospective core study (27). The individuals included in this study were selected based on the criteria of availability and quality of the biological samples for RNA studies. The study protocol was approved by the local ethics committees (UNIFESP protocol # 054/2008; FCF/USP protocol # 517), Sao Paulo, Brazil, and conducted according to good clinical practices and the Declaration of Helsinki guidelines (as revised in 2013). All subjects signed an approved written informed consent before enrollment.
Kidney recipients were treated with tacrolimus (0.1–0.2 mg/kg/day within 48 h post-transplant, and the doses were adjusted to maintain blood concentrations 8 to 15 ng/mL in the first month and afterwards 5 to 15 ng/mL). Treatment included enteric-coated mycophenolate sodium (1,440 mg/day) and prednisone (0.5 mg/kg initial dose, maximum of 30 mg, tapered to 20 mg at day 7, with a subsequent reduction of 5 mg/week to reach the dose of 5 mg/day by day 45). Some patients also received induction with basiliximab and methylprednisolone treatment before graft reperfusion in accordance with local practice.
Clinical data and laboratory tests
Blood and urine samples were obtained in pre-defined times (before and one and three months after kidney transplantation) for laboratory tests and tacrolimus monitoring. Blood samples were also obtained in PAXgene™ blood RNA tubes (Qiagen GmbH, Hilden, Germany) for RNA extraction.
Creatinine, urea, glucose, total cholesterol, HDL cholesterol and triglycerides were measured by automated methods. The eGFR was calculated using the four-variable MDRD formula (28). Three months after the transplant, plasma levels of some cytokines such as interferon-γ (IFN-γ), interleukin (IL)-2, IL-4, IL-10 and IL-17 were measured by immunoassays Milliplex™ MAP for Luminex® xMAP™ technology (Merck Millipore, Massachusetts, USA).
Tacrolimus blood concentration was measured by an immunoassay using the Abbott Architect analyzer (Abbott Diagnostics, Illinois, USA). The tacrolimus C/D [ng/(mL·mg)] was calculated for each patient.
The biopsy-confirmed acute rejection (BCAR) was performed by the local pathologist according to the Banff 2003 classification (29). The delayed graft function (DGF) was considered to be the dialysis performed in 7 days after transplantation.
RNA extraction and quantification
Total RNA was extracted from peripheral blood using the extraction system PAXgene™ Blood RNA Kit according to the manufacturer’s protocol (Qiagen GmbH, Hilden, Germany). RNA concentration and purity were analyzed using NanoDrop ND-1000 photometer and Invitrogen Qubit® 2.0 fluorometer (Thermo Fisher Scientific Inc. Foster City, USA). RNA integrity was assessed by micro-capillary electrophoresis using RNA 6000 Nano Kit and Agilent Bioanalyzer 2100 (Agilent Technologies Inc., Santa Clara, USA). Samples with RNA integrity number (RIN) lower than 5.0 were not included. RNA samples were stored at −80 °C.
Analysis of mRNA expression by real time PCR
The cDNA was synthesized from total RNA using random primers and Invitrogen SuperScript II reverse transcriptase (Thermo Fisher Scientific Inc., Foster City, CA, USA) following the manufacturer protocol.
mRNA expression was measured by quantitative real-time PCR (qPCR) using predesigned TaqMan® Gene expression assays (PPP3CA Hs00174223_m1, PPP3CB Hs00917458_m1, MTOR Hs00234508_m1, FKBP1A Hs00356621_g1, FKBP1B Hs00997682_m1, and FKBP5 Hs01561006_m) and Applied Biosystems 7500 Fast Real-Time PCR system (Thermo Fisher Scientific Inc., Foster City, USA). All cDNA samples were tested in duplicate.
Six reference genes were tested (UBC, GAPD, B2M, HPRTI, SDHA and HMBS) and analyzed using the GeNorm software (http://medgen.ugent.be/genorm). UBC and B2M were the most stable reference genes in the experimental conditions, and used further to normalize mRNA expression of the studied genes. The differential mRNA expression was calculated using the formula 2-ΔCT.
