
Identification of molecular features correlating with tumor immunity in gastric cancer by multi-omics data analysis
Introduction
Gastric cancer (GC) is the second leading cause of cancer deaths in the world and particularly prevails in Asian countries (1). Traditional therapeutic strategies (surgery, chemotherapy, radiotherapy, and targeted therapy) often have limited efficacy against advanced GCs (2). Recently, with the advance of immunotherapy in treating various advanced malignancies including the cancers with mismatch repair deficiency (3-7), immunotherapy is becoming a potentially promising treatment strategy for GC, as evidenced by that pembrolizumab and nivolumab (immune checkpoint inhibitors) are being clinically used to treat microsatellite-instable gastrointestinal (GI) cancers (7). However, currently, immunotherapeutic strategies are beneficial to only a subset of cancer patients, with a large proportion of patients failing to respond to such therapies (8). To this end, certain genetic or genomic features have been identified that are associated with cancer immunotherapy response, such as PD-L1 expression (9), tumor mutation burden (TMB) (10), deficient mismatch repair (dMMR) (11), and TP53 mutations (12). In addition, it has been shown that the tumor immune microenvironment (TIME) plays a crucial role in determining the cancer immunotherapy response (13). The “hot” tumors with dense T cell infiltration in TIME are more likely to respond to immunotherapy than the “cold” tumors lack of immune cell infiltration (14). To improve the cancer therapy response, the combination of different immunotherapeutic strategies and the combination of immunotherapy with traditional therapeutic methods have been widely investigated (15-19).
Benefit from the rapid advances in genomics technologies, many cancer genomics data are publicly available, such as TCGA (https://portal.gdc.cancer.gov/) and ICGC (https://dcc.icgc.org/). Several studies have explored tumor immunity in GC using publicly available cancer genomic datasets (20-22). Zeng et al. identified three GC subtypes based on the tumor microenvironment (TME) cell infiltration patterns (20). Our previous study revealed that the TP53 mutation contributed to an immune-suppressive TME in GC (21). Park et al. classified GC patients into three subtypes with significant prognostic differences and found that the GC subtype with the best clinical outcome was enriched in the PD-L1 signaling pathway (22).
Despite these prior explorations (20-22), the identification of genetic and genomic features significantly correlating with GC immunity and immunotherapy response is worth further investigation. For example, although the Epstein-Barr virus positive (EBV+) and the microsatellite instability (MSI) are associated with a high GC immunity and an active immunotherapy response, many EBV- and microsatellite-stable (MSS) GCs equally exhibit highly enriched immune signatures and favorable immunotherapy response (21,22). This suggests that there are other factors correlating with GC immunity and immunotherapy response. In this study, we aimed to explore the associations of various molecular features with GC immunity based on multi-omics datasets from the TCGA GC cohort (23) and the Asian Cancer Research Group (ACRG) GC cohort (24). These molecular features included genes, microRNAs (miRNAs), long non-coding RNAs (lncRNAs), proteins, and pathways. We identified the molecular features that were significantly associated with GC immunity and immunotherapy response. These molecular features may have clinical implications for GC immunotherapy. We present the following article in accordance with the MDAR reporting checklist (available at http://dx.doi.org/10.21037/atm-20-922).
Methods
Datasets
We obtained multi-omics (gene somatic mutation, gene expression, protein expression, miRNA expression, and lncRNA expression) and clinical data for the TCGA GC cohort from the GDC data portal (https://portal.gdc.cancer.gov/). The multi-omics (gene somatic mutation and gene expression) and clinical data for the ACRG GC cohort were from the associated publication by ACRG (24). The gene somatic mutation and clinical data for a GI cancer cohort [Samstein cohort (25)] with immunotherapy were from the associated publication (25). In addition, we obtained cancer-associated pathways and their gene sets from KEGG (26).
Quantification of immune signature enrichment levels in GC samples
We analyzed three immune signatures, including CD8+ T cells, immune cytolytic activity (ICA), and PD-L1 expression. The enrichment level of an immune signature in a GC sample was quantified as the mean expression level of immune signature marker genes. The marker genes of three immune signatures included CD8A for CD8+ T cells, GZMA and PRF1 for ICA, and PD-L1 for PD-L1.
Associations of immune signature enrichment levels with various molecular features in GC
We identified the genes whose mutations were significantly associated with immune signature enrichment levels (ISELs) using the Mann-Whitney U test and the genes, proteins, miRNAs, and lncRNAs whose expression levels were significantly correlated with ISELs using the Pearson’s correlation test. We quantified the activity of a pathway in a GC sample by the single-sample gene-set enrichment analysis (ssGSEA) (27) score of the gene set in the pathway and identified the cancer-associated pathways whose activity was significantly associated with ISELs using the Spearman’s correlation test. The false discovery rate (FDR) (28) was used to adjust for multiple tests.
For each GC sample, we defined its TMB as the total somatic mutation count in the sample and its tumor aneuploidy level (TAL) as the tumor ploidy score calculated by ABSOLUTE (29). We divided all GC samples into the high-TMB/TAL (> median) group and the low-TMB/TAL (< median) group and compared their ISELs using the Mann-Whitney U test.
