
Dairy consumption and hepatocellular carcinoma risk
Introduction
Hepatocellular carcinoma (HCC) mortality rates have increased over recent decades (1-4). Major risk factors for HCC are chronic liver disease and cirrhosis due to hepatitis B virus (HBV) and/or hepatitis C virus (HCV) infection, alcoholic liver disease, non-alcoholic fatty liver disease (NAFLD), steatohepatitis, intake of aflatoxin-contaminated food, type 2 diabetes mellitus (T2DM) and obesity (1-6). Although increased dairy product intake has been associated with increased risk of several cancers (7-11), epidemiological studies on dairy and milk consumption in HCC are sparse. Five older case-control studies with small patient numbers presented conflicting results (12-16). Recently, Yang et al. (17) assessed the associations of dairy products with the risk of HCC development among 51,418 men and 93,427 women in the Health Professionals Follow-Up Study and the Nurses’ Health Study. After adjustment for known HCC risk factors, higher total dairy intake was associated with an increased risk of HCC. In accordance with the association of milk consumption and T2DM (18-20), there was an inverse association between yogurt intake and HCC risk (17). The European Prospective Investigation into Cancer and Nutrition (EPIC) cohort (477,206 participants) demonstrated a significant positive HCC risk association with total dairy product intake, milk and cheese, but not yogurt (21). The Guangzhou Biobank Cohort Study (18,214 participants) showed that moderate (250–750 mL/week) versus high milk consumption (>750 mL/week) significantly increased the mortality rate for all cancers and raised the mortality rate of liver cancers from 3.8 to 7.0 per 10,000 person-years, respectively (22).
In 2017, the per capita milk consumption was 65.2 L in the United States and 52.2 L in Germany, respectively (23,24). Since the 1950s, the annual consumption of cheese, a rich source of branched-chain amino acids (BCAAs), has increased four- to five-fold in Western societies (25,26).
It is the intention of this review to present molecular mechanisms that may explain dairy-induced hepatocancerogenesis. For this purpose, it is important to keep in mind that milk represents a postnatal signaling system evolutionarily restricted to the breastfeeding period of mammals for the promotion of cell proliferation and growth (27-29).
Insulin-like growth factor 1 (IGF-1)
Circulating IGF-1 concentrations have been associated with higher cancer risk (30,31). A cross-sectional EPIC analysis of 4,731 men and women demonstrated that each 1 standard deviation (SD) increment increase in total dairy and dairy protein was associated with an increase in IGF-1 concentrations of 2.5% and 2.4%, respectively (32). A meta-analysis reported that circulating IGF-1 increased by 13.8 ng/mL in the milk intervention group compared with the control group (33). A recent study in Germany showed that each 400 g increment in daily dairy intake in adults was associated with 16.8 ng/mL higher IGF-1 serum concentrations, whereas each daily 200 g increment in milk was associated with 10.0 ng/mL higher IGF-1 (34). Remarkably, whey protein intake preferentially increased serum insulin levels, whereas casein raised serum IGF-1 levels (35).
The liver is the primary organ releasing IGF-1 into the systemic circulation (36,37). Dietary amino acids (AAs) and insulin induce hepatic IGF-1 expression and secretion (38,39) (Figure 1). Increasing AA concentrations enhance hepatic expression of IGF-1, peroxisome proliferator-activated receptor-γ (PPARγ) and mechanistic target of rapamaycin (mTOR) (38). Low protein diets are associated with lower cancer incidence and mortality rates in humans. Protein restriction inhibits tumor growth via attenuation of IGF-1/mTORC1 signaling (40). Individuals aged 50–65 with a high protein intake had a four-fold increase in the risk of cancer death during a follow-up of 18 years (41). Post-initiation development of aflatoxin B1 (AFB1)-initiated preneoplastic foci in Fischer 344 rat liver could be prevented by decreasing casein intake (42). Dietary casein reduction from 22% to 6% markedly inhibited hepatic tumor formation in HBV-transfected mice (43). A low casein diet also suppressed HBV-induced liver injury (44).
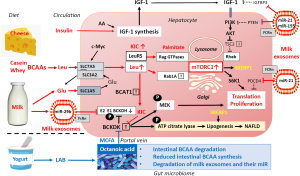
IGF-1/IGF-1 receptor signaling plays a critical role in hepatocarcinogenesis (45-51), whereas blocking of IGF-1 receptor pathway is regarded as a treatment option in HCC (52-60). Increased systemic IGF-1 levels and enhanced hepatic IGF-1 signaling characterize the initiation stage of HCC, whereas overt HCC with compromised liver function and liver cirrhosis is associated with decreased hepatic IGF-1 synthesis and low serum IGF-1 (45,61-68), a marker for liver reserve capacity in HCC patients (69).
BCAAs
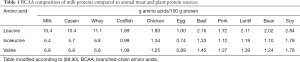
The nutrient-sensitive kinase mechanistic target of rapamycin complex 1 (mTORC1) is overactivated in many cancers including HCC (70-79). Not only insulin and IGF-1 activate mTORC1 but also AAs (80,81), preferentially the BCAAs leucine, isoleucine and valine (82-88). In comparison with other animal- and plant-derived protein-rich nutrient sources, milk proteins are highly enriched in BCAAs (89) (Table 1).
Full table
Solute carrier family 7 member 5 (SLC7A5) plays a key role for leucine uptake in cancer cells (91,92). Oncogenic MYC stimulates SLC7A5 expression promoting BCAA import for tumorigenesis (93). High SLC7A5 expression in HCC is related with tumor size, tumor stage and shortened survival time (94). SLC7A5 expression is associated with increased expression of SLC3A2 (95). Canine HCCs exhibit 28 times higher SLC7A5 expression than normal hepatocytes (96). SLC7A5 also plays a critical role in hepatic cancer stem cells (HCSCs) (97). Cellular uptake of glutamine via SLC1A5 and its subsequent efflux in the presence of BCAAs is the rate-limiting step that activates mTORC1 (98). The bidirectional transporter SLC7A5/SLC3A2 regulates the simultaneous efflux of glutamine and concomitant influx of leucine into cells (Figure 1) (98). Glutamine abundance thus improves leucine uptake and BCAA-mediated mTORC1 activation, a potential explanation for the high glutamine content of milk proteins (90). The leucine and glutamine content is increased in HCC tissue and is higher in high-grade compared with low-grade HCCs (99). HCC is thus a leucine addict like prostate cancer (100,101), another common milk-induced cancer of Western societies (7-9,11).
Chronic activation of mTORC1 is sufficient to cause HCC in mice and represents the key molecular link between HCC risk and dietary factors (71). Long-term cow’s milk consumption in young mice increased liver BCAA levels and activated mTORC1-S6K1 (102). Hepatic mTORC1 promotes the expression of lipogenic genes and leads to the development of HCC (72,79). mTORC1 upregulates sterol regulatory element-binding protein 1 (SREBP1), which contributes to NAFLD (79).
In HCC tissues, the expression level of branched-chain amino acid transaminase 1 (BCAT1), which catalyzes the production of branched-chain α-ketoacids (BCKAs) from BCAAs, is significantly increased compared with adjacent tissues (103). Patients with high BCAT1 expression possess a lower overall survival than those with low BCAT1 expression (103). Leucine and its conversion product α-ketoisocaproate (KIC) are sensed by leucyl-tRNA synthase (LeuRS) (Figure 1) (104). LeuRS acts as a GTPase-activating protein (GAP) for RagD GTPase activating mTORC1 (105). In high fat-diet (HFD)-treated diet-induced obesity (DIO) mice, BCAA supplementation increased hepatic BCAA and BCKA levels and induced severe hepatic insulin resistance (IR) (104).
Rag small GTPases are critical mediators of AA-mTORC1 assembly and mTORC1 activation at the lysosomal compartment (106,107). AAs are also capable of activating mTORC1 in the absence of Rag GTPases dependent on Rab1A (108-110). In colorectal cancer (CRC), AAs stimulate Rab1A interaction with mTORC1, whereby Rab1A promotes the formation and activation of the Rheb-mTORC1 complex at the Golgi (110). In accordance with CRC, overexpressed Rab1A in HCC has been identified as a crucial driver for AA-mediated mTORC1 activation in hepatocancerogenesis (Figure 1) (111).
The activity of BCAA catabolic enzymes is suppressed in HCC resulting in increased BCAA levels activating mTORC1 (112,113). Either downregulation of expression and/or changes in post-translational modifications, e.g., hyperphosphorylation of BCKA dehydrogenase (BCKDH) by BCKDH kinase (BCKDK) impairs BCAA catabolism leading to BCAA accumulation with chronic activation of mTORC1 in HCC (112,113). In accordance with HCC, upregulated BCKDK has also been observed in CRC (114). Hepatic overexpression of BCKDK increased the activity of ATP-citrate lyase activating de novo lipogenesis (115) linking disturbed hepatic BCAA catabolism to steatosis hepatis and HCC.
HBV and HCV
Hepatic overexpression of HBV X protein (HBx) activates AKT and mTORC1 signaling (116,117) and induces the expression of α-fetoprotein (AFP), which attenuates the function of phosphatase and tensin homolog (PTEN) leading to increased AKT-mTORC1 signaling in HBx-transfected human liver cells and HCC (118). This AFP-mediated mechanism also promotes the initiation of HCC progenitor/stem cells (119). In addition, HBx induces microRNA (miR)-181a further targeting PTEN (120).
HCV activates mTORC1 via upregulation of the SLC7A5/SLC3A2 complex augmenting cellular leucine influx (121). Thus, persistent milk consumption synergizes with HBV-/HCV-overstimulated hepatic mTORC1 signaling.
IR and obesity
Aberrant liver metabolism promotes IR, a hallmark of NAFLD and T2DM. NAFLD patients with hepatic IR generally share a high risk of impaired fasting glucose associated with early T2DM. Many patients with T2DM experience NAFLD, non-alcoholic steatohepatitis (NASH), and severe complications such as cirrhosis and HCC (122). Notably, high intake of milk, but not meat, increased serum insulin levels and IR in 8-year-old boys (123). Positive associations between full-fat dairy, non-fermented dairy products and milk with pre-diabetes and newly diagnosed T2DM has been reported in the prospective Dutch Lifeline Cohort Study including 112,086 adults (19). There is a confirmed association between elevated BCAA plasma levels and IR (25,124-133). A genetic link between obesity-associated IR and impaired BCAA catabolic gene expression in human and mouse models has been reported (134). In genetically obese (ob/ob) mice, the deficiency of BCKDH resulted in an accumulation of BCAAs and BCKAs, which both activate mTORC1 (134). Restoring BCAA catabolic flux via inhibition of BCKDK reduced BCAA and BCKA abundance and markedly attenuated IR in ob/ob mice. Similar outcomes were achieved by reducing protein intake, whereas increasing BCAA intake did the opposite. Thus, compromised BCAA catabolism is a common hallmark of IR, T2DM and HCC (112,113,134), metabolic deviations overactivating mTORC1 (18,89). In mice, BCAA tissue levels were directly related to liver tumor development and tumor size associated with dietary BCAA intake and mTORC1 activity (112).
Single nucleotide polymorphisms of fat mass and obesity-associated gene (FTO) are related to body mass index and obesity. FTO is a N6-methyladenosine (m6A) demethylase that acts as the most common mRNA modification in normal and tumor cells linking obesity to cancer (135). In murine and human cell lines, total AA deprivation reduced FTO expression (136). FTO expression is regulated by essential AAs and couples AA levels to mTORC1 signaling through a demethylation-dependent mechanism. FTO is thus an AA sensor promoting growth and translation (137,138). It has been hypothesized that milk functions as an epigenetic amplifier of FTO-mediated transcription (139). Remarkably, FTO levels are upregulated in HCC tissue and correlate to a poorer prognosis, whereas FTO knockdown suppresses proliferation and in vivo tumor growth (140). FTO triggers the demethylation of pyruvate kinase 2 (PKM2) mRNA accelerating its translation. Overexpression of PKM2 activates mTORC1 signaling through phosphorylation-mediated inactivation of the mTORC1 inhibitor AKT1 substrate 1 (AKT1S1) (141). In addition, m6A methylation of the 3' ε-stem loop results in destabilization of HBV transcripts, suggesting that m6A has regulatory function for HBV RNA (142). Milk-mediated activation of FTO may thus accelerate HBV RNA transcription.
Milk fat
During late-stage NAFLD, fibrotic and cancerous cells can proliferate. HCC cells and normal hepatocytes are exposed to high concentrations of fatty acids from both surrounding tissue and circulating lipid sources. The saturated fatty acid palmitic acid (PA) exerts lipotoxic effects in activated human hepatic stellate cells (HSCs) and epithelial hepatoma cells (143). An HFD rich in PA is associated with lower insulin sensitivity (144). The National Institutes of Health-AARP Diet and Health Study confirmed an association between saturated fat intake and HCC (145). However, total amounts of triacylglycerols stored in hepatocytes are not the major determinant of lipotoxicity, whereas specific lipid classes in particular PA, cholesterol, lysophosphatidylcholine and ceramides damage liver cells (146,147).
Fatty acid transport protein 4 (FATP4) is a minor FATP in the liver. PA activation of FATP4 triggers hepatocellular apoptosis via altered phospholipid composition and steatosis by acylation into complex lipids (148). In HFD-treated DIO mice, BCAA supplementation increased plasma free fatty acid levels (104). O-GlcNAc transferase (OGT), which upregulates PA synthesis, is involved in metabolic reprogramming and IR and plays a key role in NAFLD-associated HCC (149).
Bovine milk contains 3.5% to 5% total lipid secreted by mammary gland epithelial cells as milk fat globules (MFGs). About 98% of milk lipids are triacylglycerols. PA is the predominant fatty acid of MFG triacylglycerols (150,151), which are hydrolyzed in the intestine and transported via chylomicrons into the systemic circulation. Upregulated PA absorption by activation of its hepatic transporters is evident in NASH (152). Notably, PA activates mTORC1 by enhancing its recruitment onto lysosomal membranes (153-155). Thus, milk fat-derived dietary PA may contribute to hepatic mTORC1 activation, which is involved in the pathogenesis of NASH (156-158).
Exosomal MFG-EGF factor 8 (MFG-E8) and TGF-β
Milk contains extracellular vesicles (EVs) that modulate numerous biological processes (159-161). EV subsets of milk can be separated by ultracentrifugation (161). The 100,000 ×g fraction contains the milk exosomes (50–100 nm) (162). Milk EVs including exosomes deliver RNAs, miRs and proteins protected by a lipid bilayer membrane, which confers resistance against their intestinal degradation (163).