Analysis of miRNA expression by PCR array
Ingenuity Pathway Analysis (IPA) software (Qiagen, www.qiagenbioinformatics.com) was used to select miRNAs that target the mRNAs of genes involved in the mTOR and calcineurin pathways with validated results in human leukocytes experiments. Six miRNAs targeting PPP3CA (hsa-miR-30a-5p, hsa-miR-145-5p), PPP3CB (hsa-miR-10b-5p), MTOR (hsa-miR-99a-5p, hsa-miR-100-5p), and FKBP1A (hsa-miR-103a-3p) were selected. The predictive regulatory miRNAs were confirmed using the miRTarBase (http://mirtarbase.mbc.nctu.edu.tw) and TargetScan (www.targetscan.org) databases.
miRNAs expression of was measured using miScript miRNA PCR Array (Qiagen, Redwood City, USA) using Applied Biosystems 7500 Fast Real-Time PCR equipment (Thermo Fisher Scientific Inc., Foster City, USA). Briefly, cDNA was produced from total RNA by reverse transcription reaction using miScript II RT kit and amplified using QuantiTect SYBR Green PCR kit (Qiagen). The raw data were analyzed using the miScript miRNA PCR Array Web-based software (Qiagen). The expression values were normalized with four endogenous snRNAs: SNORD61, SNORD68, SNORD95, and RNU6B/RNU6-2. The differential miRNA expression was calculated using the formula 2−ΔCT.
Analysis of gene polymorphisms
The polymorphisms PPP3CA rs3730251 (c.249G>A) and MTOR rs1135172 (c.1437T>C), rs1064261 (c.2997C>T) and rs1057079 (c.4731G>A) were analyzed by qPCR using pre-designed TaqMan SNP genotyping assays (Thermo Fisher Scientific Inc., Foster City, USA). Details about DNA extraction, genotyping assays, and qPCR procedures and quality control were previously described (21).
Statistical analyses
Categorical variables are shown as percentage and were compared by chi-square or Fisher’s exact test. Continuous variables are shown as median and interquartile range and were compared by Mann-Whitney U test or Kruskal-Wallis test and Dunn’s test for multiple comparisons. Paired data of metabolic profile and blood concentration of immunosuppressive drugs were analyzed by paired t-test or Friedman Repeated Measures ANOVA on Ranks and Dunn’s test for multiple comparisons.
The correlation of laboratory variables (eGFR, serum creatinine, IFN-γ, IL-2, IL-4 and IL-17 plasma levels) with mRNA and miRNA expression, at 3-month post-transplant, was tested using Spearman correlation test.
Statistical analyses were carried out using SPSS for Windows version 22 (SPSS Inc., Illinois, USA) and GraphPad Prism® version 5.00 for Windows (GraphPad Software Inc., California, USA). Significance level was set at P<0.05.
Results
Characteristics of the kidney recipients
Demographic and clinical characteristics of the kidney transplant recipients are shown in the Table 1. Subjects were mainly men (66.7%) and Caucasians (55.6%), with a median age of 48.0 years old. Hypertension (11.1%) and diabetes (22.2%) were the main known causes of the ESRD. The donors were also mainly men (55.6%) and Caucasian (66.7%), with a median age of 44.0 years old. Transplantation of kidney from deceased donors was 47.2% in this study. The median cold ischemia time was 19.6 h, for deceased donors only. The cumulative incidence of graft DGF was 19.4%. During the three-month of follow up, six patients (16.7%) had one episode of treated BCAR. The subjects who developed or not BCAR had similar demographic and clinical characteristics (P>0.05, Table S1).

Full table
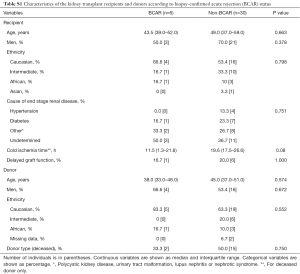
Full table
Median tacrolimus dose decreased from the day 7 post-transplant (11.5 mg/day) to the first and third month (4.5 and 3.0 mg/day, respectively, P<0.001) (Table 2). The blood concentration of tacrolimus reduced from 10.1 ng/mL in the day 7 to 5.4 ng/mL in the third month (P<0.001). The concentration for dose-administered (C/D) of tacrolimus increased from the day 7 [0.8 ng/(mL·mg)] to the first and third months post-transplant [1.7 ng/(mL·mg), P<0.001]. The mycophenolate sodium doses did not change significantly with time (1,440 mg/day) but prednisone doses were reduced, as expected (30.0 to 5.0 mg/day from day 7 to month 3, P<0.001).