Gene-set enrichment analysis
We used GSEA (30) to identify the KEGG (26) pathways that were significantly associated with the genes having an important expression correlation with ISELs using a threshold of FDR <0.05.
Survival analysis
We compared the overall survival (OS) between gene-mutated and gene-wildtype cancers in the three cohorts (TCGA-GC, ACRG-GC, and Samstein). We used Kaplan-Meier survival curves to show the survival time differences and the log-rank test to assess the significance of survival time differences.
Results
Identification of genes whose mutations are associated with antitumor immune response in GC
Based on TCGA-GC cohort, we identified eight genes whose mutations were significantly associated with heightened CD8+ T cell infiltration levels in GC (Mann-Whitney U test, FDR <0.05) (Figure 1A). The eight genes included PIK3CA, ABCB4, SOGA2, ARID1B, CDH1, ABCA12, TENM2, and YLPM1. In contrast, TP53 mutations were associated with reduced CD8+ T cell infiltration levels in GC (Mann-Whitney U test, FDR =0.042). This is consistent with our previous study showing that TP53 mutations inhibited GC immunity (21). Moreover, we identified 106 genes whose mutations were associated with elevated immune cytolytic activity (ICA) in GC and one gene (TP53) whose mutations were associated with reduced ICA (Mann-Whitney U test, FDR <0.05) (https://cdn.amegroups.cn/static/application/3378c85657fe63fe5b598b068b3f0bbb/10.21037atm-20-922-1.pdf). The 106 genes included the aforementioned eight genes whose mutations were positively associated with CD8+ T cell infiltration levels in GC. Notably, some well-recognized cancer-associated genes were presented in the list of 106 genes, including tumor suppressor genes ARID1A, ATM, CDH1, FBXW7, and PTEN, oncogenes CIC, CREBBP, MLL, NUP98, PIK3CA, and RNF213, and protein kinase encoding genes ERBB3, MTOR, and WNK1. These results indicate that the mutations in a wide range of genes are associated with the enhanced antitumor immunity in GC. However, the mutation of TP53, one of the most important tumor suppressor genes, appears to suppress antitumor immunity in GC. The associations between the mutations of genes (ARID1A, PIK3CA, and TP53) and GC immunity were confirmed to be significant in ACRG-GC cohort (Mann-Whitney U test, FDR <0.05).
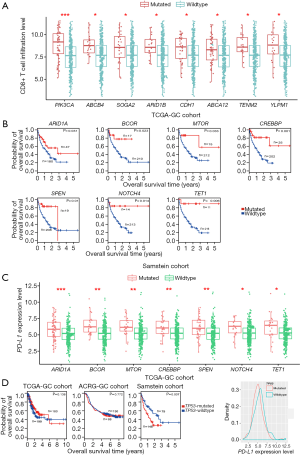
We identified 180 genes whose mutations were associated with elevated PD-L1 expression in GC (https://cdn.amegroups.cn/static/application/3378c85657fe63fe5b598b068b3f0bbb/10.21037atm-20-922-1.pdf). Interestingly, a large proportion (86%) of the genes whose mutations were associated with elevated ICA were contained in the set of 180 genes. This indicates that the mutations in many genes may result in increased antitumor immune response and enhanced protumor immunosuppression simultaneously.
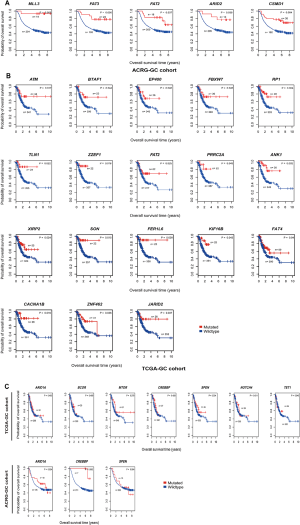
We further explored the associations between the mutations of these genes and OS in TCGA-GC, ACRG-GC, and Samstein cohorts. We found that the mutations of MLL3, FAT2, FAT3, ARID2, and CSMD1 were consistently associated with better OS in ACRG-GC cohort (log-rank test, P<0.05) (Figure S1A). In TCGA-GC cohort, the mutations of 18 genes were associated with better OS (Figure S1B). The positive associations between the mutations of these genes and OS could be attributed to the elevated antitumor immune activity in the GCs harboring these mutations. Interestingly, we found seven genes (ARID1A, BCOR, MTOR, CREBBP, SPEN, NOTCH4, and TET1) whose mutations were associated with better OS in Samstein cohort but did not show a significant correlation with OS in either of TCGA- and ACRG-GC cohorts (Figure 1B and Figure S1C). A possible explanation of this disparity could be that the GI cancer patients with the mutations of these genes were more responsive to anti-PD-1/PD-L1 immunotherapy than those without such mutations. This explanation was bolstered by the fact that the mutations of these genes were consistently associated with the elevated expression of PD-L1 in TCGA-GC cohort (Figure 1C). Interestingly, the TP53-mutated GI cancer patients had a significantly worse OS than the TP53-wildtype patients in Samstein cohort while the TP53 mutation had no a significant correlation with OS in either TCGA- or ACRG-GC cohort (Figure 1D). Again, this could be attributed to the more active response to anti-PD-1/PD-L1 immunotherapy in TP53-wildtype GI cancer patients since the TP53 mutation was associated with reduced PD-L1 expression in GC (Figure 1D).