Milk exosomes are bioavailable and distribute their cargo across species boundaries (164). They accumulate in liver following suckling, oral gavage and intravenous administration to mice and pigs (164). MFG-E8 is a major component of MFGs and milk exosomes (165-167). MFG-E8 (also called lactadherin) promotes proliferation of hepatocytes through the phosphatidylinositol-3 kinase (PI3K)/AKT/mTORC1 pathway and is a key regulator of cancer cell invasion, migration, and proliferation (168,169). MFG-E8 expression is higher in HCC cells compared with normal liver tissue (170). Overexpressed MFG-E8 promotes proliferation and migration of HCC cells, whereas MFG-E8-neutralizing antibodies inhibited HCC cell proliferation and migration (170).
The development of HCC is also associated with alterations in transforming growth factor-β (TGF-β) signaling (171-175). Increased numbers of TGF-β+ regulatory T-cells (Tregs) have been detected in the peripheral blood of HCC patients (174). Ki-67 expression is also associated with TGF-β1 and deteriorates the prognosis of patients with HBV-related HCC (175). The Th17/Treg ratio is an independent factor influencing the occurrence of HCC in HBV-infected patients (176).
Milk exosomes transport TGF-β to the milk recipient (177). In mice, bovine milk exosomes promoted Th17 differentiation, which could be suppressed by anti-TGF-β antibodies (177). Thus, milk exosomes via MFG-E8 may stimulate hepatocyte proliferation and via TGF-β impair immunological surveillance.
Exosomal miRs
Exosomes and their miR cargo play an important role in HCC development and treatment (178-186). HCC-derived exosomal miR-21 contributes to tumor progression by converting HSCs to cancer-associated fibroblasts (CAFs) (187). HBx upregulates miR-21, which downregulates programmed cell death 4 (PDCD4) in HCC (188). HCC cell apoptosis was suppressed through HBx-induced miR-21 by targeting interleukin 12 (IL-12) (189,190). MiR-21 compromises IL-12-mediated anti-cancer activity including activation of natural killer (NK) cells and cytotoxic T lymphocytes (191). The secretion of miR-21-enriched exosomes by HBV-infected hepatocytes is thus a potential viral escape mechanism from host innate immune responses (192,193).
It is conceivable that milk-derived exosomes and their miR cargo modify hepatocyte transcriptional activity. After oral or retroorbital administration, bovine milk exosomes accumulated in the liver of mice (164,194) and apparently transfer their miRs into hepatocytes (195). Part of bovine milk exosome uptake is mediated by bovine immunoglobulin G (IgG), which binds to human neonatal Fc receptor (FcRn) (196) that is highly expressed in adult human liver (197).
MiR-21
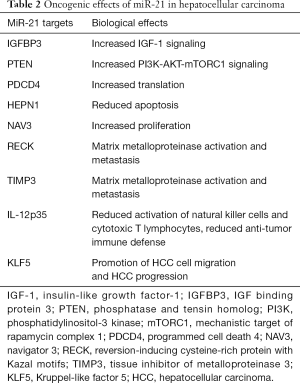
MiR-21 is a well-established oncomiR whose steady-state tissue levels are commonly increased in many malignancies including HCC (198-206). MiR-21 promotes migration and invasion by the miR-21-PDCD4-AP-1 feedback loop in human HCC (198). MiR-21 activates mTORC1 and mTORC1-dependent translation via targeting PTEN and PDCD4 (207-209). Cancer susceptibility gene HEPN1 is frequently silenced in HCC, whereas exogenous HEPN1 exhibits antiproliferative effect on HepG2 cells, suggesting that silencing of HEPN1 may be associated with hepatocancerogenesis (210). MiR-21 promotes cell proliferation and migration in human HCC by targeting HEPN1 and Navigator 3 (NAV3) (199,211) (Table 2).
Full table
Circulating miR-21 serves as a biomarker for HCC and correlates with distant metastasis (199,211-214). The 5-year overall survival of a high miR-21 expression group was significantly shorter than that of a group with low miR-21 expression (215,216). Furthermore, overexpression of miR-21 is associated with HCC recurrence in patients with HBV-mediated HCC undergoing liver transplantation (217). High-mobility group box 1 (HMGB1) induces miR-21 in HCC repressing matrix metalloproteinase (MMP) inhibitors reversion-inducing cysteine-rich protein with Kazal motifs (RECK) and tissue inhibitor of metalloproteinase 3 (TIMP3), which regulate HCC progression and metastases (218). Thus, upregulated miR-21 promotes various steps of hepatocancerogenesis (219-224). In contrast, degradation of miR-21 or treatment with anti-miR-21 suppressed HCC growth, induced apoptosis and reduced resistance to sorafenib and cisplatin (225-228).
Importantly, miR-21 is a signature miR of commercial milk (229,230) and a consistent component of milk exosomes (230). In human volunteers, increased plasma miR-21 levels have been reported 6 hours after milk consumption (231). Because human and bovine miR-21 sequences are identical (232), milk-derived exosomal miR-21 and HCC-induced exosomal miR-21 may synergistically promote hepatocancerogenesis (219-223,233).
Commercial milk obtained from persistently pregnant cows contains androgenic hormones (234,235). Mean milk androstenedione concentrations of pregnant cows are three-fold higher compared to non-pregnant cows (236,237). High affinity binding sites of androgens have been characterized in primary HCC cells (238). Androgen receptor activation by androst-5-ene-3,17-dione, androst-5-ene-3β,17β-diol, dihydrotestosterone, and 5α-androstane-3β increased miR-21 transcription in HepG2 cells (239). Thus, milk-derived androgenic steroids may enhance oncogenic miR-21 expression.
MiR-155
HBV and HCV are involved in HCC pathogenesis and affect the expression of miRs (240). MiR-155 is an important oncomiR driving HCC (241-243). The expression of miR-155 is upregulated in tissues and serum of patients with HCC (241-244). HBV via suppression of zinc finger and homeodomain protein 2 (ZHX2), also known as α-fetoprotein regulator 1, promotes proliferation of HCC through miR-155 activation (245). Overexpressed miR-155 in HCV-induced HCC activates Wnt signaling (246,247). Epstein-Barr virus (EBV) infection, also observed in HCC (248), as well increased the expression of miR-155 (249,250).
Both miR-21 and miR-155 directly target PTEN and thereby activate AKT-mTORC1 signaling (251,252). HCC exosome-mediated transfer of miR-155 contributes to HCC cell proliferation by targeting PTEN (252). Other targets of miR-155 in HCC are forkhead box O3A (FoxO3a) (253,254), collagen triple helix repeat containing 1 (CTHRC1) (255), AT-rich interactive domain 2 (ARID2) (256), sex-determining region Y box 6 (SOX6) (257), F-box and WD40 domain protein 7 (FBXW7) (258), and Kruppel-like factor 5 (KLF5) (259).
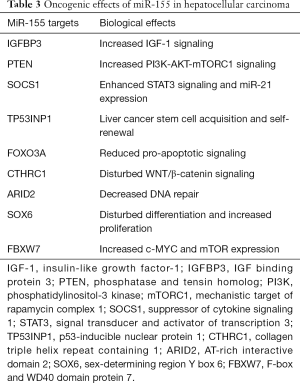
FoxO3a exerts antitumor properties in HCC, inducing the expression of proapoptotic genes, or interfering with signaling cascades commonly altered in HCC such as Wnt/β-catenin, PI3K/AKT/mTORC1 or MAPKs pathways (260). MiR-155 overexpression increases metastasis- and antiapoptosis-related protein expression and decreases proapoptosis-related protein expression (255,256). As shown in an HCC cell line, ARID2 knockout disrupts DNA repair processes resulting in susceptibility to carcinogens and potential hypermutation (261). Decreased expression of SOX6, which plays critical roles in cell fate determination, differentiation and proliferation, confers a poor prognosis in HCC (262). Because FBXW7 mediates ubiquitination-dependent degradation of c-MYC and mTOR (263,264), miR-155-mediated suppression of FBXW7 is likely to affect HCC tumorigenesis. MiR-155 in synergy with miR-21 suppresses IGF binding protein 3 (IGFBP3) activating IGF/IGFR signaling in HCC (265,266) (Figure 1). TGF-β1, a component of milk exosomes (177), promotes the expression of miR-155. Increased levels of miR-155 in HCC cells accelerate epithelial-mesenchymal transition (EMT), activate PI3K-serum/glucocorticoid-regulated kinase 3 (SGK3)-β-catenin signaling, promote cellular invasion and migration (267,268). Notably, TGF-β1 promotes the expression miR-155 in HCC (267). MiR-155 is also upregulated in diffuse large B-cell lymphoma (269,270), another malignancy related to milk consumption (10). A relationship between serum miR-155 and telomerase expression has been observed in HCC (271). MiR-155 downregulates suppressor of cytokine signaling 1 (SOCS1), resulting in activation of STAT3 and STAT3-induced transcription of miR-21 (272,273). MiR-155 targets tumor protein p53-inducible nuclear protein 1 (TP53INP1) regulating liver CSC acquisition and self-renewal (274). Under hypoxic conditions, HCC cells secrete miR-155-enriched exosomes enhancing angiogenesis in endothelial cells (275). Finally, miR-155 has been identified as a biomarker for tumor recurrence and survival of HCC patients following orthotopic liver transplantation (276) (Table 3).
Full table
The role of EVs in mediating HCC progression and metastasis as well as HCC therapy is in the focus of recent research (178,277). Importantly, miR-155 is enriched in exosomes released from HCC cells and exosome-derived miR-155 is transferred into targeted cells increasing HCC cell proliferation (251,252). HCV proteins associate with the membrane tetraspanin CD81 for HCV infection of hepatocytes (278). HCV envelope glycoprotein E2 binds to CD81 associated with exosomes that were captured from the plasma of HCV-infected patients (279). In analogy to HCV, HBx can tweak the exosome biogenesis machinery both by enhancing neutral sphingomyelinase 2 activity as well as by interacting with exosomal neutral sphingomyelinase 2, CD9 and CD81 (280). Enhanced secretion of exosomes by HBx-expressing cells has been confirmed with co-localization of HBx with CD9 and CD63 (280).
Milk exosomes also contain CD9, CD63 and CD81, all dominant milk exosome membrane proteins (165,166) that may augment HBV and HCV transmission. In analogy with HBV- and HCV-induced exosome secretion, milk exosome-derived miR-21 and miR-155 (229,230,281-284) via milk exosome uptake may modify hepatocyte gene transcription promoting HCC.
Notably, the liver is the major organ for the uptake of orally administered milk exosomes (164,284). Thus, not only HCC-derived exosomal miR-155 but also milk exosome-derived miR-155 may converge in exosome-induced hepatocancerogenesis (285). Of interest, the combined panel of miR-155, miR-96, miR-99a and AFP has a higher sensitivity and specificity for the diagnosis of HCC when compared with each single marker (244).
MiR-29b
MiR-29b is a bioavailable bovine milk miR, which increases in plasma and peripheral blood mononuclear cells of healthy volunteers 4 to 6 hours after milk consumption (195). MiR-29b plays a key role in hepatic IR (18,89) and is related to impaired hepatic BCAA catabolism (286-288). Human and bovine miR-29b exhibit identical sequences (mirbase.org). Intriguingly, miR-29b inhibits BCAA catabolism via targeting the E2 core component (DBT) of BCKDH (Figure 1) (289). In this regard, milk-derived miR-29b may augment compromised BCAA catabolism in HCC (112,113). Milk miR-29b-mediated suppression of BCAA catabolism during the breastfeeding period might conserve BCAAs for mTORC1-dependent growth and BCAA incorporation into structural and functional proteins such as albumin (290).
Milk exosomes: potential carriers of oncogenic viruses
Host exosome pathways are hijacked by viruses such as HBV, HCV, and EBV (291-297). Circulating virus particles may interact with exosomal tetraspanins of milk (165,166), a potential mechanism promoting cellular virus uptake. The detection of a novel HCV-like virus (BovHepV) in domestic cows with liver tropism is of concern (298,299), because it is not yet known whether BovHepV is able to enter milk exosomes and modify hepatic gene expression of milk consumers.
Furthermore, virus-like small circular single-stranded DNA (ssDNA) molecules have been isolated from commercial milk (300-302). These replication-competent genomic DNA sequences called bovine meat and milk factors (BMMF1 and BMMF2) are regarded as a specific class of infectious agents spanning between bacterial plasmid and circular ssDNA viruses with similarities to the genome structure and replication of hepatitis deltavirus (303). The possibility that these potentially oncogenic DNA sequences are transported via milk exosomes is under investigation (Harald zur Hausen, personal communication).
Milk exosomes for drug delivery
There are recent efforts to use exosomes for drug transfer as anti-cancer agents (277,304-307) and HCC treatment (180,308,309). Some investigators promote cow milk-derived exosomes as potential carriers for drug delivery (196,284,310,311). However, it should be kept in mind that milk exosomes per se carry oncogenic miRs (miR-21, miR-155) and TGF-β that should not augment oncogenic signaling cascades (160).
Aflatoxin
Ruminants metabolize AFB1 ingested by contaminated food to aflatoxin M1 (AFM1), the hydroxylated mycotoxin, which is excreted into milk (312,313). There is concern about an increase of AFM1 concentrations in milk of maize-fed cows due to the climate change (314). The International Agency for Research on Cancer classified AFB1 and AFM1 as human carcinogens of group 1Jeny (315-317). AFM1 is relatively stable during pasteurization, storage and processing (318-320). More than 50% of HCC patients from high aflatoxin exposure areas harbor a codon 249 G to T transversion in the p53 tumor suppressor gene, which is found to be consistent with the mutagenic specificity of AFB1 observed in vitro (321). This is of concern as milk signaling per se attenuates p53 activity by various mechanisms including AKT-mediated activation of mouse double minute 2 (MDM2) (322,323). As demonstrated with 1H NMR spectroscopy, AFB1 exposure of rats elevated hepatic BCAA levels (324). In accordance, AFM1 exposure of HepG2 cells enhanced cellular BCAA levels (325). Consumption of alcohol (2,000 kcal added to the diet) in human alcoholics for 2 to 4 weeks as well increased BCAA levels (326). In baboons, chronic alcohol intake over years increased plasma leucine concentrations and enhanced plasma BCAA levels with steatosis hepatis as well as hepatitis-fibrosis (Table 4) (326).