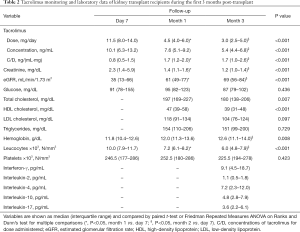
Full table
Kidney function and other laboratory variables of the kidney recipients improved from day 7 to month 3, as expected (Table 2). Serum creatinine and number of leucocytes were reduced, whereas the estimated glomerular filtration rate (eGFR) and hemoglobin were increased in the first and/or third month of therapy (P<0.05). Compared to month 1, total cholesterol and high-density lipoprotein (HDL) cholesterol levels were lower in the third month (P<0.05). Descriptive data of cytokines plasma levels at the third month post-transplant are also shown in the Table 2.
Circulating mRNAs differentially expressed
mRNA expression of PPP3CA and MTOR in peripheral blood was reduced after one (PPP3CA 28%) and three (PPP3CA 41% and MTOR 25%) months of immunosuppressive therapy, compared to pre-transplant (PreTx) (P<0.05) (Figure 1). The expression of the genes encoding calcineurin beta (PPP3CB) and FK506-binding proteins (FKBP1A, FKBP1B and FKBP5) did not change during treatment (P>0.05).
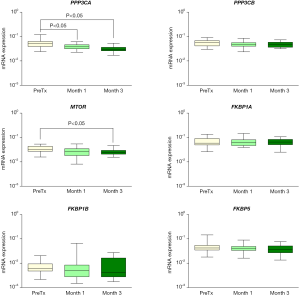
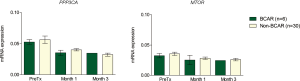
As an exploratory analysis, the deregulation of PPP3CA and MTOR expression was independent on the BCAR condition (P>0.05, Figure S1). However, at the first month, BCAR patients showed higher FKBP1B expression than non-BCAR group (median normalized mRNA expression of 0.009 versus 0.004, respectively, P=0.026).
Circulating miRNAs differentially expressed
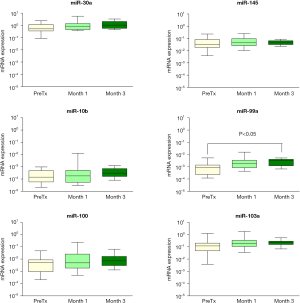
Expression of miRNAs targeting PPP3CA (miR-30a, miR-145), PPP3CB (miR-10b) MTOR (miR-99a, miR-100), and FKBP1A (miR-103a) was analyzed in peripheral blood of kidney recipients. The results of this analysis showed that miR-99a levels were about two-fold increase in the third month of therapy compared to PreTx (P<0.05), whereas the miR-100, miR-145, miR-30a, miR-10b, and miR-103a expression remained unchanged (P>0.05) (Figure 2). The expression of the miRNAs did not differ between BCAR and non-BCAR groups (P>0.05) (data not shown).
Influence of PPP3CA and MTOR variants on mRNA expression
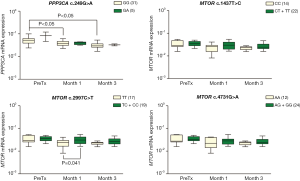
As shown in the Figure 3, PPP3CA expression was downregulated at 1 and 3 months post-transplant in patients carrying PPP3CA c.249GG genotype (P<0.05) but not in GA genotype carriers (P>0.05, Figure 3). However, this result should be analyzed with caution, because only five kidney recipients were carriers of PPP3CA c.249GA genotype in this sample. Analysis of MTOR c.2997C>T showed that patients carrying the TT genotype had lower MTOR mRNA expression than the C allele carriers (TC+CC genotype) at month 1 post-transplant (median 0.023 versus 0.031, respectively, P=0.041) (Figure 3).