Therefore, the identification of genes whose mutations were significantly associated with tumor immune signatures may provide predictive biomarkers for immunotherapy response in GC.
Identification of genes whose expression levels are associated with antitumor immune response in GC
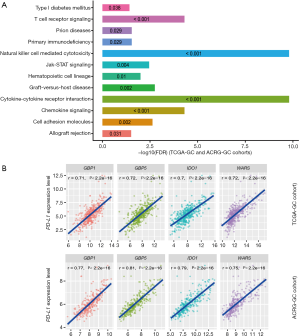
We found 136 and 123 genes whose expression levels exhibited a strong positive correlation with CD8+ T cell enrichment levels in TCGA-GC cohort and ACRG-GC cohort, respectively (Pearson correlation coefficient r>0.7) (https://cdn.amegroups.cn/static/application/4b0111ff02b66c45bc28328104e88994/10.21037atm-20-922-2.pdf). A total of 76 genes showed a strong positive expression correlation with CD8+ T cell enrichment levels in both cohorts (https://cdn.amegroups.cn/static/application/4b0111ff02b66c45bc28328104e88994/10.21037atm-20-922-2.pdf), most of which were immune relevant, such as three granzyme genes (GZMA, GZMH, and GZMK), and numerous cytokine and cytokine receptor genes, including CXCL9, CXCL13, CXCR6, CCL5, IL2RB, IL10RA, and IL21R. GSEA (30) identified 12 KEGG (26) pathways significantly associated with the 76 genes, including cytokine-cytokine receptor interaction, natural killer cell mediated cytotoxicity, chemokine signaling, T cell receptor signaling, graft-versus-host disease, cell adhesion molecules, Jak-STAT signaling, hematopoietic cell lineage, primary immunodeficiency, prion diseases, allograft rejection, and type I diabetes mellitus (Figure 2A). Evidently, all these pathways are immune relevant.
We identified 146, 734, and 83 genes whose expression levels had a significant inverse correlation with CD8+ T cell enrichment levels in TCGA-GC cohort, ACRG-GC cohort, and both cohorts, respectively (r<−0.3) (https://cdn.amegroups.cn/static/application/4b0111ff02b66c45bc28328104e88994/10.21037atm-20-922-2.pdf). Among the 83 genes, six (GUCY2C, MAP3K9, NEK3, PAK6, STK35, and WNK2) encodes protein kinases. Considering that kinase inhibitors are a class of intensively investigated anticancer drugs that have been widely used in clinics or clinical trials (31), these protein kinases could be potentially useful targets for cancer therapy in that inhibiting them may promote antitumor immune response. GSEA identified one KEGG pathway (steroid biosynthesis) significantly associated with the 83 genes, suggesting that the steroid biosynthesis pathway activity may have an inverse correlation with antitumor immune response in GC.
Likewise, we found 59 and 57 genes having a strong positive and a significant inverse expression correlation with ICA in both cohorts, respectively, and identified 16 immune-related pathways associated with the 59 genes (https://cdn.amegroups.cn/static/application/4b0111ff02b66c45bc28328104e88994/10.21037atm-20-922-2.pdf). Furthermore, we found four genes (GBP1, GBP5, IDO1, and WARS) whose expression levels exhibited a strong positive correlation with PD-L1 expression levels (Figure 2B). The associations of these genes with PD-L1 and GC immunity have been investigated (32). In addition, we identified 80 genes whose expression levels had a significant negative correlation with PD-L1 expression levels (https://cdn.amegroups.cn/static/application/4b0111ff02b66c45bc28328104e88994/10.21037atm-20-922-2.pdf). GSEA revealed that the 80 genes were significantly associated with three KEGG pathways, including drug metabolism-cytochrome P450, peroxisome, and terpenoid backbone biosynthesis.
Collectively, the identification of genes with a significant expression correlation with antitumor immune response or protumor immunosuppression may provide useful intervention targets for promoting antitumor immunity and immunotherapy response in GC.
Identification of proteins whose expression levels are associated with antitumor immune response in GC
Based on TCGA-GC cohort, we identified six proteins (Caspase-7, PI3K-p85, PREX1, Lck, Bcl-2, and transglutaminase) whose expression levels exhibited a significant positive correlation with the enrichment levels of CD8+ T cells in GC (r>0.3) (Figure 3). Notably, both Caspase-7 (33) and Bcl-2 (34) are involved in the apoptosis pathway which has been positively associated with antitumor immune signatures in cancer (35). Transglutaminase has been associated with tumor development and tumor immunity (36). The associations of PI3K-p85 (37), PREX1 (38), and Lck (39) with tumor immunity have been explored. In addition, we found that the expression levels of acetyl-CoA carboxylase (ACC) showed a significant inverse correlation with CD8+ T cell enrichment levels (r=−0.3). Previous studies have shown that ACC was essential for cancer cell survival and was an important target for cancer therapy (40,41). Our result suggests that ACC may promote cancer development by inhibiting antitumor immune response. Moreover, we found that ACC showed a significant inverse expression correlation with ICA in GC (r=−0.33), bolstering the notion that ACC suppresses antitumor immunity.