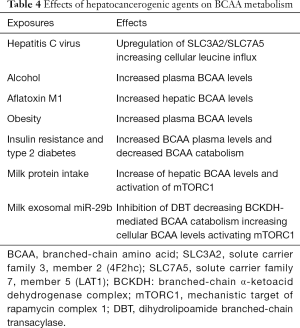
Full table
Liver cirrhosis and advanced HCC
In contrast to the unnoticed induction phase of BCAA-mTORC1-driven hepatocancerogenesis, direct supplementation of BCAAs in patients with advanced liver cirrhosis may reduce profibrotic signaling and prevent progressive liver failure (327). In patients with overt HCC, BCAA supplementation improved overall and disease-specific survival in those patients with low BCAA to tyrosine ratios despite having normal albumin levels (328). Overexpression of platelet-derived growth factor C (PDGF-C) promotes liver fibrosis, which is preceded by activation and proliferation of HSCs (329). In PDGF-C transgenic mice, an anti-fibrotic effect of BCAAs has been observed (330). In HSCs, BCAAs restored TGF-β1-stimulated expression of profibrotic genes, whereas in hepatocytes, BCAAs restored TGF-β1-induced apoptosis, lipogenesis, and Wnt/β-catenin signaling, and inhibited the transformation of WB-F344 rat liver epithelial stem-like cells. The inhibitory effect of BCAA on TGF-β1 signaling was mTORC1-dependent, suggesting a negative feedback regulation from mTORC1 to TGF-β1 (330). TGF-β1 activates HSC proliferation and primes HSCs for extracellular matrix deposition and scar contraction (331). In HFD-treated mice, HCC induction by the hepatic carcinogen diethylnitrosamine (DEN) resulted in mTORC1 inhibition, increase of IL-6, activation of STAT3 and HCC development (332). In a rat model of DEN-induced liver fibrosis and HCC, a diet containing either 3% casein, 3% or 6% BCAAs for 13 weeks beginning 6 weeks after DEN administration demonstrated less dysplastic foci and less numbers of HCC in the BCAA groups at 16 weeks of DEN administration compared to the casein group (333). However, at 19 weeks of DEN, there was a trend to higher HCC numbers in the BCAA groups compared to casein controls (333). In contrast, attenuation of mTORC1 activity by low-dose oral rapamycin reduced fibrogenesis, improved liver function, and prolonged survival in rats with bile duct ligation-induced liver cirrhosis (334). Obviously, the type of model, target cells and stage of HCC development are of importance for HCC-promoting or HCC-preventing effects of BCAAs.
Nevertheless, BCAA-mediated activation of hepatic mTORC1 is generally accepted to promote HCC development, progression and spreading (78,79,335,336). Activation of mTOR is more intense in the tumor edge, reinforcing its role in HCC proliferation and spreading (336). In addition, mTOR is overexpressed in multinodular HCC and is associated with increased post-liver transplantation tumor recurrence rates (336). mTORC1 upregulates SREBP1 via crosstalk with the STAT5 pathway, which contributes to NAFLD-related HCC pathogenesis (79). In HBV-associated tumorigenesis, the mTORC1 signal cascade also plays an important role in promoting de novo lipogenesis through activation of SREBP1 and ATP-citrate lyase (337). Loss-of-function of tuberin (TSC2), a key negative regulator of mTORC1, is common in HCC (338). Finally, the majority of therapeutic interventions in HCC aim at decreasing AKT-mTORC1 signaling (339-342).
Inverse relation between yogurt intake and HCC risk
Epidemiological studies confirmed an inverse relationship between yogurt consumption and the risk of HCC (17,21) as well as T2DM (20,343,344), two related pathologies exhibiting disturbed BCAA catabolism (112,113,132-134). The combination of Streptococcus thermophiles and Lactobacillus delbrueckii subsp. bulgaricus is the classic starter in yogurt production. Lactic acid bacteria (LAB) degrade milk’s natural signaling capacity. LAB modify intestinal and systemic BCAA homeostasis, degrade milk proteins, and produce a-ketoacids including KIC (345-347), a physiological inhibitor of BCKDK (348). Probiotic yogurts also contain the strong BCKDK inhibitor octanoic acid (349,350). Via the portal vein, octanoic acid is taken up by the liver (Figure 1) (351). In rats, oral administration of octanoic acid activated hepatic BCKDH via suppression of BCKDK activity reducing BCAA plasma concentrations (350). Lactobacillus delbrueckii subsp. bulgaricus and Streptococcus thermophilus increase the intestinal ratio of Bacteroidetes to Firmicutes associated with increased butyrate production (352-354). The butyrate-derivative phenylbutyrate (PB) is an allosteric inhibitor of BCKDK and reduces BCAA plasma concentrations (355). Activated BCKDK promotes CRC via direct MEK phosphorylation (356). A cancer-preventive effect of PB has been observed in two liver carcinoma cell lines (Bel-7402, HepG2) (357-360). Furthermore, LAB-mediated fermentation of milk decreased the size of milk exosomes, exosome protein and miR-21 as well as miR-29b content (361). Yogurt intake increased the density of LAB and C. perfringens and reduced Bacteroides (362). Bacteroides vulgatus is a major producer of intestinal BCAAs that has been related to IR (363).
Thus, the HCC- and T2DM-preventive effect of yogurt consumption may reside in LAB-mediated improvements of intestinal and systemic BCAA homeostasis via suppression of intestinal BCAA synthesis and hepatic BCKDK resulting in upregulated BCKDH activity and reduced plasma BCAA levels (Figure 1).
Summary and conclusions
On the basis of epidemiological and translational evidence, it can be concluded that total dairy, milk and butter are critical risk factors of HCC. Increased intake of milk protein-derived BCAAs and saturated milk fat promote hepatic IGF-1 and mTORC1 activation. HCC-related deviations on BCAA catabolism combined with extensive BCAA intake promote excessive mTORC1 activation. Milk-derived exosomes and their oncogenic miRs apparently synergize with oncogenic signaling of HCC-derived exosomes in hepatocancerogenesis.
For HCC prevention, dietary reduction of BCAAs should be pursued in accordance with recommendations for the prevention of IR and T2DM (25,364). Notably, the most common antidiabetic drug metformin exhibits potential for the prevention and adjuvant treatment of HCC (365-368). Metformin inhibits mTORC1 (369,370), suppresses BCAT2 associated with a reduction of KIC (371), and reduces circulating BCAA levels (372), thus operates in the opposite direction of milk signaling (28,373).
The Canadian Dietary Guidelines recommend an animal protein-reduced and vegetable-accentuated diet (374), which apparently decreases the risk of HCC (375,376) including milk-related overall and cancer mortality (8,22,377-380). Milk exosomes and their oncogenic miRs have to be removed from the human food chain (160) as substantial evidence implies that milk exosome-derived oncogenic miRs promote HCC in synergy with HBV-/HCV-induced hepatic exosomes. Thus, accumulating evidence adds milk, cheese and butter with the exception of yogurt to the list of dairy-related risk factors of HCC (Table 5).
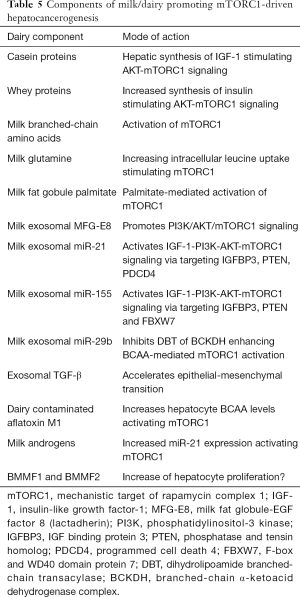
Full table
Acknowledgments
Funding: None.
Footnote
Provenance and Peer Review: This article was commissioned by the Guest Editors (Ralf Weiskirchen and Wolfgang Stremmel) for the series “Unresolved Basic Issues in Hepatology” published in Annals of Translational Medicine. The article was sent for external peer review organized by the Guest Editors and the editorial office.
Conflicts of Interest: The author has completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/atm-2020-ubih-06). The series “Unresolved Basic Issues in Hepatology” was commissioned by the editorial office without any funding or sponsorship. The author has no other conflicts of interest to declare.
Ethical Statement: The author is accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work is appropriately investigated and resolved.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Bertuccio P, Turati F, Carioli G, et al. Global trends and predictions in hepatocellular carcinoma mortality. J Hepatol 2017;67:302-9. [Crossref] [PubMed]
- Chacko S, Samanta S. Hepatocellular carcinoma: A life-threatening disease. Biomed Pharmacother 2016;84:1679-88. [Crossref] [PubMed]
- Yang JD, Hainaut P, Gores GJ, et al. A global view of hepatocellular carcinoma: trends, risk, prevention and management. Nat Rev Gastroenterol Hepatol 2019;16:589-604. [Crossref] [PubMed]
- Altekruse SF, McGlynn KA, Reichman ME. Hepatocellular carcinoma incidence, mortality, and survival trends in the United States from 1975 to 2005. J Clin Oncol 2009;27:1485-91. [Crossref] [PubMed]
- Baecker A, Liu X, La Vecchia C, et al. Worldwide incidence of hepatocellular carcinoma cases attributable to major risk factors. Eur J Cancer Prev 2018;27:205-12. [Crossref] [PubMed]
- Sagnelli E, Macera M, Russo A, et al. Epidemiological and etiological variations in hepatocellular carcinoma. Infection 2020;48:7-17. [Crossref] [PubMed]
- Song Y, Chavarro JE, Cao Y, et al. Whole milk intake is associated with prostate cancer-specific mortality among U.S. male physicians. J Nutr 2013;143:189-96. [Crossref] [PubMed]
- Lu W, Chen H, Niu Y, et al. Dairy products intake and cancer mortality risk: a meta- analysis of 11 population-based cohort studies. Nutr J 2016;15:91. [Crossref] [PubMed]
- Tat D, Kenfield SA, Cowan JE, et al. Milk and other dairy foods in relation to prostate cancer recurrence: Data from the cancer of the prostate strategic urologic research endeavor (CaPSURE™). Prostate 2018;78:32-9. [Crossref] [PubMed]
- Wang J, Li X, Zhang D. Dairy product consumption and risk of Non-Hodgkin lymphoma: A meta-analysis. Nutrients 2016;8:120. [Crossref] [PubMed]
- Melnik BC, John SM, Carrera-Bastos P, et al. The impact of cow's milk-mediated mTORC1-signaling in the initiation and progression of prostate cancer. Nutr Metab (Lond) 2012;9:74. [Crossref] [PubMed]
- Fukuda K, Shibata A, Hirohata I, et al. A hospital-based case-control study on hepatocellular carcinoma in Fukuoka and Saga prefectures, northern Kyushu, Japan. Jpn J Cancer Res 1993;84:708-14. [Crossref] [PubMed]
- Kuper H, Tzonou A, Lagiou P, et al. Diet and hepatocellular carcinoma: a case-control study in Greece. Nutr Cancer 2000;38:6-12. [Crossref] [PubMed]
- Talamini R, Polesel J, Montella M, et al. Food groups and risk of hepatocellular carcinoma: A multicenter case-control study in Italy. Int J Cancer 2006;119:2916-21. [Crossref] [PubMed]
- La Vecchia C, Negri E, Decarli A, et al. Risk factors for hepatocellular carcinoma in northern Italy. Int J Cancer 1988;42:872-6. [Crossref] [PubMed]
- Yu SZ, Huang XE, Koide T, et al. Hepatitis B and C viruses infection, lifestyle and genetic polymorphisms as risk factors for hepatocellular carcinoma in Haimen, China. Jpn J Cancer Res 2002;93:1287-92. [Crossref] [PubMed]
- Yang W, Sui J, Ma Y, et al. A prospective study of dairy product intake and the risk of hepatocellular carcinoma in U.S. men and women. Int J Cancer 2020;146:1241-9. [Crossref] [PubMed]
- Melnik BC. The pathogenic role of persistent milk signaling in mTORC1- and milk- microRNA-driven type 2 diabetes mellitus. Curr Diabetes Rev 2015;11:46-62. [Crossref] [PubMed]
- Brouwer-Brolsma EM, Sluik D, Singh-Povel CM, et al. Dairy product consumption is associated with pre-diabetes and newly diagnosed type 2 diabetes in the Lifelines Cohort Study. Br J Nutr 2018;119:442-5. [Crossref] [PubMed]
- Sluijs I, Forouhi NG, Beulens JW, et al. The amount and type of dairy product intake and incident type 2 diabetes: results from the EPIC-InterAct Study. Am J Clin Nutr 2012;96:382-90. [Crossref] [PubMed]
- Duarte-Salles T, Fedirko V, Stepien M, et al. Dairy products and risk of hepatocellular carcinoma: the European Prospective Investigation into Cancer and Nutrition. Int J Cancer 2014;135:1662-72. [Crossref] [PubMed]
- Wang XJ, Jiang CQ, Zhang WS, et al. Milk consumption and risk of mortality from all-cause, cardiovascular disease and cancer in older people. Clin Nutr 2020;39:3442-51. [Crossref] [PubMed]
- Available online: https://www.statista.com/statistics/448642/per-capita-consumption-of-milk-in-the-us/
- https://milchindustrie.de/wpcontent/uploads/2018/11/ProkopfDeutschland_Mopro_2010-2018x_Homepage.pdf
- Melnik BC. Leucine signaling in the pathogenesis of type 2 diabetes and obesity. World J Diabetes 2012;3:38-53. [Crossref] [PubMed]
- Available online: https://www.statista.com/statistics/868231/global-annual-consumption-of-cheese- by-country/
- Melnik BC, John SM, Schmitz G. Milk is not just food but most likely a genetic transfection system activating mTORC1 signaling for postnatal growth. Nutr J 2013;12:103. [Crossref] [PubMed]