Correlation of mRNA or miRNA expression with laboratory variables
The main results of the correlation analyses at the 3rd month post-transplant are shown in Table 3. Positive correlations were found between eGFR and miR-30a, and serum creatinine and FKBP1A mRNA (P<0.05). Moreover, plasma IFN-γ was positively correlated with PPP3CA mRNA, IL-2 with miR-10b and miR-100 expression, and IL-17 with PPP3CB mRNA (P<0.05). On the other hand, IL-4 levels were inversely correlated with miR-10b expression (P=0.009).

Full table
Discussion
This study investigated the expression of genes related to calcineurin and mTOR signaling pathways and their regulatory miRNAs in peripheral blood of kidney recipients. The association of MTOR and PPP3CA variants with variability in mRNA expression was also analyzed.
In the first three months post-transplant, the mRNA expression of PPP3CA and MTOR reduced in peripheral blood. PPP3CA encodes the A subunit of the calcineurin, a calcium and calmodulin-dependent phosphatase, which binds the immunophilin FKBP12 forming a complex that activates signal transduction and transcription of IL2 and other cytokines genes involved in T-cell proliferation and differentiation. Tacrolimus inhibits the formation of the complex calcineurin-FKBP12. MTOR encodes a serine/threonine kinase of the PI3K/AKT signaling pathway, which also binds FKBP12. Activation of this pathway triggers a series of intracellular processes resulting in cell growth, proliferation and differentiation (14).
Reduction of calcineurin activity in peripheral blood by tacrolimus-based therapy was observed previously in renal transplant recipients within the first 15 days of therapy (30). Calcineurin activity measured by NFAT1 translocation to the nuclei of peripheral blood mononuclear cells (PBMC) was also reduced by tacrolimus in patients with end-stage liver disease waiting for transplantation (31).
Downregulation of PPP3CA and MTOR mRNA may be caused by genetic or epigenetic mechanisms which can contribute to the immunosuppressive effect of tacrolimus on cell proliferation and differentiation (16,25). Moreover, other drugs contained in the immunosuppressive regimen may be also modulating this effect. In rats treated with mycophenolate mofetil, the hippocampal mRNA and protein levels of mTOR was found to be decreased (32).
The synonymous variant PPP3CA c.249G>A and the missense variant MTOR c.2997C>T were associated with variability of gene expression in peripheral blood of the kidney recipients. Besides their influence on gene expression, these polymorphisms may alter the interactions of tacrolimus with molecules within the calcineurin and mTOR signaling pathway. The role of these variants in the regulation of gene expression and in the interaction of these molecules with tacrolimus has yet to be investigated by functional studies.
In this study, FKBP1A, FKBP1B and FKBP5 mRNA expression has not changed in the early phase after transplantation. Interestingly FKBP1A mRNA expression was positively correlated with serum creatinine at the 3-month therapy. In addition, FKBP1B was upregulated in patients with acute rejection. These results are suggestive that increased mRNA levels of FKBP1A and FKBP1B are associated with to worse kidney allograft function and increased risk of acute rejection.
Expression of six miRNAs was also evaluated in peripheral blood of kidney recipients within three months after transplantation. miR-30a, miR-145, miR-10b and others miRNAs bind to the 3'UTR of the PPP3CA and PPP3CB mRNA regulating their expression post-transcriptionally. Previous studies reported that reduced expression of miR-30a was associated with acute rejection and poor prognosis of graft function in kidney transplantation (33,34). Downregulation of miR-145 in biopsy samples was shown to be associated with the risk for acute kidney transplant rejection (35). miR-145 was also shown to be downregulated in blood cells of kidney recipients with interstitial fibrosis/tubular atrophy, antibody-mediated graft rejection, and reduced allograft survival (36).