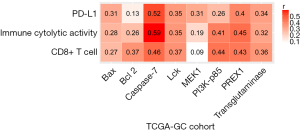
Furthermore, we found six proteins (Caspase-7, PREX1, Lck, transglutaminase, Bax, and MEK1) whose expression levels had a significant positive correlation with PD-L1 expression levels in GC (r>0.3) (Figure 3). Among them, the elevated expression of Caspase-7, PREX1, Lck, and transglutaminase was also significantly associated with highly enriched CD8+ T cells and ICA in GC. The positive correlation between the expression of these proteins and PD-L1 expression indicates that their upregulation could be a predictor for the active response to anti-PD-1/PD-L1 immunotherapy in GC since PD-L1 expression is a biomarker for predicting the anti-PD-1/PD-L1 immunotherapy response in cancer (42).
All together, the identification of proteins significantly correlating with antitumor immune signatures may provide predictive biomarkers or intervention targets for improving GC immunotherapy.
Identification of miRNAs whose expression levels are associated with antitumor immune response in GC
Based on TCGA-GC cohort, we identified 19 and 8 miRNAs whose expression levels had a significant positive and negative correlation with the enrichment levels of CD8+ T cells in GC, respectively (|r|≥0.3) (Figure 4). 14 out of the 19 miRNAs also had a significant positive expression correlation with ICA in GC (r≥0.3) (https://cdn.amegroups.cn/static/application/a7d3d9d3c316a9cc2278b8b6881156f4/10.21037atm-20-922-3.pdf). A literature review showed that many of these miRNAs were associated with tumor immune response, such as hsa-miR-150, 155, 142, 342, 146, 101, 511, 29 (43). Notably, hsa-miR-155-5p expression levels were significantly associated with CD8+ T cell infiltration levels and ICA in GC (r>0.5). The positive association between hsa-miR-155 expression and tumor immunity has been confirmed in a recent study (44). The hsa-miR-150 genes hsa-miR-150-5p and hsa-miR-150-3p exhibited a significant positive expression correlation with CD8+ T cell infiltration levels and ICA in GC (r>0.5). Previous studies have shown that hsa-miR-150 played a key role in tumor pathogenesis and tumor immunity (45). hsa-miR-342 was importantly associated with tumor immunity (46). Our data showed that the expression levels of hsa-miR-342 had a significant positive correlation with CD8+ T cell enrichment levels and ICA in GC (r>0.5).
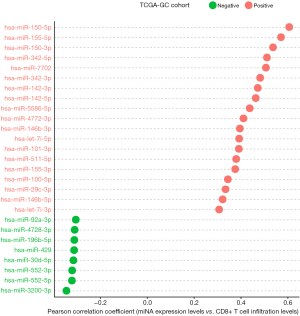
We found 15 miRNAs whose expression levels were significantly and positively associated with PD-L1 expression levels in GC (r≥0.3). The 15 miRNAs included hsa-let-7i-3p, hsa-let-7i-5p, hsa-miR-142-3p, hsa-miR-142-5p, hsa-miR-146b-3p, hsa-miR-146b-5p, hsa-miR-150-3p, hsa-miR-155-3p, hsa-miR-155-5p, hsa-miR-342-3p, hsa-miR-342-5p, hsa-miR-4772-3p, hsa-miR-4772-5p, hsa-miR-511-5p, and hsa-miR-7702 (https://cdn.amegroups.cn/static/application/a7d3d9d3c316a9cc2278b8b6881156f4/10.21037atm-20-922-3.pdf). Previous studies have shown that many of these miRNAs regulated PD-L1 expression on tumor cells (47).
Collectively, the identification of miRNAs correlating with antitumor immune signatures or tumor immunosuppressive signatures may provide potential miRNA biomarkers for GC immunotherapy.
Identification of LncRNAs whose expression levels are associated with antitumor immune response in GC
LncRNAs play important roles in immune regulation and tumor immunity (48-50). Based on TCGA-GC cohort, we identified 107 and 19 lncRNAs with a significant positive and negative expression correlation with the enrichment levels of CD8+ T cells in GC, respectively (|r|≥0.3) (https://cdn.amegroups.cn/static/application/98131eca64c3c7dbb54af3a6732c4f4b/10.21037atm-20-922-4.pdf). Notably, AC022706.1 had the strongest expression correlation with CD8+ T cell enrichment levels in GC (r=0.64) (Figure 5). It also had a strong expression correlation with ICA in GC (r=0.58) (Figure 5). LINC01871 had the second strongest expression correlation with CD8+ T cell enrichment levels (r=0.63) and the strongest expression correlation with ICA in GC (r=0.71) (Figure 5). The expression levels of LINC01871 were also significantly correlated with PD-L1 expression levels in a positive direction in GC (r=0.59) (Figure 5). AC006033.2 had the third strongest expression correlation with CD8+ T cell enrichment levels (r=0.60), the second strongest expression correlation with ICA in GC (r=0.59), and the third strongest expression correlation with PD-L1 expression in GC (r=0.54) (Figure 5).