- Melnik BC. Milk--A nutrient system of mammalian evolution promoting mTORC1- dependent translation. Int J Mol Sci 2015;16:17048-87. [Crossref] [PubMed]
- Melnik BC, Schmitz G. Milk's role as an epigenetic regulator in health and disease. Diseases 2017;5:12. [Crossref] [PubMed]
- Young NJ, Metcalfe C, Gunnell D, et al. A cross-sectional analysis of the association between diet and insulin-like growth factor (IGF)-I, IGF-II, IGF-binding protein (IGFBP)-2, and IGFBP-3 in men in the United Kingdom. Cancer Causes Control 2012;23:907-17. [Crossref] [PubMed]
- Harrison S, Lennon R, Holly J, et al. Does milk intake promote prostate cancer initiation or progression via effects on insulin-like growth factors (IGFs)? A systematic review and meta-analysis. Cancer Causes Control 2017;28:497-528. [Crossref] [PubMed]
- Crowe FL, Key TJ, Allen NE, et al. The association between diet and serum concentrations of IGF-I, IGFBP-1, IGFBP-2, and IGFBP-3 in the European Prospective Investigation into Cancer and Nutrition. Cancer Epidemiol Biomarkers Prev 2009;18:1333-40. [Crossref] [PubMed]
- Qin LQ, He K, Xu JY. Milk consumption and circulating insulin-like growth factor-I level: a systematic literature review. Int J Food Sci Nutr 2009;60:330-40. [Crossref] [PubMed]
- Romo Ventura E, Konigorski S, Rohrmann S, et al. Association of dietary intake of milk and dairy products with blood concentrations of insulin-like growth factor 1 (IGF-1) in Bavarian adults. Eur J Nutr 2020;59:1413-20. [Crossref] [PubMed]
- Hoppe C, Mølgaard C, Dalum C, et al. Differential effects of casein versus whey on fasting plasma levels of insulin, IGF-1 and IGF-1/IGFBP-3: results from a randomized 7-day supplementation study in prepubertal boys. Eur J Clin Nutr 2009;63:1076-83. [Crossref] [PubMed]
- Daughaday WH, Rotwein P. Insulin-like growth factors I and II. Peptide, messenger ribonucleic acid and gene structures, serum, and tissue concentrations. Endocr Rev 1989;10:68-91. [Crossref] [PubMed]
- Ohlsson C, Mohan S, Sjögren K, et al. The role of liver-derived insulin-like growth factor-I. Endocr Rev 2009;30:494-535. [Crossref] [PubMed]
- Wan X, Wang S, Xu J, et al. Dietary protein-induced hepatic IGF-1 secretion mediated by PPARγ activation. PLoS One 2017;12:e0173174 [Crossref] [PubMed]
- Pao CI, Farmer PK, Begovic S, et al. Regulation of insulin-like growth factor-I (IGF-I) and IGF-binding protein 1 gene transcription by hormones and provision of amino acids in rat hepatocytes. Mol Endocrinol 1993;7:1561-8. [PubMed]
- Yin J, Ren W, Huang X, et al. Protein restriction and cancer. Biochim Biophys Acta Rev Cancer 2018;1869:256-62. [Crossref] [PubMed]
- Levine ME, Suarez JA, Brandhorst S, et al. Low protein intake is associated with a major reduction in IGF-1, cancer, and overall mortality in the 65 and younger but not older population. Cell Metab 2014;19:407-17. [Crossref] [PubMed]
- Schulsinger DA, Root MM, Campbell TC. Effect of dietary protein quality on development of aflatoxin B1-induced hepatic preneoplastic lesions. J Natl Cancer Inst 1989;81:1241-5. [Crossref] [PubMed]
- Cheng Z, Hu J, King J, et al. Inhibition of hepatocellular carcinoma development in hepatitis B virus transfected mice by low dietary casein. Hepatology 1997;26:1351-4. [PubMed]
- Hu JF, Cheng Z, Chisari FV, et al. Repression of hepatitis B virus (HBV) transgene and HBV-induced liver injury by low protein diet. Oncogene 1997;15:2795-801. [Crossref] [PubMed]
- Adamek A, Kasprzak A. Insulin-like growth factor (IGF) system in liver diseases. Int J Mol Sci 2018;19:1308. [Crossref] [PubMed]
- Price JA, Kovach SJ, Johnson T, et al. Insulin-like growth factor I is a comitogen for hepatocyte growth factor in a rat model of hepatocellular carcinoma. Hepatology 2002;36:1089-97. [Crossref] [PubMed]
- Ma Y, Han CC, Li Y, et al. Insulin-like growth factor-binding protein-3 inhibits IGF- 1-induced proliferation of human hepatocellular carcinoma cells by controlling bFGF and PDGF autocrine/paracrine loops. Biochem Biophys Res Commun 2016;478:964-9. [Crossref] [PubMed]
- Lei T, Ling X. IGF-1 promotes the growth and metastasis of hepatocellular carcinoma via the inhibition of proteasome-mediated cathepsin B degradation. World J Gastroenterol 2015;21:10137-49. [Crossref] [PubMed]
- Zhao C, Wang Q, Wang B, et al. IGF-1 induces the epithelial-mesenchymal transition via Stat5 in hepatocellular carcinoma. Oncotarget 2017;8:111922-30. [Crossref] [PubMed]
- Liu F, Sun Y, Liu B, et al. Insulin-like growth factor-1 induces epithelial-mesenchymal transition in hepatocellular carcinoma by activating survivin. Oncol Rep 2018;40:952-8. [Crossref] [PubMed]
- Nielsen KO, Mirza AH, Kaur S, et al. Hepatitis B virus suppresses the secretion of insulin-like growth factor binding protein 1 to facilitate anti-apoptotic IGF-1 effects in HepG2 cells. Exp Cell Res 2018;370:399-408. [Crossref] [PubMed]
- Al-Gayyar MMH, Bagalagel A, Noor AO, et al. The therapeutic effects of nicotinamide in hepatocellular carcinoma through blocking IGF-1 and effecting the balance between Nrf2 and PKB. Biomed Pharmacother 2019;112:108653 [Crossref] [PubMed]
- Zhang YW, Yan DL, Wang W, et al. Knockdown of insulin-like growth factor I receptor inhibits the growth and enhances chemo-sensitivity of liver cancer cells. Curr Cancer Drug Targets 2012;12:74-84. [Crossref] [PubMed]
- Yan XD, Yao M, Wang L, et al. Overexpression of insulin-like growth factor-I receptor as a pertinent biomarker for hepatocytes malignant transformation. World J Gastroenterol 2013;19:6084-92. [Crossref] [PubMed]
- Yao M, Wang L, Yang J, et al. IGF-I receptor as an emerging potential molecular- targeted for hepatocellular carcinoma in vitro and in vivo. Tumour Biol 2016;37:14677-86. [Crossref] [PubMed]
- Pivonello C, Negri M, De Martino MC, et al. The dual targeting of insulin and insulin- like growth factor 1 receptor enhances the mTOR inhibitor-mediated antitumor efficacy in hepatocellular carcinoma. Oncotarget 2016;7:9718-31. [Crossref] [PubMed]
- Bie CQ, Liu XY, Cao MR, et al. Lentivirus-mediated RNAi knockdown of insulin-like growth factor-1 receptor inhibits the growth and invasion of hepatocellular carcinoma via down-regulating midkine expression. Oncotarget 2016;7:79305-18. [Crossref] [PubMed]
- Wang F, Bank T, Malnassy G, et al. Inhibition of insulin-like growth factor 1 receptor enhances the efficacy of sorafenib in inhibiting hepatocellular carcinoma cell growth and survival. Hepatol Commun 2018;2:732-46. [Crossref] [PubMed]
- Zhang Z, Lei B, Chai W, et al. Increased expression of insulin-like growth factor-1 receptor predicts poor prognosis in patients with hepatocellular carcinoma. Medicine (Baltimore) 2019;98:e17680 [Crossref] [PubMed]
- Wang L, Yao M, Zheng W, et al. Insulin-like growth factor I receptor: A novel target for hepatocellular carcinoma gene therapy. Mini Rev Med Chem 2019;19:272-80. [Crossref] [PubMed]
- Adamek A, Kasprzak A, Mikoś H, et al. The insulin-like growth factor-1 and expression of its binding protein-3 in chronic hepatitis C and hepatocellular carcinoma. Oncol Rep 2013;30:1337-45. [Crossref] [PubMed]
- Elmashad N, Ibrahim WS, Mayah WW, et al. Predictive value of serum insulin-like growth factor-1 in hepatocellular carcinoma. Asian Pac J Cancer Prev 2015;16:613-9. [Crossref] [PubMed]
- Takahashi Y. The role of growth hormone and insulin-like growth factor-I in the liver. Int J Mol Sci 2017;18:1447. [Crossref] [PubMed]
- Lee DH, Lee JH, Jung YJ, et al. Validation of a modified Child-Turcotte-Pugh classification system utilizing insulin-like growth factor-1 for patients with hepatocellular carcinoma in an HBV endemic area. PLoS One 2017;12:e0170394 [Crossref] [PubMed]
- Yao Y, Mao W, Dong M, et al. Serum insulin-like growth factor-1 (IGF-1): a novel prognostic factor for early recurrence of hepatocellular carcinoma (HCC). Clin Lab 2017;63:261-70. [Crossref] [PubMed]
- Wang J, Li YC, Deng M, et al. Serum insulin-like growth factor-1 and its binding protein 3 as prognostic factors for the incidence, progression, and outcome of hepatocellular carcinoma: a systematic review and meta-analysis. Oncotarget 2017;8:81098-108. [Crossref] [PubMed]
- Huber Y, Bierling F, Labenz C, et al. Validation of insulin-like growth factor-1 as a prognostic parameter in patients with hepatocellular carcinoma in a European cohort. BMC Cancer 2018;18:774. [Crossref] [PubMed]
- Shen L, Xu L, Zhang J, et al. Preoperative serum insulin-like growth factor 1 level as a prognostic factor in patients undergoing hepatic resection for hepatocellular carcinoma. J Interferon Cytokine Res 2018;38:153-60. [Crossref] [PubMed]
- Abdel-Wahab R, Shehata S, Hassan MM, et al. Type I insulin-like growth factor as a liver reserve assessment tool in hepatocellular carcinoma. J Hepatocell Carcinoma 2015;2:131-42. [PubMed]
- Whittaker S, Marais R, Zhu AX. The role of signaling pathways in the development and treatment of hepatocellular carcinoma. Oncogene 2010;29:4989-5005. [Crossref] [PubMed]
- Menon S, Yecies JL, Zhang HH, et al. Chronic activation of mTOR complex 1 is sufficient to cause hepatocellular carcinoma in mice. Sci Signal 2012;5:ra24. [Crossref] [PubMed]
- Calvisi DF, Wang C, Ho C, et al. Increased lipogenesis, induced by AKT-mTORC1-RPS6 signaling, promotes development of human hepatocellular carcinoma. Gastroenterology 2011;140:1071-83. [Crossref] [PubMed]
- Wang C, Cigliano A, Delogu S, et al. Functional crosstalk between AKT/mTOR and Ras/MAPK pathways in hepatocarcinogenesis: implications for the treatment of human liver cancer. Cell Cycle 2013;12:1999-2010. [Crossref] [PubMed]
- Samarin J, Laketa V, Malz M, et al. PI3K/AKT/mTOR-dependent stabilization of oncogenic far-upstream element binding proteins in hepatocellular carcinoma cells. Hepatology 2016;63:813-26. [Crossref] [PubMed]
- Singal SS, Nygard K, Dhruv MR, et al. Co-localization of insulin-like growth factor binding protein-1, casein kinase-2β, and mechanistic target of rapamycin in human hepatocellular carcinoma cells as demonstrated by dual immunofluorescence and in situ proximity ligation assay. Am J Pathol 2018;188:111-24. [Crossref] [PubMed]
- Liu P, Ge M, Hu J, et al. A functional mammalian target of rapamycin complex 1 signaling is indispensable for c-Myc-driven hepatocarcinogenesis. Hepatology 2017;66:167-81. [Crossref] [PubMed]
- Sun H, Yang W, Tian Y, et al. An inflammatory-CCRK circuitry drives mTORC1- dependent metabolic and immunosuppressive reprogramming in obesity-associated hepatocellular carcinoma. Nat Commun 2018;9:5214. [Crossref] [PubMed]
- Akula SM, Abrams SL, Steelman LS, et al. RAS/RAF/MEK/ERK, PI3K/PTEN/AKT/ mTORC1 and TP53 pathways and regulatory miRs as therapeutic targets in hepatocellular carcinoma. Expert Opin Ther Targets 2019;23:915-29. [Crossref] [PubMed]
- Li T, Weng J, Zhang Y, et al. mTOR direct crosstalk with STAT5 promotes de novo lipid synthesis and induces hepatocellular carcinoma. Cell Death Dis 2019;10:619. [Crossref] [PubMed]
- Foster KG, Fingar DC. Mammalian target of rapamycin (mTOR): conducting the cellular signaling symphony. J Biol Chem 2010;285:14071-7. [Crossref] [PubMed]
- Zheng X, Liang Y, He Q, et al. Current models of mammalian target of rapamycin complex 1 (mTORC1) activation by growth factors and amino acids. Int J Mol Sci 2014;15:20753-69. [Crossref] [PubMed]
- Moberg M, Apró W, Ekblom B, et al. Activation of mTORC1 by leucine is potentiated by branched-chain amino acids and even more so by essential amino acids following resistance exercise. Am J Physiol Cell Physiol 2016;310:C874-84. [Crossref] [PubMed]
- Jewell JL, Kim YC, Russell RC, et al. Metabolism. Differential regulation of mTORC1 by leucine and glutamine. Science 2015;347:194-8. [Crossref] [PubMed]
- Wolfson RL, Sabatini DM. The dawn of the age of amino acid sensors for the mTORC1 pathway. Cell Metab 2017;26:301-9. [Crossref] [PubMed]
- Nguyen TP, Frank AR, Jewell JL. Amino acid and small GTPase regulation of mTORC1. Cell Logist 2017;7:e1378794 [Crossref] [PubMed]
- Mossmann D, Park S, Hall MN. mTOR signalling and cellular metabolism are mutual determinants in cancer. Nat Rev Cancer 2018;18:744-57. [Crossref] [PubMed]
- Zhuang Y, Wang XX, He J, et al. Recent advances in understanding of amino acid signaling to mTORC1 activation. Front Biosci (Landmark Ed) 2019;24:971-82. [Crossref] [PubMed]
- Li XZ, Yan XH. Sensors for the mTORC1 pathway regulated by amino acids. J Zhejiang Univ Sci B 2019;20:699-712. [Crossref] [PubMed]