The expression of mir-30a and miR-145 in peripheral blood remained stable and it was not associated with BCAR within the 3-first months of treatment. These miRNAs are likely to be more informative as biomarkers for long-term allograft rejection. Interestingly, miR-30a expression was positively correlated with eGFR values, which is suggestive that increased levels of this miRNA in peripheral blood maybe associated with better allograft renal function at early stage post-transplant.
miR-10b is considered a potentially oncogenic miRNA and increased expression of miR-10b was reported in oral, pancreatic and other neoplasms (37,38). Urinary expression of miR-10b and miR-210 was reported to be downregulated in renal allograft recipients with acute rejection (39,40). However, the expression of miR-10b or the target PPP3CB mRNA was not altered by exposure to immunosuppressive therapy or BCAR in this sample.
miR-99a, which targets MTOR mRNA, was upregulated in this study. This result is consistent with the downregulation of the MTOR expression at 3-month post-transplant. It has been reported that miR-99a and miR-100 have inhibitory effects on proliferation of breast and cervical cancer cells and on bladder carcinogenesis by targeting mTOR signaling pathway (41-43). Overexpression of miR-99a and miR-125b was shown to downregulate the activation and cytotoxicity of human circulating γδ T cells, which express T-cell receptors composed of γ and δ chains (44). Despite of small subset of T cells in peripheral blood, the γδ T cells display broad functional abilities and constitute an active and dynamic component of the solid organ transplant (45).
Expression of miR-99a, miR-100 and other miRNAs in serum was reported to be upregulated in kidney recipients with acute rejection compared to patients with continuous stable kidney function (46). Despite of the positive correlation between miRNA-99 and miR-100 expression and plasma IL-2, which has been associated with immunosuppressive monitoring and response in kidney recipients (47), the deregulation of these miRNAs was not associated with acute rejection in this sample.
Expression of miR-103a in peripheral blood, as its target FKBP1A mRNA, did not differ during the initial 3-month of the immunosuppressive therapy. Other studies have indicated that increased miR-103a expression was associated with suppression of cell proliferation, migration, and invasion in gastric cancer and malignant mesothelioma (48,49). Expression of miR-103a and other four miRNAs in blood cells were downregulated in kidney recipients with severe T cell-mediated vascular rejection (50).
The expression of MTOR and PPP3CA mRNA can also be modulated by other miRNAs during the early phase of immunosuppressive therapy. Therefore, further studies with larger samples and wider range miRNAs are necessary to investigate the modulators of the genes involved in the calcineurin and mTOR signaling pathways in kidney transplant models.
The main limitations of this study were the number of the highly selected kidney transplant recipients with a low immunological risk for rejection and the time of follow up. However, this work presents innovative data on the pharmacogenomics and epigenomics of kidney transplantation, contributing to the knowledge of possible mechanisms capable of influencing the therapeutic response in renal transplant patients.
In conclusion, the expression of MTOR and PPP3CA mRNA is downregulated and miR-99a is upregulated in peripheral blood of kidney recipients in the early phase of tacrolimus-based therapy, and PPP3CA and MTOR variants influenced mRNA expression. These molecules may have potential application as biomarker candidates for pharmacotherapy follow up.
Acknowledgments
The authors thank Cristina M. Fajardo, Antony B.C. Salazar and Nagila Oliveira for technical support and assistance in patient selection and data collection. VB was a recipient of fellowship from CAPES, Brazil. MHH and RDCH are recipients of fellowships from CNPq, Brazil. FDVG was a recipient of fellowship from FAPESP, Brazil.