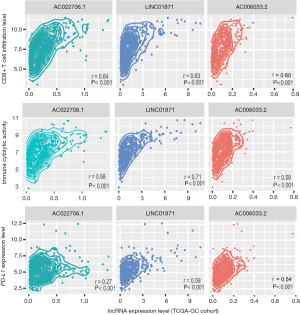
We identified 58 and 79 lncRNAs with a significant positive and negative expression correlation with ICA in GC, respectively (|r|≥0.3) (https://cdn.amegroups.cn/static/application/98131eca64c3c7dbb54af3a6732c4f4b/10.21037atm-20-922-4.pdf). Moreover, we found 58 and 17 lncRNAs whose expression levels were positively and negatively associated with PD-L1 expression levels in GC, respectively (|r|≥0.3) (https://cdn.amegroups.cn/static/application/98131eca64c3c7dbb54af3a6732c4f4b/10.21037atm-20-922-4.pdf).
Because the associations between lncRNAs and tumor immunity have not been sufficiently explored, the identification of lncRNAs associated with antitumor immune signatures may provide useful lncRNA candidates for furthering such exploration.
Identification of cancer-associated pathways whose activity is associated with antitumor immune response in GC
Based on TCGA-GC cohort, we identified nine cancer-associated pathways whose activity was positively correlated with CD8+ T cell infiltration levels in GC [Spearman correlation coefficient (ρ) ≥0.3] (Figure 6). The nine pathways included focal adhesion, mTOR signaling, PI3K-AKT signaling, MAPK signaling, HIF-1 signaling, VEGF signaling, apoptosis, Jak-STAT signaling, and cytokine-cytokine receptor interaction. The positive association between a majority of these pathways (such as focal adhesion, MAPK signaling, VEGF signaling, apoptosis, and Jak-STAT signaling) and GC immunity has been revealed in our previous study (21). The HIF-1 signaling pathway is hyperactivated in various cancers and is associated with the activation of tumor‐associated immune signatures (51). This is consistent with our result that there was a significant positive correlation between this pathway and CD8+ T cell infiltration in GC. Moreover, our previous study (35) showed that tumor glycolysis had a significant positive association with immune signatures in a wide variety of cancers. This is consistent with that the glycolysis-stimulating HIF-1 signaling was positively associated with immune signatures in GC. In addition, our results showed that the mTOR signaling pathway was associated with CD8+ T cell infiltration levels in GC. This is in line with previous studies showing that mTOR signaling played a significant role in regulating tumor microenvironment, thereby affecting tumor immunity (52). We further identified a number of cancer-associated pathways which had a positive correlation with ICA and PD-L1 expression levels in TCGA- and ACRG-GC cohorts, including p53 signaling, HIF-1 signaling, apoptosis, Jak-STAT signaling, and cytokine-cytokine receptor interaction (https://cdn.amegroups.cn/static/application/a41ae079640a0bc9bd35cf88aec82e8d/10.21037atm-20-922-5.pdf).
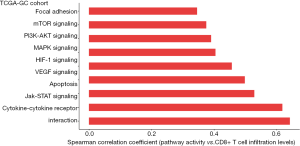
Collectively, the identification of antitumor immunity-associated pathways provided potential intervention targets for promoting antitumor immunity and immunotherapy response in GC.