- Melnik BC, Schmitz G. The impact of persistent milk consumption in the pathogenesis of type 2 diabetes mellitus. Functional Foods in Health and Disease 2019;9:629-47. [Crossref]
- Lenders CM, Liu S, Wilmore DW, et al. Evaluation of a novel food composition database that includes glutamine and other amino acids derived from gene sequencing data. Eur J Clin Nutr 2009;63:1433-9. [Crossref] [PubMed]
- Wang Q, Holst J. L-type amino acid transport and cancer: targeting the mTORC1 pathway to inhibit neoplasia. Am J Cancer Res 2015;5:1281-94. [PubMed]
- Salisbury TB, Arthur S. The Regulation and Function of the L-Type Amino Acid Transporter 1 (LAT1) in Cancer. Int J Mol Sci 2018;19:2373. [Crossref] [PubMed]
- Yue M, Jiang J, Gao P, et al. Oncogenic MYC activates a feedforward regulatory loop promoting essential amino acid metabolism and tumorigenesis. Cell Rep 2017;21:3819-32. [Crossref] [PubMed]
- Li J, Qiang J, Chen SF, et al. The impact of L-type amino acid transporter 1 (LAT1) in human hepatocellular carcinoma. Tumour Biol 2013;34:2977-81. [Crossref] [PubMed]
- Namikawa M, Kakizaki S, Kaira K, et al. Expression of amino acid transporters (LAT1, ASCT2 and xCT) as clinical significance in hepatocellular carcinoma. Hepatol Res 2015;45:1014-22. [Crossref] [PubMed]
- Ogihara K, Naya Y, Sato R, et al. Analysis of L-type amino acid transporter in canine hepatocellular carcinoma. J Vet Med Sci 2015;77:527-34. [Crossref] [PubMed]
- Mohamed RH, Abu-Shahba N, Mahmoud M, et al. Co-regulatory network of oncosuppressor miRNAs and transcription factors for pathology of human hepatic cancer stem cells (HCSC). Sci Rep 2019;9:5564. [Crossref] [PubMed]
- Nicklin P, Bergman P, Zhang B, et al. Bidirectional transport of amino acids regulate mTOR and autophagy. Cell 2009;136:521-34. [Crossref] [PubMed]
- Yang Y, Li C, Nie X, et al. Metabonomic studies of human hepatocellular carcinoma using high-resolution magic-angle spinning 1H NMR spectroscopy in conjunction with multivariate data analysis. J Proteome Res 2007;6:2605-14. [Crossref] [PubMed]
- Wang Q, Bailey CG, Ng C, et al. Androgen receptor and nutrient signaling pathways coordinate the demand for increased amino acid transport during prostate cancer progression. Cancer Res 2011;71:7525-36. [Crossref] [PubMed]
- Yanagisawa N, Satoh T, Hana K, et al. L-amino acid transporter 1 may be a prognostic marker for local progression of prostatic cancer under expectant management. Cancer Biomark 2015;15:365-74. [Crossref] [PubMed]
- Bar Yamin H, Barnea M, Genzer Y, et al. Long-term commercial cow's milk consumption and its effects on metabolic parameters associated with obesity in young mice. Mol Nutr Food Res 2014;58:1061-8. [Crossref] [PubMed]
- Zou H, Liao M, Xu W, et al. Data mining of the expression and regulatory role of BCAT1 in hepatocellular carcinoma. Oncol Lett 2019;18:5879-88. [Crossref] [PubMed]
- Zhao H, Zhang F, Sun D, et al. Branched-chain amino acids exacerbate obesity- related hepatic glucose and lipid metabolic disorders via attenuating Akt2 signaling. Diabetes 2020;69:1164-77. [Crossref] [PubMed]
- Han JM, Jeong SJ, Park MC, et al. Leucyl-tRNA synthetase is an intracellular leucine sensor for the mTORC1-signaling pathway. Cell 2012;149:410-24. [Crossref] [PubMed]
- Kim E, Goraksha-Hicks P, Li L, et al. Regulation of TORC1 by Rag GTPases in nutrient response. Nat Cell Biol 2008;10:935-45. [Crossref] [PubMed]
- Sancak Y, Peterson TR, Shaul YD, et al. The Rag GTPases bind raptor and mediate amino acid signaling to mTORC1. Science 2008;320:1496-501. [Crossref] [PubMed]
- Thomas JD, Zhang YJ, Wei YH, et al. Rab1A is an mTORC1 activator and a colorectal oncogene. Cancer Cell 2014;26:754-69. [Crossref] [PubMed]
- Abraham RT. Cell biology. Making sense of amino acid sensing. Science 2015;347:128-9. [Crossref] [PubMed]
- Sanchez-Gurmaches J, Guertin DA. mTORC1 gRABs the Golgi. Cancer Cell 2014;26:601-3. [Crossref] [PubMed]
- Xu BH, Li XX, Yang Y, et al. Aberrant amino acid signaling promotes growth and metastasis of hepatocellular carcinomas through Rab1A-dependent activation of mTORC1 by Rab1A. Oncotarget 2015;6:20813-28. [Crossref] [PubMed]
- Ericksen RE, Lim SL, McDonnell E, et al. Loss of BCAA Catabolism during Carcinogenesis Enhances mTORC1 Activity and Promotes Tumor Development and Progression. Cell Metab 2019;29:1151-1165.e6. [Crossref] [PubMed]
- Ericksen RE, Han W. Malignant manipulaTORs of metabolism: suppressing BCAA catabolism to enhance mTORC1 activity. Mol Cell Oncol 2019;6:1585171 [Crossref] [PubMed]
- Tian Q, Yuan P, Quan C, et al. Phosphorylation of BCKDK of BCAA catabolism at Y246 by Src promotes metastasis of colorectal cancer. Oncogene 2020;39:3980-96. [Crossref] [PubMed]
- White PJ, McGarrah RW, Grimsrud PA, et al. The BCKDH Kinase and Phosphatase Integrate BCAA and Lipid Metabolism via Regulation of ATP-Citrate Lyase. Cell Metab 2018;27:1281-1293.e7. [Crossref] [PubMed]
- Bagga S, Rawat S, Ajenjo M, et al. Hepatitis B virus (HBV) X protein-mediated regulation of hepatocyte metabolic pathways affects viral replication. Virology 2016;498:9-22. [Crossref] [PubMed]
- Wang X, Huo B, Liu J, et al. Hepatitis B virus X reduces hepatocyte apoptosis and promotes cell cycle progression through the Akt/mTOR pathway in vivo. Gene 2019;691:87-95. [Crossref] [PubMed]
- Zhu M, Guo J, Li W, et al. Hepatitis B virus X protein induces expression of alpha- fetoprotein and activates PI3K/mTOR signaling pathway in liver cells. Oncotarget 2015;6:12196-208. [Crossref] [PubMed]
- Zhu M, Li W, Lu Y, et al. HBx drives alpha fetoprotein expression to promote initiation of liver cancer stem cells through activating PI3K/AKT signal pathway. Int J Cancer 2017;140:1346-55. [Crossref] [PubMed]
- Tian Y, Xiao X, Gong X, et al. HBx promotes cell proliferation by disturbing the cross-talk between miR-181a and PTEN. Sci Rep 2017;7:40089. [Crossref] [PubMed]
- Nguyen NNT, Lim YS, Nguyen LP, et al. Hepatitis C virus modulates solute carrier family 3 member 2 for viral propagation. Sci Rep 2018;8:15486. [Crossref] [PubMed]
- Mu W, Cheng XF, Liu Y, et al. Potential nexus of non-alcoholic fatty liver disease and type 2 diabetes mellitus: insulin resistance between hepatic and peripheral tissues. Front Pharmacol 2019;9:1566. [Crossref] [PubMed]
- Hoppe C, Mølgaard C, Vaag A, et al. High intakes of milk, but not meat, increase s- insulin and insulin resistance in 8-year-old boys. Eur J Clin Nutr 2005;59:393-8. [Crossref] [PubMed]
- Felig P, Marliss E, Cahill GFJ Jr. Plasma amino acid levels and insulin secretion in obesity. N Engl J Med 1969;281:811-6. [Crossref] [PubMed]
- Newgard CB, An J, Bain JR, et al. A branched-chain amino acid-related metabolic signature that differentiates obese and lean humans and contributes to insulin resistance. Cell Metab 2009;9:311-26. [Crossref] [PubMed]
- Lynch CJ, Adams SH. Branched-chain amino acids in metabolic signalling and insulin resistance. Nat Rev Endocrinol 2014;10:723-36. [Crossref] [PubMed]
- Lu J, Xie G, Jia W, et al. Insulin resistance and the metabolism of branched-chain amino acids. Front Med 2013;7:53-9. [Crossref] [PubMed]
- She P, Olson KC, Kadota Y, et al. Leucine and protein metabolism in obese Zucker rats. PLoS One 2013;8:e59443 [Crossref] [PubMed]
- Menni C, Fauman E, Erte I, et al. Biomarkers for type 2 diabetes and impaired fasting glucose using a nontargeted metabolomics approach. Diabetes 2013;62:4270-6. [Crossref] [PubMed]
- Lu Y, Wang Y, Ong CN, et al. Metabolic signatures and risk of type 2 diabetes in a Chinese population: an untargeted metabolomics study using both LC-MS and GC- MS. Diabetologia 2016;59:2349-59. [Crossref] [PubMed]
- Chen T, Ni Y, Ma X, et al. Branched-chain and aromatic amino acid profiles and diabetes risk in Chinese populations. Sci Rep 2016;6:20594. [Crossref] [PubMed]
- White PJ, Lapworth AL, An J, et al. Branched-chain amino acid restriction in Zucker- fatty rats improves muscle insulin sensitivity by enhancing efficiency of fatty acid oxidation and acyl-glycine export. Mol Metab 2016;5:538-51. [Crossref] [PubMed]
- Lotta LA, Scott RA, Sharp SJ, et al. Genetic predisposition to an impaired metabolism of the branched-chain amino acids and risk of type 2 diabetes: A Mendelian randomisation analysis. PLoS Med 2016;13:e1002179 [Crossref] [PubMed]
- Zhou M, Jing S, Wu CY, et al. Targeting BCAA catabolism to treat obesity-associated insulin resistance. Diabetes 2019;68:1730-46. [Crossref] [PubMed]
- Chen J, Du B. Novel positioning from obesity to cancer: FTO, an m6A RNA demethylase, regulates tumour progression. J Cancer Res Clin Oncol 2019;145:19-29. [Crossref] [PubMed]
- Cheung MK, Gulati P, O'Rahilly S, et al. FTO expression is regulated by availability of essential amino acids. Int J Obes (Lond) 2013;37:744-7. [Crossref] [PubMed]
- Yeo GS. The role of the FTO (Fat Mass and Obesity Related) locus in regulating body size and composition. Mol Cell Endocrinol 2014;397:34-41. [Crossref] [PubMed]
- Gulati P, Cheung MK, Antrobus R, et al. Role for the obesity-related FTO gene in the cellular sensing of amino acids. Proc Natl Acad Sci U S A 2013;110:2557-62. [Crossref] [PubMed]
- Melnik BC. Milk: an epigenetic amplifier of FTO-mediated transcription? Implications for Western diseases. J Transl Med 2015;13:385. [Crossref] [PubMed]
- Li J, Zhu L, Shi Y, et al. m6A demethylase FTO promotes hepatocellular carcinoma tumorigenesis via mediating PKM2 demethylation. Am J Transl Res 2019;11:6084-92. [PubMed]
- He CL, Bian YY, Xue Y, et al. Pyruvate kinase M2 activates mTORC1 by phosphorylating AKT1S1. Sci Rep 2016;6:21524. [Crossref] [PubMed]
- Imam H, Khan M, Gokhale NS, et al. N6-methyladenosine modification of hepatitis B virus RNA differentially regulates the viral life cycle. Proc Natl Acad Sci U S A 2018;115:8829-34. [Crossref] [PubMed]
- Hetherington AM, Sawyez CG, Zilberman E, et al. Differential lipotoxic effects of palmitate and oleate in activated human hepatic stellate cells and epithelial hepatoma cells. Cell Physiol Biochem 2016;39:1648-62. [Crossref] [PubMed]
- Silva Figueiredo P, Carla Inada A, Marcelino G, et al. Fatty acids consumption: The role metabolic aspects involved in obesity and its associated disorders. Nutrients 2017;9:1158. [Crossref] [PubMed]
- Freedman ND, Cross AJ, McGlynn KA, et al. Association of meat and fat intake with liver disease and hepatocellular carcinoma in the NIH-AARP cohort. J Natl Cancer Inst 2010;102:1354-65. [Crossref] [PubMed]
- Hirsova P, Ibrahim SH, Gores GJ, et al. Lipotoxic lethal and sublethal stress signaling in hepatocytes: relevance to NASH pathogenesis. J Lipid Res 2016;57:1758-70. Erratum in: J Lipid Res 2017 Jan;58(1):299. doi: 10.1194/jlr.R066357ERR. [Crossref] [PubMed]
- Marra F, Svegliati-Baroni G. Lipotoxicity and the gut-liver axis in NASH pathogenesis. J Hepatol 2018;68:280-95. [Crossref] [PubMed]
- Seeßle J, Liebisch G, Schmitz G, et al. Palmitate activation by fatty acid transport protein 4 as a model system for hepatocellular apoptosis and steatosis. Biochim Biophys Acta 2015;1851:549-65. [Crossref] [PubMed]
- Xu W, Zhang X, Wu JL, et al. O-GlcNAc transferase promotes fatty liver-associated liver cancer through inducing palmitic acid and activating endoplasmic reticulum stress. J Hepatol 2017;67:310-20. [Crossref] [PubMed]
- Jensen RG, Ferris AM, Lammi-Keefe CJ. The composition of milk fat. J Dairy Sci 1991;74:3228-43. [Crossref] [PubMed]
- Jensen RG. The composition of bovine milk lipids: January 1995 to December 2000. J Dairy Sci 2002;85:295-350. [Crossref] [PubMed]
- Utsunomiya H, Yamamoto Y, Takeshita E, et al. Upregulated absorption of dietary palmitic acids with changes in intestinal transporters in non-alcoholic steatohepatitis (NASH). J Gastroenterol 2017;52:940-54. [Crossref] [PubMed]
- Yasuda M, Tanaka Y, Kume S, et al. Fatty acids are novel nutrient factors to regulate mTORC1 lysosomal localization and apoptosis in podocytes. Biochim Biophys Acta 2014;1842:1097-108. [Crossref] [PubMed]
- Kumar S, Tikoo K. Independent role of PP2A and mTORc1 in palmitate induced podocyte death. Biochimie 2015;112:73-84. [Crossref] [PubMed]
- Kwon B, Querfurth HW. Palmitate activates mTOR/p70S6K through AMPK inhibition and hypophosphorylation of raptor in skeletal muscle cells: Reversal by oleate is similar to metformin. Biochimie 2015;118:141-50. [Crossref] [PubMed]
- Engin A. Non-alcoholic fatty liver disease. Adv Exp Med Biol 2017;960:443-67. [Crossref] [PubMed]
- Bakan I, Laplante M. Connecting mTORC1 signaling to SREBP-1 activation. Curr Opin Lipidol 2012;23:226-34. [Crossref] [PubMed]