Funding: This study was funded by Sao Paulo Research Foundation (FAPESP, grants #2011/10039-6 and #2016/13118-8), Sao Paulo, Brazil. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Footnote
Reporting Checklist: The authors have completed the STROBE reporting checklist. Available at http://dx.doi.org/10.21037/atm-20-1757
Data Sharing Statement: Available at http://dx.doi.org/10.21037/atm-20-1757
Peer Review File: Available at http://dx.doi.org/10.21037/atm-20-1757
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/atm-20-1757). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. The study conducted according to good clinical practices and the Declaration of Helsinki guidelines (as revised in 2013). The study protocol was approved by the ethics committees of the Federal University of Sao Paulo (UNIFESP, # 054/2008) and School of Pharmaceutical Sciences of the University of Sao Paulo (FCF/USP, # 517), Sao Paulo, Brazil. Written informed consent was obtained from each participant for the collection of clinical data.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Webster AC, Nagler EV, Morton RL, et al. Chronic Kidney Disease. Lancet 2017;389:1238-52. [Crossref] [PubMed]
- Robinson BM, Akizawa T, Jager KJ, et al. Factors affecting outcomes in patients reaching end-stage kidney disease worldwide: differences in access to renal replacement therapy, modality use, and haemodialysis practices. Lancet 2016;388:294-306. [Crossref] [PubMed]
- Carminatti M, Tedesco-Silva H, Silva Fernandes NM, et al. Chronic kidney disease progression in kidney transplant recipients: A focus on traditional risk factors. Nephrology 2019;24:141-7. [Crossref] [PubMed]
- Lentine KL, Kasiske BL, Levey AS, et al. Summary of Kidney Disease: Improving Global Outcomes (KDIGO) Clinical Practice Guideline on the Evaluation and Care of Living Kidney Donors. Transplantation 2017;101:1783-92. [Crossref] [PubMed]
- Wong TC, Lo CM, Fung JYY. Emerging drugs for prevention of T-cell mediated rejection in liver and kidney transplantation. Expert Opin Emerg Drugs 2017;22:123-36. [Crossref] [PubMed]
- Rusnak F, Mertz P. Calcineurin: Form and function. Physiol Rev 2000;80:1483-521. [Crossref] [PubMed]
- Navarro-Villarán E, Tinoco J, Jiménez G, et al. Differential antitumoral properties and renal-associated tissue damage induced by tacrolimus and mammalian target of rapamycin inhibitors in hepatocarcinoma: In vitro and in vivo studies. PLoS One 2016;11:e0160979. [Crossref] [PubMed]
- Kolos JM, Voll AM, Bauder M, et al. FKBP Ligands—Where We Are and Where to Go? Front Pharmacol 2018;9:1425. [Crossref] [PubMed]
- Tong M, Jiang Y. FK506-Binding Proteins and Their Diverse Functions. Curr Mol Pharmacol 2015;9:48-65. [Crossref] [PubMed]
- Andrews LM, Li Y, De Winter BCM, et al. Pharmacokinetic considerations related to therapeutic drug monitoring of tacrolimus in kidney transplant patients. Expert Opin Drug Metab Toxicol 2017;13:1225-36. [Crossref] [PubMed]
- Wang J, Li K, Zhang X, et al. The correlation between the expression of genes involved in drug metabolism and the blood level of tacrolimus in liver transplant receipts. Sci Rep 2017;7:3429. [Crossref] [PubMed]
- Cusinato DAC, Lacchini R, Romao EA, et al. Relationship of CYP3A5 genotype and ABCB1 diplotype to tacrolimus disposition in Brazilian kidney transplant patients. Br J Clin Pharmacol 2014;78:364-72. [Crossref] [PubMed]
- Genvigir FDV, Salgado PC, Felipe CR, et al. Influence of the CYP3A4/5 genetic score and ABCB1 polymorphisms on tacrolimus exposure and renal function in Brazilian kidney transplant patients. Pharmacogenet Genomics 2016;26:462-72. [Crossref] [PubMed]