Discussion
Our bioinformatics analysis identified a number of genes, miRNAs, lncRNAs, proteins, and pathways whose mutations, expression, or activity were significantly associated with antitumor immunity in GC. Of note, we found some important oncogenes and tumor suppressor genes whose mutations were appreciably associated with antitumor immunity in GC, including PIK3CA, MTOR, RNF213, TP53, ARID1A, PTEN, ATM, and CDH1. These significant associations may partially explain why the mutations of some of these genes were associated with OS in GC. Interestingly, we found seven genes (ARID1A, BCOR, MTOR, CREBBP, SPEN, NOTCH4, and TET1) whose mutations were associated with better OS in GI cancer patients receiving anti-PD-1/PD-L1 immunotherapy but were not associated with OS in GC patients not receiving such a therapy (Figure 1B). A potential mechanism underlying this difference is the elevated expression of PD-L1 in the cancer patients with the mutation of these genes. To date, many studies have revealed that PD-L1 expression is a predictive biomarker for anti-PD-1/PD-L1 immunotherapy in GC patients (53,54). A clinical trial study (KEYNOTE-059) revealed that pembrolizumab, an anti-PD-1 monoclonal antibody, had encouraging antitumor activity in PD-L1-positive advanced gastric/gastroesophageal cancer patients (53). Jin et al. reported that a PD-L1-positive unresectable locally advanced GC with MSI exhibited pathological complete response to a single dose of anti-PD-1 immunotherapy in combination with chemotherapy (54). Hence, the identification of gene mutations associated with tumor immunity may furnish biomarkers for the optimal stratification of GC patients responsive to immunotherapy. Likewise, we identified some important genes and proteins whose expression levels were significantly associated with antitumor immunity in GC and thus may serve as biomarkers or intervention targets for promoting GC immunotherapy. For example, ACC exhibited a significant inverse expression correlation with CD8+ T cell infiltration levels and ICA in GC, suggesting that inhibiting ACC could enhance antitumor immunity in GC. Furthermore, we identified a number of miRNAs and lncRNAs having a significant expression correlation with GC immunity, such as the miRNAs hsa-miR-150, 155, 142, 342, 146, 101, 511, 29 and the lncRNAs AC022706.1, LINC01871, and AC006033.2. These non-coding RNAs could be potentially useful biomarkers for GC prognosis and immunotherapy. Finally, we identified numerous cancer-associated pathways whose activity was significantly associated with antitumor immunity in GC, including the mTOR, PI3K-AKT, MAPK, HIF-1, and VEGF signaling pathways. These pathways were mainly involved in cell proliferation, growth and migration, metabolism, and immune response.
TMB often has a positive association with tumor immunity and immunotherapy response (25,55) while TALs often inversely correlate with them (56). As expected, we observed a significant positive correlation of TMB with PD-L1 expression levels in TCGA-GC cohort (Student’s t test, P=0.03) (Figure 7A) and a significant negative correlation of TALs with CD8+ T cell enrichment levels, ICA, and PD-L1 expression levels in TCGA- and ACRG-GC cohorts (Student’s t-test, P<0.001) (Figure 7B). This indicates that TMB and TALs are useful biomarkers for predicting GC patients responsive to immunotherapy.
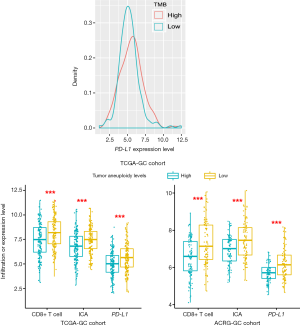
In conclusion, the molecular features significantly associated with GC immunity could be useful biomarkers for stratifying GC patients responsive to immunotherapy or potential intervention targets for promoting antitumor immunity and immunotherapy response in GC.
Acknowledgments
We thank Mr. Zehang Jiang for help in data analysis.
Funding: This work was supported by the China Pharmaceutical University (grant number 3150120001 to XW).
Footnote
Reporting Checklist: The authors have completed the MDAR reporting checklist. Available at http://dx.doi.org/10.21037/atm-20-922
Conflicts of Interest: Both authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/atm-20-922). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Bray F, Ferlay J, Soerjomataram I, et al. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin 2018;68:394-424. [Crossref] [PubMed]
- Orditura M, Galizia G, Sforza V, et al. Treatment of gastric cancer. World J Gastroenterol 2014;20:1635-49. [Crossref] [PubMed]
- Van Allen EM, Miao D, Schilling B, et al. Genomic correlates of response to CTLA-4 blockade in metastatic melanoma. Science 2015;350:207-11. [Crossref] [PubMed]