- Yecies JL, Zhang HH, Menon S, et al. Akt stimulates hepatic SREBP1c and lipogenesis through parallel mTORC1-dependent and independent pathways. Cell Metab 2011;14:21-32. Erratum in: Cell Metab 2011 Aug 3;14(2):280. [Crossref] [PubMed]
- Zempleni J, Aguilar-Lozano A, Sadri M, et al. Biological activities of extracellular vesicles and their cargos from bovine and human milk in humans and implications for infants. J Nutr 2017;147:3-10. [Crossref] [PubMed]
- Melnik BC, Schmitz G. Exosomes of pasteurized milk: potential pathogens of Western diseases. J Transl Med 2019;17:3. [Crossref] [PubMed]
- Benmoussa A, Laugier J, Beauparlant CJ, et al. Complexity of the microRNA transcriptome of cow milk and milk-derived extracellular vesicles isolated via differential ultracentrifugation. J Dairy Sci 2020;103:16-29. [Crossref] [PubMed]
- Yamada T, Inoshima Y, Matsuda T, et al. Comparison of methods for isolating exosomes from bovine milk. J Vet Med Sci 2012;74:1523-25. [Crossref] [PubMed]
- Benmoussa A, Lee CH, Laffont B, et al. Commercial dairy cow milk microRNAs resist digestion under simulated gastrointestinal tract conditions. J Nutr 2016;146:2206-15. [Crossref] [PubMed]
- Manca S, Upadhyaya B, Mutai E, et al. Milk exosomes are bioavailable and distinct microRNA cargos have unique tissue distribution patterns. Sci Rep 2018;8:11321. [Crossref] [PubMed]
- Reinhardt TA, Lippolis JD, Nonnecke BJ, et al. Bovine milk exosome proteome. J Proteomics 2012;75:1486-92. [Crossref] [PubMed]
- Benmoussa A, Gotti C, Bourassa S, et al. Identification of protein markers for extracellular vesicle (EV) subsets in cow's milk. J Proteomics 2019;192:78-88. [Crossref] [PubMed]
- Blans K, Hansen MS, Sørensen LV, et al. Pellet-free isolation of human and bovine milk extracellular vesicles by size-exclusion chromatography. J Extracell Vesicles 2017;6:1294340 [Crossref] [PubMed]
- Li H, Zhang T, Wang K, et al. MFGE8 protects against CCl4 -induced liver injury by reducing apoptosis and promoting proliferation of hepatocytes. J Cell Physiol. 2019; Epub ahead of print. [Crossref] [PubMed]
- Li H, Xu W, Ma Y, et al. Milk fat globule membrane protein promotes C2C12 cell proliferation through the PI3K/Akt signaling pathway. Int J Biol Macromol 2018;114:1305-14. [Crossref] [PubMed]
- Ko DS, Kim SH, Park JY, et al. Milk fat globule-EGF factor 8 contributes to progression of hepatocellular carcinoma. Cancers (Basel) 2020;12:403. [Crossref] [PubMed]
- Chen J, Zaidi S, Rao S, et al. Analysis of genomes and transcriptomes of hepatocellular carcinomas identifies mutations and gene expression changes in the transforming growth factor-β pathway. Gastroenterology 2018;154:195-210. [Crossref] [PubMed]
- Saad El-Din S, Fouad H, Rashed LA, et al. Impact of mesenchymal stem cells and vitamin D on transforming growth factor beta signaling pathway in hepatocellular carcinoma in rats. Asian Pac J Cancer Prev 2018;19:905-12. [PubMed]
- Arrese M, Hernandez A, Astete L, et al. TGF-β and hepatocellular carcinoma: When a friend becomes an enemy. Curr Protein Pept Sci 2018;19:1172-9. [Crossref] [PubMed]
- An Y, Gao S, Zhao WC, et al. Transforming growth factor-β and peripheral regulatory cells are negatively correlated with the overall survival of hepatocellular carcinoma. World J Gastroenterol 2018;24:2733-40. [Crossref] [PubMed]
- Yang C, Su H, Liao X, et al. Marker of proliferation Ki-67 expression is associated with transforming growth factor beta 1 and can predict the prognosis of patients with hepatic B virus-related hepatocellular carcinoma. Cancer Manag Res 2018;10:679-96. [Crossref] [PubMed]
- Li K, Liu H, Guo T. Th17/Treg imbalance is an indicator of liver cirrhosis process and a risk factor for HCC occurrence in HBV patients. Clin Res Hepatol Gastroenterol 2017;41:399-407. [Crossref] [PubMed]
- Pieters BC, Arntz OJ, Bennink MB, et al. Commercial cow milk contains physically stable extracellular vesicles expressing immunoregulatory TGF-β. PLoS One 2015;10:e0121123 [Crossref] [PubMed]
- He M, Qin H, Poon TC, et al. Hepatocellular carcinoma-derived exosomes promote motility of immortalized hepatocyte through transfer of oncogenic proteins and RNAs. Carcinogenesis 2015;36:1008-18. [Crossref] [PubMed]
- Wu Z, Zeng Q, Cao K, et al. Exosomes: small vesicles with big roles in hepatocellular carcinoma. Oncotarget 2016;7:60687-97. [Crossref] [PubMed]
- Pan JH, Zhou H, Zhao XX, et al. Role of exosomes and exosomal microRNAs in hepatocellular carcinoma: Potential in diagnosis and antitumour treatments Int J Mol Med 2018;41:1809-16. [Crossref] [PubMed]
- Liu H, Li B. The functional role of exosome in hepatocellular carcinoma. J Cancer Res Clin Oncol 2018;144:2085-95. [Crossref] [PubMed]
- Chen L, Guo P, He Y, et al. HCC-derived exosomes elicit HCC progression and recurrence by epithelial-mesenchymal transition through MAPK/ERK signalling pathway. Cell Death Dis 2018;9:513. [Crossref] [PubMed]
- Han Q, Zhao H, Jiang Y, et al. HCC-derived exosomes: Critical player and target for cancer immune escape. Cells 2019;8:558. [Crossref] [PubMed]
- Abudoureyimu M, Zhou H, Zhi Y, et al. Recent progress in the emerging role of exosome in hepatocellular carcinoma. Cell Prolif 2019;52:e12541 [Crossref] [PubMed]
- Sasaki R, Kanda T, Yokosuka O, et al. Exosomes and hepatocellular carcinoma: From bench to bedside. Int J Mol Sci 2019;20:1406. [Crossref] [PubMed]
- Chen R, Xu X, Tao Y, et al. Exosomes in hepatocellular carcinoma: a new horizon. Cell Commun Signal 2019;17:1. [Crossref] [PubMed]
- Zhou Y, Ren H, Dai B, et al. Hepatocellular carcinoma-derived exosomal miRNA-21 contributes to tumor progression by converting hepatocyte stellate cells to cancer- associated fibroblasts. J Exp Clin Cancer Res 2018;37:324. [Crossref] [PubMed]
- Qiu X, Dong S, Qiao F, et al. HBx-mediated miR-21 upregulation represses tumor- suppressor function of PDCD4 in hepatocellular carcinoma. Oncogene 2013;32:3296-305. [Crossref] [PubMed]
- Yin D, Wang Y, Sai W, et al. HBx-induced miR-21 suppresses cell apoptosis in hepatocellular carcinoma by targeting interleukin-12. Oncol Rep 2016;36:2305-12. [Crossref] [PubMed]
- Lu TX, Munitz A, Rothenberg ME. MicroRNA-21 is up-regulated in allergic airway inflammation and regulates IL-12p35 expression. J Immunol 2009;182:4994-5002. [Crossref] [PubMed]
- Lu X. Impact of IL-12 in Cancer. Curr Cancer Drug Targets 2017;17:682-97. [Crossref] [PubMed]
- Kouwaki T, Fukushima Y, Daito T, et al. Extracellular vesicles including exosomes regulate innate immune responses to hepatitis B virus infection. Front Immunol 2016;7:335. [Crossref] [PubMed]
- Kouwaki T, Okamoto M, Tsukamoto H, et al. Extracellular vesicles deliver host and virus RNA and regulate innate immune response. Int J Mol Sci 2017;18:666. [Crossref] [PubMed]
- Kusuma RJ, Manca S, Friemel T, et al. Human vascular endothelial cells transport foreign exosomes from cow's milk by endocytosis. Am J Physiol Cell Physiol 2016;310:C800-7. [Crossref] [PubMed]
- Baier SR, Nguyen C, Xie F, et al. MicroRNAs are absorbed in biologically meaningful amounts from nutritionally relevant doses of cow milk and affect gene expression in peripheral blood mononuclear cells, HEK-293 kidney cell cultures, and mouse livers. J Nutr 2014;144:1495-500. [Crossref] [PubMed]
- Betker JL, Angle BM, Graner MW, et al. The potential of exosomes from cow milk for oral delivery. J Pharm Sci 2019;108:1496-505. [Crossref] [PubMed]
- Li T, Balthasar JP. FcRn Expression in Wildtype Mice, Transgenic Mice, and in Human Tissues. Biomolecules 2018;8:115. [Crossref] [PubMed]
- Zhu Q, Wang Z, Hu Y, et al. miR-21 promotes migration and invasion by the miR-21- PDCD4-AP-1 feedback loop in human hepatocellular carcinoma. Oncol Rep 2012;27:1660-8. [PubMed]
- Wang Z, Yang H, Ren L. MiR-21 promoted proliferation and migration in hepatocellular carcinoma through negative regulation of Navigator-3. Biochem Biophys Res Commun 2015;464:1228-34. [Crossref] [PubMed]
- Wu H, Ng R, Chen X, et al. MicroRNA-21 is a potential link between non-alcoholic fatty liver disease and hepatocellular carcinoma via modulation of the HBP1-p53- Srebp1c pathway. Version 2. Gut 2016;65:1850-60. [Crossref] [PubMed]
- Hu S, Tao R, Wang S, et al. MicroRNA-21 promotes cell proliferation in human hepatocellular carcinoma partly by targeting HEPN1. Tumour Biol 2015;36:5467-72. [Crossref] [PubMed]
- Amr KS, Ezzat WM, Elhosary YA, et al. The potential role of miRNAs 21 and 199-a in early diagnosis of hepatocellular carcinoma. Gene 2016;575:66-70. [Crossref] [PubMed]
- Zhang Y, Wei C, Guo CC, et al. Prognostic value of microRNAs in hepatocellular carcinoma: a meta-analysis. Oncotarget 2017;8:107237-57. [Crossref] [PubMed]
- Koenig AB, Barajas JM, Guerrero MJ, et al. A comprehensive analysis of argonaute- CLIP data identifies novel, conserved and species-specific targets of miR- 21 in human liver and hepatocellular carcinoma. Int J Mol Sci 2018;19:851. [Crossref] [PubMed]
- Yoon JS, Kim G, Lee YR, et al. Clinical significance of microRNA-21 expression in disease progression of patients with hepatocellular carcinoma. Biomark Med 2018;12:1105-14. [Crossref] [PubMed]
- Bharali D, Banerjee BD, Bharadwaj M, et al. Expression analysis of microRNA-21 and microRNA-122 in hepatocellular carcinoma. J Clin Exp Hepatol 2019;9:294-301. [Crossref] [PubMed]
- Meng F, Henson R, Wehbe-Janek H, et al. MicroRNA-21 regulates expression of the PTEN tumor suppressor gene in human hepatocellular cancer. Gastroenterology 2007;133:647-58. [Crossref] [PubMed]
- Bao L, Yan Y, Xu C, et al. MicroRNA-21 suppresses PTEN and hSulf-1 expression and promotes hepatocellular carcinoma progression through AKT/ERK pathways. Cancer Lett 2013;337:226-36. [Crossref] [PubMed]
- Zhou L, Yang ZX, Song WJ, et al. MicroRNA-21 regulates the migration and invasion of a stem-like population in hepatocellular carcinoma. Int J Oncol 2013;43:661-9. [Crossref] [PubMed]
- Moh MC, Lee LH, Yang X, et al. HEPN1, a novel gene that is frequently down- regulated in hepatocellular carcinoma, suppresses cell growth and induces apoptosis in HepG2 cells. J Hepatol 2003;39:580-6. [Crossref] [PubMed]
- Kamel RR, Amr KS, Afify M, et al. Relation between microRNAs and apoptosis in hepatocellular carcinoma. Open Access Maced J Med Sci 2016;4:31-7. [Crossref] [PubMed]
- Zhuang C, Jiang W, Huang D, et al. Serum miR-21, miR-26a and miR-101 as potential biomarkers of hepatocellular carcinoma. Clin Res Hepatol Gastroenterol 2016;40:386-96. [Crossref] [PubMed]
- Guo X, Lv X, Lv X, et al. Circulating miR-21 serves as a serum biomarker for hepatocellular carcinoma and correlated with distant metastasis. Oncotarget 2017;8:44050-8. [Crossref] [PubMed]
- Qu J, Yang J, Chen M, et al. MicroRNA-21 as a diagnostic marker for hepatocellular carcinoma: A systematic review and meta-analysis. Pak J Med Sci 2019;35:1466-71. [Crossref] [PubMed]
- Wang WY, Zhang HF, Wang L, et al. miR-21 expression predicts prognosis in hepatocellular carcinoma. Clin Res Hepatol Gastroenterol 2014;38:715-9. [Crossref] [PubMed]
- Yi PS, Li JS. High expression of miR-21 is not a predictor of poor prognosis in all patients with hepatocellular carcinoma. Mol Clin Oncol 2018;8:733-9. [PubMed]
- Dundar HZ, Aksoy F, Aksoy SA, et al. Overexpression of miR-21 is associated with recurrence in patients with hepatitis B virus-mediated hepatocellular carcinoma undergoing liver transplantation. Transplant Proc 2019;51:1157-61. [Crossref] [PubMed]
- Chen M, Liu Y, Varley P, et al. High-mobility group box 1 promotes hepatocellular carcinoma progression through miR-21-mediated matrix metalloproteinase activity. Cancer Res 2015;75:1645-56. [Crossref] [PubMed]
- Tian XP, Wang CY, Jin XH, et al. Acidic microenvironment up-regulates exosomal miR-21 and miR-10b in early-stage hepatocellular carcinoma to promote cancer cell proliferation and metastasis. Theranostics 2019;9:1965-79. [Crossref] [PubMed]
- Cao LQ, Yang XW, Chen YB, et al. Exosomal miR-21 regulates the TETs/PTENp1/PTEN pathway to promote hepatocellular carcinoma growth. Mol Cancer 2019;18:148. [Crossref] [PubMed]
- Pu C, Huang H, Wang Z, et al. Extracellular vesicle-associated mir-21 and mir-144 are markedly elevated in serum of patients with hepatocellular carcinoma. Front Physiol 2018;9:930. [Crossref] [PubMed]
- Zhong XZ, Deng Y, Chen G, et al. Investigation of the clinical significance and molecular mechanism of miR-21-5p in hepatocellular carcinoma: A systematic review based on 24 studies and bioinformatics investigation. Oncol Lett 2019;17:230-46. [PubMed]