- Genvigir FDV, Nishikawa AM, Felipe CR, et al. Influence of ABCC2, CYP2C8, and CYP2J2 Polymorphisms on Tacrolimus and Mycophenolate Sodium-Based Treatment in Brazilian Kidney Transplant Recipients. Pharmacotherapy 2017;37:535-45. [Crossref] [PubMed]
- Yu M, Liu M, Zhang W, et al. Pharmacokinetics, Pharmacodynamics and Pharmacogenetics of Tacrolimus in Kidney Transplantation. Curr Drug Metab 2018;19:513-22. [Crossref] [PubMed]
- Zhang X, Lin G, Tan L, et al. Current progress of tacrolimus dosing in solid organ transplant recipients: Pharmacogenetic considerations. Biomed Pharmacother 2018;102:107-14. [Crossref] [PubMed]
- Genvigir FDV, Campos-Salazar AB, Felipe CR, et al. CYP3A5*3 and CYP2C8*3 variants influence exposure and clinical outcomes of tacrolimus-based therapy. Pharmacogenomics 2020;21:7-21. [Crossref] [PubMed]
- Wu Z, Xu Q, Qiu X, et al. FOXP3 rs3761548 polymorphism is associated with tacrolimus-induced acute nephrotoxicity in renal transplant patients. Eur J Clin Pharmacol 2017;73:39-47. [Crossref] [PubMed]
- Wu Z, Xu Q, Qiu X, et al. FKBP1A rs6041749 polymorphism is associated with allograft function in renal transplant patients. Eur J Clin Pharmacol 2019;75:33-40. [Crossref] [PubMed]
- Salgado PC, Genvigir FDV, Felipe CR, et al. Association of the PPP3CA c.249G>A variant with clinical outcomes of tacrolimus-based therapy in kidney transplant recipients. Pharmgenomics Pers Med 2017;10:101-6. [Crossref] [PubMed]
- Campos-Salazar AB, Genvigir FDV, Felipe CR, et al. Polymorphisms in mTOR and calcineurin signaling pathways are associated with long-term clinical outcomes in kidney transplant recipients. Front Pharmacol 2018;9:1296. [Crossref] [PubMed]
- Mas VR, Le TH, Maluf DG. Epigenetics in kidney transplantation: Current evidence, predictions, and future research directions. Transplantation 2016;100:23-38. [Crossref] [PubMed]
- Zhou M, Hara H, Dai Y, et al. Circulating Organ-Specific MicroRNAs Serve as Biomarkers in Organ-Specific Diseases: Implications for Organ Allo- and Xeno-Transplantation. Int J Mol Sci 2016;17:E1232. [Crossref] [PubMed]
- Saliminejad K, Khorram Khorshid HR, Soleymani Fard S, et al. An overview of microRNAs: Biology, functions, therapeutics, and analysis methods. J Cell Physiol 2019;234:5451-65. [Crossref] [PubMed]
- Janszky N, Süsal C. Circulating and urinary microRNAs as possible biomarkers in kidney transplantation. Transplant Rev (Orlando) 2018;32:110-8. [Crossref] [PubMed]
- Metzinger-Le Meuth V, Fourdinier O, Charnaux N, et al. The expanding roles of microRNAs in kidney pathophysiology. Nephrol Dial Transplant 2019;34:7-15. [Crossref] [PubMed]
- Silva HT, Felipe CR, Garcia VD, et al. Planned randomized conversion from tacrolimus to sirolimus-based immunosuppressive regimen in de novo kidney transplant recipients. Am J Transplant 2013;13:3155-63. [Crossref] [PubMed]
- Levey AS, Bosch JP, Lewis JB, et al. A more accurate method to estimate glomerular filtration rate from serum creatinine: A new prediction equation. Ann Intern Med 1999;130:461-70. [Crossref] [PubMed]
- Racusen LC, Colvin RB, Solez K, et al. Antibody-mediated rejection criteria - an addition to the Banff 97 classification of renal allograft rejection. Am J Transplant 2003;3:708-14. [Crossref] [PubMed]
- Mortensen DM, Koefoed-Nielsen PB, Jørgensen KA. Calcineurin Activity in Tacrolimus-Treated Renal Transplant Patients Early After and 5 Years After Transplantation. Transplant Proc 2006;38:2651-3. [Crossref] [PubMed]
- Noceti O, Pouché L, Esperó PN, et al. Activity of the calcineurin pathway in patients on the liver transplantation waiting list: Factors of variability and response to tacrolimus inhibition. Clin Chem 2017;63:1734-44. [Crossref] [PubMed]