- Herbst RS, Baas P, Kim D-W, et al. Pembrolizumab versus docetaxel for previously treated, PD-L1-positive, advanced non-small-cell lung cancer (KEYNOTE-010): a randomised controlled trial. Lancet 2016;387:1540-50. [Crossref] [PubMed]
- Bauml J, Seiwert TY, Pfister DG, et al. Pembrolizumab for Platinum- and Cetuximab-Refractory Head and Neck Cancer: Results From a Single-Arm, Phase II Study. J Clin Oncol 2017;35:1542-9. [Crossref] [PubMed]
- Tomita Y, Fukasawa S, Shinohara N, et al. Nivolumab versus everolimus in advanced renal cell carcinoma: Japanese subgroup analysis from the CheckMate 025 study. Jpn J Clin Oncol 2017;47:639-46. [Crossref] [PubMed]
- Ruiz-Banobre J, Goel A. DNA Mismatch Repair Deficiency and Immune Checkpoint Inhibitors in Gastrointestinal Cancers. Gastroenterology 2019;156:890-903. [Crossref] [PubMed]
- Braun DA, Burke KP, Van Allen EM. Genomic Approaches to Understanding Response and Resistance to Immunotherapy. Clin Cancer Res 2016;22:5642-50. [Crossref] [PubMed]
- Taube JM, Klein A, Brahmer JR, et al. Association of PD-1, PD-1 ligands, and other features of the tumor immune microenvironment with response to anti-PD-1 therapy. Clin Cancer Res 2014;20:5064-74. [Crossref] [PubMed]
- Goodman AM, Kato S, Bazhenova L, et al. Tumor Mutational Burden as an Independent Predictor of Response to Immunotherapy in Diverse Cancers. Mol Cancer Ther 2017;16:2598-608. [Crossref] [PubMed]
- Le DT, Uram JN, Wang H, et al. PD-1 Blockade in Tumors with Mismatch-Repair Deficiency. N Engl J Med 2015;372:2509-20. [Crossref] [PubMed]
- Dong ZY, Zhong WZ, Zhang XC, et al. Potential Predictive Value of TP53 and KRAS Mutation Status for Response to PD-1 Blockade Immunotherapy in Lung Adenocarcinoma. Clin Cancer Res 2017;23:3012-24. [Crossref] [PubMed]
- Binnewies M, Roberts EW, Kersten K, et al. Understanding the tumor immune microenvironment (TIME) for effective therapy. Nat Med 2018;24:541-50. [Crossref] [PubMed]
- Haanen J. Converting Cold into Hot Tumors by Combining Immunotherapies. Cell 2017;170:1055-6. [Crossref] [PubMed]
- Wang B, Zhang W, Jankovic V, et al. Combination cancer immunotherapy targeting PD-1 and GITR can rescue CD8(+) T cell dysfunction and maintain memory phenotype. Sci Immunol 2018;3:eaat7061. [Crossref] [PubMed]
- Bourgeois-Daigneault MC, Roy DG, Aitken AS, et al. Neoadjuvant oncolytic virotherapy before surgery sensitizes triple-negative breast cancer to immune checkpoint therapy. Sci Transl Med 2018;10:eaao1641.
- Bommareddy PK, Aspromonte S, Zloza A, et al. MEK inhibition enhances oncolytic virus immunotherapy through increased tumor cell killing and T cell activation. Sci Transl Med 2018;10:eaau0417. [Crossref] [PubMed]
- Goel S, DeCristo MJ, Watt AC, et al. CDK4/6 inhibition triggers anti-tumour immunity. Nature 2017;548:471-5. [Crossref] [PubMed]
- Li M, Liu Z, Wang X. Exploration of the combination of PLK1 inhibition with immunotherapy in cancer treatment. Journal of Oncology 2018;2018:13. [Crossref] [PubMed]
- Zeng D, Li M, Zhou R, et al. Tumor microenvironment characterization in gastric cancer identifies prognostic and immunotherapeutically relevant gene signatures. Cancer Immunol Res 2019;7:737-50. [Crossref] [PubMed]
- Jiang Z, Liu Z, Li M, et al. Immunogenomics Analysis Reveals that TP53 Mutations Inhibit Tumor Immunity in Gastric Cancer. Transl Oncol 2018;11:1171-87. [Crossref] [PubMed]
- Park C, Cho J, Lee J, et al. Host immune response index in gastric cancer identified by comprehensive analyses of tumor immunity. Oncoimmunology 2017;6:e1356150. [Crossref] [PubMed]
- Cancer Genome Atlas Research N. Comprehensive molecular characterization of gastric adenocarcinoma. Nature 2014;513:202-9. [Crossref] [PubMed]
- Cristescu R, Lee J, Nebozhyn M, et al. Molecular analysis of gastric cancer identifies subtypes associated with distinct clinical outcomes. Nat Med 2015;21:449-56. [Crossref] [PubMed]
- Samstein RM, Lee CH, Shoushtari AN, et al. Tumor mutational load predicts survival after immunotherapy across multiple cancer types. Nat Genet 2019;51:202-6. [Crossref] [PubMed]
- Kanehisa M, Furumichi M, Tanabe M, et al. KEGG: new perspectives on genomes, pathways, diseases and drugs. Nucleic Acids Res 2017;45:D353-61. [Crossref] [PubMed]
- Hanzelmann S, Castelo R, Guinney J. GSVA: gene set variation analysis for microarray and RNA-seq data. BMC Bioinformatics 2013;14:7. [Crossref] [PubMed]
- Benjamini Y, Hochberg Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J R Stat Soc Series B Stat Methodol 1995;57:289-300.