- Zhang T, Yang Z, Kusumanchi P, et al. Critical role of microRNA-21 in the pathogenesis of liver diseases. Front Med (Lausanne) 2020;7:7. [Crossref] [PubMed]
- Najafi Z, Sharifi M, Javadi G. Degradation of miR-21 induces apoptosis and inhibits cell proliferation in human hepatocellular carcinoma. Cancer Gene Ther 2015;22:530-5. [Crossref] [PubMed]
- Wagenaar TR, Zabludoff S, Ahn SM, et al. Anti-miR-21 suppresses hepatocellular carcinoma growth via broad transcriptional network deregulation. Mol Cancer Res 2015;13:1009-21. [Crossref] [PubMed]
- He C, Dong X, Zhai B, et al. MiR-21 mediates sorafenib resistance of hepatocellular carcinoma cells by inhibiting autophagy via the PTEN/Akt pathway. Oncotarget 2015;6:28867-81. [Crossref] [PubMed]
- Zhang J, Jiao J, Cermelli S, et al. miR-21 inhibition reduces liver fibrosis and prevents tumor development by inducing apoptosis of CD24+ progenitor cells. Cancer Res 2015;75:1859-67. [Crossref] [PubMed]
- Chen S, Yang C, Sun C, et al. miR-21-5p suppressed the sensitivity of hepatocellular carcinoma cells to cisplatin by targeting FASLG. DNA Cell Biol 2019;38:865-73. [Crossref] [PubMed]
- Chen X, Gao C, Li H, et al. Identification and characterization of microRNAs in raw milk during different periods of lactation, commercial fluid, and powdered milk products. Cell Res 2010;20:1128-37. [Crossref] [PubMed]
- Baddela VS, Nayan V, Rani P, et al. Physicochemical biomolecular insights into buffalo milk-derived nanovesicles. Appl Biochem Biotechnol 2016;178:544-57. [Crossref] [PubMed]
- Wang L, Sadri M, Giraud D, et al. RNase H2-dependent polymerase chain reaction and elimination of confounders in sample collection, storage, and analysis strengthen evidence that microRNAs in bovine milk are bioavailable in humans. J Nutr 2018;148:153-9. [Crossref] [PubMed]
- Fromm B, Tosar JP, Lu Y, et al. Human and cow have identical miR-21-5p and miR- 30a-5p sequences, which are likely unsuited to study dietary uptake from cow milk. J Nutr 2018;148:1506-7. [Crossref] [PubMed]
- Zhou Y, Xue R, Wang J, et al. Puerarin inhibits hepatocellular carcinoma invasion and metastasis through miR-21-mediated PTEN/AKT signaling to suppress the epithelial- mesenchymal transition. Braz J Med Biol Res 2020;53:e8882 [Crossref] [PubMed]
- Danby FW. Acne, dairy and cancer: The 5alpha-P link. Dermatoendocrinol 2009;1:12-6. [Crossref] [PubMed]
- Danby FW. Nutrition and acne. Clin Dermatol 2010;28:598-604. [Crossref] [PubMed]
- Regal P, Cepeda A, Fente C. Development of an LC-MS/MS method to quantify sex hormones in bovine milk and influence of pregnancy in their levels. Food Addit Contam Part A Chem Anal Control Expo Risk Assess 2012;29:770-9. [Crossref] [PubMed]
- Goyon A, Cai JZ, Kraehenbuehl K, et al. Determination of steroid hormones in bovine milk by LC-MS/MS and their levels in Swiss Holstein cow milk. Food Addit Contam Part A Chem Anal Control Expo Risk Assess 2016;33:804-16. [Crossref] [PubMed]
- Wilkinson ML, Iqbal MJ, Williams R. Characterisation of high affinity binding sites of androgens in primary hepatocellular carcinoma. Clin Chim Acta 1985;152:105-13. [Crossref] [PubMed]
- Teng Y, Litchfield LM, Ivanova MM, et al. Dehydroepiandrosterone-induces miR-21 transcription in HepG2 cells through estrogen receptor β and androgen receptor. Mol Cell Endocrinol 2014;392:23-36. [Crossref] [PubMed]
- Sadri Nahand J, Bokharaei-Salim F, Salmaninejad A, et al. microRNAs: Key players in virus-associated hepatocellular carcinoma. J Cell Physiol 2019;234:12188-225. [Crossref] [PubMed]
- Sidorkiewicz M, Grek M, Jozwiak B, et al. Expression of microRNA-155 precursor in peripheral blood mononuclear cells from hepatitis C patients after antiviral treatment. Acta Virol 2010;54:75-8. [Crossref] [PubMed]
- Ji J, Xu M, Tu J, et al. MiR-155 and its functional variant rs767649 contribute to the susceptibility and survival of hepatocellular carcinoma. Oncotarget 2016;7:60303-9. [Crossref] [PubMed]
- Guan C, Yang F, He X, et al. Clinical significance of microRNA-155 expression in hepatocellular carcinoma. Oncol Lett 2016;11:1574-80. [Crossref] [PubMed]
- Ning S, Liu H, Gao B, et al. miR-155, miR-96 and miR-99a as potential diagnostic and prognostic tools for the clinical management of hepatocellular carcinoma. Oncol Lett 2019;18:3381-7. [Crossref] [PubMed]
- Song X, Tan S, Wu Z, et al. HBV suppresses ZHX2 expression to promote proliferation of HCC through miR-155 activation. Int J Cancer 2018;143:3120-30. [Crossref] [PubMed]
- Kałużna EM. MicroRNA-155 and microRNA-196b: promising biomarkers in hepatitis C virus infection? Rev Med Virol 2014;24:169-85. [Crossref] [PubMed]
- Zhang Y, Wei W, Cheng N, et al. Hepatitis C virus-induced up-regulation of microRNA-155 promotes hepatocarcinogenesis by activating Wnt signaling. Hepatology 2012;56:1631-40. [Crossref] [PubMed]
- Sugawara Y, Mizugaki Y, Uchida T, et al. Detection of Epstein-Barr virus (EBV) in hepatocellular carcinoma tissue: a novel EBV latency characterized by the absence of EBV-encoded small RNA expression. Virology 1999;256:196-202. [Crossref] [PubMed]
- Skalsky RL. Analysis of viral and cellular microRNAs in EBV-infected cells. Methods Mol Biol 2017;1532:133-46. [Crossref] [PubMed]
- Wood CD, Carvell T, Gunnell A, et al. Enhancer control of microRNA miR-155 expression in Epstein-Barr Virus-infected B cells. J Virol 2018;92:e00716-18. [Crossref] [PubMed]
- Fu X, Wen H, Jing L, et al. MicroRNA-155-5p promotes hepatocellular carcinoma progression by suppressing PTEN through the PI3K/Akt pathway. Cancer Sci 2017;108:620-31. [Crossref] [PubMed]
- Sun JF, Zhang D, Gao CJ, et al. Exosome-mediated miR-155 transfer contributes to hepatocellular carcinoma cell proliferation by targeting PTEN. Med Sci Monit Basic Res 2019;25:218-28. [Crossref] [PubMed]
- Liao WW, Zhang C, Liu FR, et al. Effects of miR-155 on proliferation and apoptosis by regulating FoxO3a/BIM in liver cancer cell line HCCLM3. Eur Rev Med Pharmacol Sci 2018;22:1277-85. [PubMed]
- Qian F, Hu Q, Tian Y, et al. ING4 suppresses hepatocellular carcinoma via a NF-κB/miR-155/FOXO3a signaling axis. Int J Biol Sci 2019;15:369-85. [Crossref] [PubMed]
- Chen G, Wang D, Zhao X, et al. miR-155-5p modulates malignant behaviors of hepatocellular carcinoma by directly targeting CTHRC1 and indirectly regulating GSK-3β-involved Wnt/β-catenin signaling. Cancer Cell Int 2017;17:118. [Crossref] [PubMed]
- Zhang L, Wang W, Li X, et al. MicroRNA-155 promotes tumor growth of human hepatocellular carcinoma by targeting ARID2. Int J Oncol 2016;48:2425-34. [Crossref] [PubMed]
- Xie Q, Chen X, Lu F, et al. Aberrant expression of microRNA 155 may accelerate cell proliferation by targeting sex-determining region Y box 6 in hepatocellular carcinoma. Cancer 2012;118:2431-42. [Crossref] [PubMed]
- Tang B, Lei B, Qi G, et al. MicroRNA-155-3p promotes hepatocellular carcinoma formation by suppressing FBXW7 expression. J Exp Clin Cancer Res 2016;35:93. [Crossref] [PubMed]
- Wang J, Chu Y, Xu M, et al. miR-21 promotes cell migration and invasion of hepatocellular carcinoma by targeting KLF5. Oncol Lett 2019;17:2221-7. [PubMed]
- Carbajo-Pescador S, Mauriz JL, García-Palomo A, et al. FoxO proteins: regulation and molecular targets in liver cancer. Curr Med Chem 2014;21:1231-46. [Crossref] [PubMed]
- Oba A, Shimada S, Akiyama Y, et al. ARID2 modulates DNA damage response in human hepatocellular carcinoma cells. J Hepatol 2017;66:942-51. [Crossref] [PubMed]
- Guo X, Yang M, Gu H, et al. Decreased expression of SOX6 confers a poor prognosis in hepatocellular carcinoma. Cancer Epidemiol 2013;37:732-6. [Crossref] [PubMed]
- Welcker M, Orian A, Jin J, et al. The Fbw7 tumor suppressor regulates glycogen synthase kinase 3 phosphorylation-dependent c-Myc protein degradation. Proc Natl Acad Sci U S A 2004;101:9085-90. Erratum in: Proc Natl Acad Sci U S A 2006 Jan 10;103(2):504. Grim, Jonathan A [corrected to Grim, Jonathan E]. [Crossref] [PubMed]
- Mao JH, Kim IJ, Wu D, et al. FBXW7 targets mTOR for degradation and cooperates with PTEN in tumor suppression. Science 2008;321:1499-502. [Crossref] [PubMed]
- Yang CH, Yue J, Pfeffer SR, et al. MicroRNA-21 promotes glioblastoma tumorigenesis by down-regulating insulin-like growth factor-binding protein-3 (IGFBP3). J Biol Chem 2014;289:25079-87. [Crossref] [PubMed]
- El Tayebi HM, Waly AA, Assal RA, et al. Transcriptional activation of the IGF-II/IGF-1R axis and inhibition of IGFBP-3 by miR-155 in hepatocellular carcinoma. Oncol Lett 2015;10:3206-12. [Crossref] [PubMed]
- Li DP, Fan J, Wu YJ, et al. MiR-155 up-regulated by TGF-β promotes epithelial- mesenchymal transition, invasion and metastasis of human hepatocellular carcinoma cells in vitro. Am J Transl Res 2017;9:2956-65. [PubMed]
- Kong X, Liu F, Gao J. MiR-155 promotes epithelial-mesenchymal transition in hepatocellular carcinoma cells through the activation of PI3K/SGK3/β-catenin signaling pathways. Oncotarget 2016;7:66051-60. [Crossref] [PubMed]
- Huang X, Shen Y, Liu M, et al. Quantitative proteomics reveals that miR-155 regulates the PI3K-AKT pathway in diffuse large B-cell lymphoma. Am J Pathol 2012;181:26-33. [Crossref] [PubMed]
- Larrabeiti-Etxebarria A, Lopez-Santillan M, Santos-Zorrozua B, et al. Systematic Review of the Potential of MicroRNAs in Diffuse Large B Cell Lymphoma. Cancers (Basel) 2019;11:144. [Crossref] [PubMed]
- Ezzat WM, Amr KS, Raouf HA, et al. Relationship between serum microRNA155 and telomerase expression in hepatocellular carcinoma. Arch Med Res 2016;47:349-55. [Crossref] [PubMed]
- Yan XL, Jia YL, Chen L, et al. Hepatocellular carcinoma-associated mesenchymal stem cells promote hepatocarcinoma progression: role of the S100A4-miR155-SOCS1- MMP9 axis. Hepatology 2013;57:2274-86. [Crossref] [PubMed]
- Tscherner A, Brown AC, Stalker L, et al. STAT3 signaling stimulates miR-21 expression in bovine cumulus cells during in vitro oocyte maturation. Sci Rep 2018;8:11527. [Crossref] [PubMed]
- Liu F, Kong X, Lv L, et al. MiR-155 targets TP53INP1 to regulate liver cancer stem cell acquisition and self-renewal. FEBS Lett 2015;589:500-6. [Crossref] [PubMed]
- Matsuura Y, Wada H, Eguchi H, et al. Exosomal miR-155 derived from hepatocellular carcinoma cells under hypoxia promotes angiogenesis in endothelial cells. Dig Dis Sci 2019;64:792-802. [Crossref] [PubMed]
- Han ZB, Chen HY, Fan JW, et al. Up-regulation of microRNA-155 promotes cancer cell invasion and predicts poor survival of hepatocellular carcinoma following liver transplantation. J Cancer Res Clin Oncol 2012;138:153-61. [Crossref] [PubMed]
- Yang N, Li S, Li G, et al. The role of extracellular vesicles in mediating progression, metastasis and potential treatment of hepatocellular carcinoma. Oncotarget 2017;8:3683-95. [Crossref] [PubMed]
- Hsu M, Zhang J, Flint M, et al. Hepatitis C virus glycoproteins mediate pH-dependent cell entry of pseudotyped retroviral particles. Proc Natl Acad Sci U S A 2003;100:7271-6. [Crossref] [PubMed]
- Masciopinto F, Giovani C, Campagnoli S, et al. Association of hepatitis C virus envelope proteins with exosomes. Eur J Immunol 2004;34:2834-42. [Crossref] [PubMed]
- Kapoor NR, Chadha R, Kumar S, et al. The HBx gene of hepatitis B virus can influence hepatic microenvironment via exosomes by transferring its mRNA and protein. Virus Res 2017;240:166-74. [Crossref] [PubMed]
- Izumi H, Kosaka N, Shimizu T, et al. Bovine milk contains microRNA and messenger RNA that are stable under degradative conditions. J Dairy Sci 2012;95:4831-41. [Crossref] [PubMed]
- Sun Q, Chen X, Yu J, et al. Immune modulatory function of abundant immune-related microRNAs in microvesicles from bovine colostrum. Protein Cell 2013;4:197-210. [Crossref] [PubMed]
- Oh S, Park MR, Son SJ, et al. Comparison of total RNA isolation methods for analysis of immune-related microRNAs in market milks. Korean J Food Sci Anim Resour 2015;35:459-65. [Crossref] [PubMed]
- Munagala R, Aqil F, Jeyabalan J, Gupta RC. Bovine milk-derived exosomes for drug delivery. Cancer Lett 2016;371:48-61. [Crossref] [PubMed]
- Zhao L, Liu W, Xiao J, et al. The role of exosomes and "exosomal shuttle microRNA" in tumorigenesis and drug resistance. Cancer Lett 2015;356:339-46. [Crossref] [PubMed]
- Gaggini M, Carli F, Rosso C, et al. Altered amino acid concentrations in NAFLD: Impact of obesity and insulin resistance. Hepatology 2018;67:145-58. [Crossref] [PubMed]
- Gannon NP, Schnuck JK, Vaughan RA. BCAA Metabolism and Insulin Sensitivity - Dysregulated by Metabolic Status? Mol Nutr Food Res 2018;62:e1700756 [Crossref] [PubMed]