- Mazumder AG, Patial V, Singh D. Mycophenolate mofetil contributes to downregulation of the hippocampal interleukin type 2 and 1β mediated PI3K/AKT/mTOR pathway hyperactivation and attenuates neurobehavioral comorbidities in a rat model of temporal lobe epilepsy. Brain Behav Immun 2019;75:84-93. [Crossref] [PubMed]
- Ben-Dov IZ, Muthukumar T, Morozov P, et al. MicroRNA sequence profiles of human kidney allografts with or without tubulointerstitial fibrosis. Transplantation 2012;94:1086-94. [Crossref] [PubMed]
- Amrouche L, Rabant M, Anglicheau D. MicroRNAs as biomarkers of graft outcome. Transplant Rev (Orlando) 2014;28:111-8. [Crossref] [PubMed]
- Oghumu S, Bracewell A, Nori U, et al. Acute pyelonephritis in renal allografts-a new role for MicroRNAs? Transplantation 2014;97:559-68. [Crossref] [PubMed]
- Matz M, Heinrich F, Lorkowski C, et al. MicroRNA regulation in blood cells of renal transplanted patients with interstitial fibrosis/tubular atrophy and antibody-mediated rejection. PLoS One 2018;13:e0201925. [Crossref] [PubMed]
- Lu YC, Chen YJ, Wang HM, et al. Oncogenic function and early detection potential of miRNA-10b in oral cancer as identified by microRNA profiling. Cancer Prev Res (Phila) 2012;5:665-74. [Crossref] [PubMed]
- Ouyang H, Gore J, Deitz S, et al. microRNA-10b enhances pancreatic cancer cell invasion by suppressing TIP30 expression and promoting EGF and TGF-β actions. Oncogene 2014;33:4664-74. [Crossref] [PubMed]
- Liu X, Dong C, Jiang Z, et al. MicroRNA-10b downregulation mediates acute rejection of renal allografts by derepressing BCL2L11. Exp Cell Res 2015;333:155-63. [Crossref] [PubMed]
- Lorenzen JM, Volkmann I, Fiedler J, et al. Urinary miR-210 as a mediator of acute T-cell mediated rejection in renal allograft recipients. Am J Transplant 2011;11:2221-7. [Crossref] [PubMed]
- Hu Y, Zhu Q, Tang L. MiR-99a antitumor activity in human breast cancer cells through targeting of mTOR expression. PLoS One 2014;9:e92099. [Crossref] [PubMed]
- Wang L, Chang L, Li Z, et al. MiR-99a and -99b inhibit cervical cancer cell proliferation and invasion by targeting mTOR signaling pathway. Med Oncol 2014;31:934. [Crossref] [PubMed]
- Xu C, Zeng Q, Xu W, et al. miRNA-100 inhibits human bladder urothelial carcinogenesis by directly targeting mTOR. Mol Cancer Ther 2013;12:207-19. [Crossref] [PubMed]
- Zhu Y, Zhang S, Li Z, et al. miR-125b-5p and miR-99a-5p downregulate human γδ T-cell activation and cytotoxicity. Cell Mol Immunol 2019;16:112-25. [Crossref] [PubMed]
- Sullivan LC, Shaw EM, Stankovic S, et al. The complex existence of γδ T cells following transplantation: the good, the bad and the simply confusing. Clin Transl Immunology 2019;8:e1078. [Crossref] [PubMed]
- Tao J, Yang X, Han Z, et al. Serum MicroRNA-99a Helps Detect Acute Rejection in Renal Transplantation. Transplant Proc 2015;47:1683-7. [Crossref] [PubMed]
- Daniel V, Naujokat C, Sadeghi M, et al. Association of circulating interleukin (IL)-12- and IL-10-producing dendritic cells with time posttransplant, dose of immunosuppression, and plasma cytokines in renal-transplant recipients. Transplantation 2005;79:1498-506. [Crossref] [PubMed]
- Weber DG, Casjens S, Johnen G, et al. Combination of MIR-103a-3p and mesothelin improves the biomarker performance of malignant mesothelioma diagnosis. PLoS One 2014;9:e114483. [Crossref] [PubMed]
- Liang J, Liu X, Xue H, et al. MicroRNA-103a inhibits gastric cancer cell proliferation, migration and invasion by targeting c-Myb. Cell Prolif 2015;48:78-85. [Crossref] [PubMed]
- Matz M, Fabritius K, Lorkowski C, et al. Identification of T cell-mediated vascular rejection after kidney transplantation by the combined measurement of 5 specific MicroRNAs in blood. Transplantation 2016;100:898-907. [Crossref] [PubMed]