- Carter SL, Cibulskis K, Helman E, et al. Absolute quantification of somatic DNA alterations in human cancer. Nat Biotechnol 2012;30:413-21. [Crossref] [PubMed]
- Subramanian A, Tamayo P, Mootha VK, et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci U S A 2005;102:15545-50. [Crossref] [PubMed]
- Gharwan H, Groninger H. Kinase inhibitors and monoclonal antibodies in oncology: clinical implications. Nat Rev Clin Oncol 2016;13:209-27. [Crossref] [PubMed]
- Patil PA, Blakely AM, Lombardo KA, et al. Expression of PD-L1, indoleamine 2,3-dioxygenase and the immune microenvironment in gastric adenocarcinoma. Histopathology 2018;73:124-36. [Crossref] [PubMed]
- Lamkanfi M, Kanneganti TD. Caspase-7: a protease involved in apoptosis and inflammation. Int J Biochem Cell Biol 2010;42:21-4. [Crossref] [PubMed]
- Czabotar PE, Lessene G, Strasser A, et al. Control of apoptosis by the BCL-2 protein family: implications for physiology and therapy. Nat Rev Mol Cell Biol 2014;15:49-63. [Crossref] [PubMed]
- Jiang Z, Liu Z, Li M, et al. Increased glycolysis correlates with elevated immune activity in tumor immune microenvironment. EBioMedicine 2019;42:431-42. [Crossref] [PubMed]
- Eckert RL, Kaartinen MT, Nurminskaya M, et al. Transglutaminase regulation of cell function. Physiol Rev 2014;94:383-417. [Crossref] [PubMed]
- Dituri F, Mazzocca A, Giannelli G, et al. PI3K functions in cancer progression, anticancer immunity and immune evasion by tumors. Clin Dev Immunol 2011;2011:947858. [Crossref] [PubMed]
- Porter AP, Papaioannou A, Malliri A. Deregulation of Rho GTPases in cancer. Small GTPases 2016;7:123-38. [Crossref] [PubMed]
- Harashima N, Tanaka K, Sasatomi T, et al. Recognition of the Lck tyrosine kinase as a tumor antigen by cytotoxic T lymphocytes of cancer patients with distant metastases. Eur J Immunol 2001;31:323-32. [Crossref] [PubMed]
- Chajes V, Cambot M, Moreau K, et al. Acetyl-CoA carboxylase alpha is essential to breast cancer cell survival. Cancer Res 2006;66:5287-94. [Crossref] [PubMed]
- Wang C, Rajput S, Watabe K, et al. Acetyl-CoA carboxylase-a as a novel target for cancer therapy. Front Biosci (Schol Ed) 2010;2:515-26. [PubMed]
- Patel SP, Kurzrock R. PD-L1 Expression as a Predictive Biomarker in Cancer Immunotherapy. Mol Cancer Ther 2015;14:847-56. [Crossref] [PubMed]
- Paladini L, Fabris L, Bottai G, et al. Targeting microRNAs as key modulators of tumor immune response. J Exp Clin Cancer Res 2016;35:103. [Crossref] [PubMed]
- Ekiz HA, Huffaker TB, Grossmann AH, et al. MicroRNA-155 coordinates the immunological landscape within murine melanoma and correlates with immunity in human cancers. JCI Insight 2019;4:e126543. [Crossref] [PubMed]
- Eichmuller SB, Osen W, Mandelboim O, et al. Immune Modulatory microRNAs Involved in Tumor Attack and Tumor Immune Escape. J Natl Cancer Inst 2017;109. [Crossref] [PubMed]
- Rupaimoole R, Calin GA, Lopez-Berestein G, et al. miRNA Deregulation in Cancer Cells and the Tumor Microenvironment. Cancer Discov 2016;6:235-46. [Crossref] [PubMed]
- Wang Q, Lin W, Tang X, et al. The Roles of microRNAs in Regulating the Expression of PD-1/PD-L1 Immune Checkpoint. Int J Mol Sci 2017;18:2540. [Crossref] [PubMed]
- Heward JA, Lindsay MA. Long non-coding RNAs in the regulation of the immune response. Trends Immunol 2014;35:408-19. [Crossref] [PubMed]
- Geng H, Tan XD. Functional diversity of long non-coding RNAs in immune regulation. Genes Dis 2016;3:72-81. [Crossref] [PubMed]
- Denaro N, Merlano MC, Lo Nigro C. Long noncoding RNAs as regulators of cancer immunity. Mol Oncol 2019;13:61-73. [Crossref] [PubMed]
- Balamurugan K. HIF-1 at the crossroads of hypoxia, inflammation, and cancer. Int J Cancer 2016;138:1058-66. [Crossref] [PubMed]
- Guri Y, Nordmann TM, Roszik J. mTOR at the Transmitting and Receiving Ends in Tumor Immunity. Front Immunol 2018;9:578. [Crossref] [PubMed]
- Catenacci Daniel V, Wainberg Z, Fuchs Charles S, et al. KEYNOTE-059 cohort 3: safety and efficacy of pembrolizumab monotherapy for first-line treatment of patients (pts) with PD-L1-positive advanced gastric/gastroesophageal (G/GEJ) cancer. Ann Oncol 2017;28 Suppl 3:iii153. [Crossref]
- Jin H, Li P, Mao C, et al. Pathological Complete Response After a Single Dose of Anti-PD-1 Therapy in Combination with Chemotherapy as a First-Line Setting in an Unresectable Locally Advanced Gastric Cancer with PD-L1 Positive and Microsatellite Instability. Onco Targets Ther 2020;13:1751-6. [Crossref] [PubMed]
- He Y, Liu ZX, Jiang ZH, et al. Identification of genomic features associated with immunotherapy response in gastrointestinal cancers. World J Gastrointest Oncol 2019;11:270-80. [Crossref] [PubMed]
- Davoli T, Uno H, Wooten EC, et al. Tumor aneuploidy correlates with markers of immune evasion and with reduced response to immunotherapy. Science 2017;355:eaaf8399. [Crossref] [PubMed]