- Wang J, Liu Y, Lian K, et al. BCAA catabolic defect alters glucose metabolism in lean mice. Front Physiol 2019;10:1140. [Crossref] [PubMed]
- Mersey BD, Jin P, Danner DJ. Human microRNA (miR29b) expression controls the amount of branched chain alpha-ketoacid dehydrogenase complex in a cell. Hum Mol Genet 2005;14:3371-7. [Crossref] [PubMed]
- Ijichi C, Matsumura T, Tsuji T, et al. Branched-chain amino acids promote albumin synthesis in rat primary hepatocytes through the mTOR signal transduction system. Biochem Biophys Res Commun 2003;303:59-64. [Crossref] [PubMed]
- Meckes DG Jr. Exosomal communication goes viral. J Virol 2015;89:5200-3. [Crossref] [PubMed]
- Chahar HS, Bao X, Casola A. Exosomes and their role in the life cycle and pathogenesis of RNA viruses. Viruses 2015;7:3204-25. [Crossref] [PubMed]
- Alenquer M, Amorim MJ. Exosome biogenesis, regulation, and function in viral infection. Viruses 2015;7:5066-83. [Crossref] [PubMed]
- Yang Y, Han Q, Hou Z, et al. Exosomes mediate hepatitis B virus (HBV) transmission and NK-cell dysfunction. Cell Mol Immunol 2017;14:465-75. [Crossref] [PubMed]
- Tsai P, Lin TY, Cheng SL, et al. Differential dynamics of hepatic protein expressions with long-term cultivated hepatitis C virus infection. J Microbiol Immunol Infect 2020;53:715-23. t. [Crossref] [PubMed]
- Li S, Li S, Wu S, et al. Exosomes modulate the viral replication and host immune responses in HBV infection. Biomed Res Int 2019;2019:2103943 [Crossref] [PubMed]
- Urbanelli L, Buratta S, Tancini B, et al. The role of extracellular vesicles in viral infection and transmission. Vaccines (Basel) 2019;7:102. [Crossref] [PubMed]
- Baechlein C, Fischer N, Grundhoff A, et al. Identification of a novel Hepacivirus in domestic cattle from Germany. J Virol 2015;89:7007-15. [Crossref] [PubMed]
- Schlottau K, Wernike K, Forth L, et al. Presence of two different bovine hepacivirus clusters in Germany. Transbound Emerg Dis 2018;65:1705-11. [Crossref] [PubMed]
- zur Hausen H, de Villiers EM. Dairy cattle serum and milk factors contributing to the risk of colon and breast cancers. Int J Cancer 2015;137:959-67. [Crossref] [PubMed]
- Falida K, Eilebrecht S, Gunst K, et al. Isolation of two virus-like circular DNAs from commercially available milk samples. Genome Announc 2017;5:e00266-17 [Crossref] [PubMed]
- Zur Hausen H, Bund T, de Villiers EM. Infectious agents in bovine red meat and milk and their potential role in cancer and other chronic diseases. Curr Top Microbiol Immunol 2017;407:83-116. [Crossref] [PubMed]
- de Villiers EM, Gunst K, Chakraborty D, et al. A specific class of infectious agents isolated from bovine serum and dairy products and peritumoral colon cancer tissue. Emerg Microbes Infect 2019;8:1205-18. [Crossref] [PubMed]
- Kumar L, Verma S, Vaidya B, et al. Exosomes: Natural carriers for siRNA delivery. Curr Pharm Des 2015;21:4556-65. [Crossref] [PubMed]
- Zhou Y, Zhou G, Tian C, et al. Exosome-mediated small RNA delivery for gene therapy. Wiley Interdiscip Rev RNA 2016;7:758-71. [Crossref] [PubMed]
- Moris D, Beal EW, Chakedis J, et al. Role of exosomes in treatment of hepatocellular carcinoma. Surg Oncol 2017;26:219-28. [Crossref] [PubMed]
- Pullan JE, Confeld MI, Osborn JK, et al. Exosomes as drug carriers for cancer therapy. Mol Pharm 2019;16:1789-98. [Crossref] [PubMed]
- Li X, Li C, Zhang L, et al. The significance of exosomes in the development and treatment of hepatocellular carcinoma. Mol Cancer 2020;19:1. [Crossref] [PubMed]
- Ishiguro K, Yan IK, Lewis-Tuffin L, et al. Targeting liver cancer stem cells using engineered biological nanoparticles for the treatment of hepatocellular cancer. Hepatol Commun 2020;4:298-313. [Crossref] [PubMed]
- Vashisht M, Rani P, Onteru SK, et al. Curcumin encapsulated in milk exosomes resists human digestion and possesses enhanced intestinal permeability in vitro. Appl Biochem Biotechnol 2017;183:993-1007. [Crossref] [PubMed]
- Sanwlani R, Fonseka P, Chitti SV, et al. Milk-derived extracellular vesicles in inter- organism, cross-species communication and drug delivery. Proteomes 2020;8:11. [Crossref] [PubMed]
- Prandini A, Tansini G, Sigolo S, et al. On the occurrence of aflatoxin M1 in milk and dairy products. Food Chem Toxicol 2009;47:984-91. [Crossref] [PubMed]
- Ismail A, Akhtar S, Levin RE, et al. Aflatoxin M1: Prevalence and decontamination strategies in milk and milk products. Crit Rev Microbiol 2016;42:418-27. [PubMed]
- Van der Fels-Klerx HJ, Vermeulen LC, Gavai AK, et al. Climate change impacts on aflatoxin B1 in maize and aflatoxin M1 in milk: A case study of maize grown in Eastern Europe and imported to the Netherlands. PLoS One 2019;14:e0218956 [Crossref] [PubMed]
- Castegnaro M, Wild CP. IARC activities in mycotoxin research. Nat Toxins 1995;3:327-31. [Crossref] [PubMed]
- Magnussen A, Parsi MA. Aflatoxins, hepatocellular carcinoma and public health. World J Gastroenterol 2013;19:1508-12. [Crossref] [PubMed]
- IARC monographs on the identification of carcinogenic hazards to humans. Available online: https://monographs.iarc.fr/list-of-classifications (last updated 2020-03-03).
- Marchese S, Polo A, Ariano A, et al. Aflatoxin B1 and M1: Biological Properties and Their Involvement in Cancer Development. Toxins (Basel) 2018;10:214. [Crossref] [PubMed]
- Vaz A, Cabral Silva AC, Rodrigues P, et al. Detection Methods for Aflatoxin M1 in Dairy Products. Microorganisms 2020;8:246. [Crossref] [PubMed]
- Nguyen T, Flint S, Palmer J. Control of aflatoxin M1 in milk by novel methods: A review. Food Chem 2020;311:125984 [Crossref] [PubMed]
- Shen HM, Ong CN. Mutations of the p53 tumor suppressor gene and ras oncogenes in aflatoxin hepatocarcinogenesis. Mutat Res 1996;366:23-44. [Crossref] [PubMed]
- Melnik BC. Milk disrupts p53 and DNMT1, the guardians of the genome: implications for acne vulgaris and prostate cancer. Nutr Metab (Lond) 2017;14:55. [Crossref] [PubMed]
- Simabuco FM, Morale MG, Pavan ICB, et al. p53 and metabolism: from mechanism to therapeutics. Oncotarget 2018;9:23780-823. [Crossref] [PubMed]
- Zhang L, Ye Y, An Y, et al. Systems responses of rats to aflatoxin B1 exposure revealed with metabonomic changes in multiple biological matrices. J Proteome Res 2011;10:614-23. [Crossref] [PubMed]
- Marchese S, Sorice A, Ariano A, et al. Evaluation of Aflatoxin M1 Effects on the Metabolomic and Cytokinomic Profiling of a Hepatoblastoma Cell Line. Toxins (Basel) 2018;10:436. [Crossref] [PubMed]
- Shaw S, Lieber CS. Plasma amino acid abnormalities in the alcoholic: respective role of alcohol, nutrition, and liver injury. Gastroenterology 1978;74:677-82. [Crossref] [PubMed]
- Marchesini G, Bianchi G, Merli M, et al. Nutritional supplementation with branched- chain amino acids in advanced cirrhosis: a double-blind, randomized trial. Gastroenterology 2003;124:1792-801. [Crossref] [PubMed]
- Tada T, Kumada T, Toyoda H, et al. Impact of branched-chain amino acid granule therapy in patients with hepatocellular carcinoma who have normal albumin levels and low branched-chain amino acid to tyrosine ratios. Nutr Cancer 2019;71:1132-41. [Crossref] [PubMed]
- Campbell JS, Hughes SD, Gilbertson DG, et al. Platelet-derived growth factor C induces liver fibrosis, steatosis, and hepatocellular carcinoma. Proc Natl Acad Sci U S A 2005;102:3389-94. [Crossref] [PubMed]
- Takegoshi K, Honda M, Okada H, et al. Branched-chain amino acids prevent hepatic fibrosis and development of hepatocellular carcinoma in a non-alcoholic steatohepatitis mouse model. Oncotarget 2017;8:18191-205. [Crossref] [PubMed]
- Shah R, Reyes-Gordillo K, Arellanes-Robledo J, et al. TGF-β1 up-regulates the expression of PDGF-β receptor mRNA and induces a delayed PI3K-, AKT-, and p70(S6K)-dependent proliferative response in activated hepatic stellate cells. Alcohol Clin Exp Res 2013;37:1838-48. [Crossref] [PubMed]
- Umemura A, Park EJ, Taniguchi K, et al. Liver damage, inflammation, and enhanced tumorigenesis after persistent mTORC1 inhibition. Cell Metab 2014;20:133-44. [Crossref] [PubMed]
- Cha JH, Bae SH, Kim HL, et al. Branched-chain amino acids ameliorate fibrosis and suppress tumor growth in a rat model of hepatocellular carcinoma with liver cirrhosis. PLoS One 2013;8:e77899 [Crossref] [PubMed]
- Neef M, Ledermann M, Saegesser H, et al. Low-dose oral rapamycin treatment reduces fibrogenesis, improves liver function, and prolongs survival in rats with established liver cirrhosis. J Hepatol 2006;45:786-96. [Crossref] [PubMed]
- Ferrín G, Guerrero M, Amado V, et al. Activation of mTOR Signaling Pathway in Hepatocellular Carcinoma. Int J Mol Sci 2020;21:1266. [Crossref] [PubMed]
- Guerrero M, Ferrín G, Rodríguez-Perálvarez M, et al. mTOR expression in liver transplant candidates with hepatocellular carcinoma: Impact on histological features and tumour recurrence. Int J Mol Sci 2019;20:336. [Crossref] [PubMed]
- Teng CF, Wu HC, Hsieh WC, et al. Activation of ATP citrate lyase by mTOR signal induces disturbed lipid metabolism in hepatitis B virus pre-S2 mutant tumorigenesis. J Virol 2015;89:605-14. [Crossref] [PubMed]
- Huynh H, Hao HX, Chan SL, et al. Loss of tuberous sclerosis complex 2 (TSC2) is frequent in hepatocellular carcinoma and predicts response to mTORC1 inhibitor everolimus. Mol Cancer Ther 2015;14:1224-35. [Crossref] [PubMed]
- Ashworth RE, Wu J. Mammalian target of rapamycin inhibition in hepatocellular carcinoma. World J Hepatol 2014;6:776-82. [Crossref] [PubMed]
- Liao H, Huang Y, Guo B, et al. Dramatic antitumor effects of the dual mTORC1 and mTORC2 inhibitor AZD2014 in hepatocellular carcinoma. Am J Cancer Res 2014;5:125-39. [PubMed]
- Engl T, Rutz J, Maxeiner S, et al. mTOR inhibition reduces growth and adhesion of hepatocellular carcinoma cells in vitro. Mol Med Rep 2017;16:7064-71. [Crossref] [PubMed]
- Apostolidis L, Pfeiffenberger J, Gotthardt D, et al. Survival of hepatocellular carcinoma patients treated with sorafenib beyond progression. Gastrointest Tumors 2018;5:38-46. [Crossref] [PubMed]
- Fan M, Li Y, Wang C, et al. Dietary protein consumption and the risk of type 2 diabetes: A dose-response meta-analysis of prospective studies. Nutrients 2019;11:2783. [Crossref]
- Drouin-Chartier JP, Li Y, Ardisson Korat AV, et al. Changes in dairy product consumption and risk of type 2 diabetes: results from 3 large prospective cohorts of US men and women. Am J Clin Nutr 2019;110:1201-12. [Crossref] [PubMed]
- van Kranenburg R, Kleerebezema M, van Hylckama Vliega J, et al. Flavour formation from amino acids by lactic acid bacteria: predictions from genome sequence analysis. Int Dairy J 2002;12:111-21. [Crossref]
- Smit G, Smit BA, Engels WJ. Flavour formation by lactic acid bacteria and biochemical flavour profiling of cheese products. FEMS Microbiol Rev 2005;29:591-610. [Crossref] [PubMed]
- Murakami T, Matsuo M, Shimizu A, et al. Dissociation of branched-chain alpha-keto acid dehydrogenase kinase (BDK) from branched-chain alpha-keto acid dehydrogenase complex (BCKDC) by BDK inhibitors. J Nutr Sci Vitaminol (Tokyo) 2005;51:48-50. [Crossref] [PubMed]
- Paxton R, Harris RA. Regulation of branched-chain alpha-ketoacid dehydrogenase kinase. Arch Biochem Biophys 1984;231:48-57. [Crossref] [PubMed]
- Sarwar A, Aziz T, Al-Dalali S, et al. Physicochemical and Microbiological Properties of Synbiotic Yogurt Made with Probiotic Yeast Saccharomyces boulardii in Combination with Inulin. Foods 2019;8:468. [Crossref] [PubMed]
- Kadota Y, Toyoda T, Hayashi-Kato M, et al. Octanoic acid promotes branched-chain amino acid catabolisms via the inhibition of hepatic branched-chain alpha-keto acid dehydrogenase kinase in rats. Metabolism 2015;64:1157-64. [Crossref] [PubMed]
- Guillot E, Vaugelade P, Lemarchal P, et al. Intestinal absorption and liver uptake of medium-chain fatty acids in non-anaesthetized pigs. Br J Nutr 1993;69:431-42. [Crossref] [PubMed]
- Chriett S, Pirola L. Essential roles of four-carbon backbone chemicals in the control of metabolism. World J Biol Chem 2015;6:223-30. [Crossref] [PubMed]
- Usui Y, Kimura Y, Satoh T, et al. Effects of long-term intake of a yogurt fermented with Lactobacillus delbrueckii subsp. bulgaricus 2038 and Streptococcus thermophilus 1131 on mice. Int Immunol 2018;30:319-31. [Crossref] [PubMed]
- Kok CR, Hutkins R. Yogurt and other fermented foods as sources of health-promoting bacteria. Nutr Rev 2018;76:4-15. [Crossref] [PubMed]
- Tso SC, Qi X, Gui WJ, et al. Structure-based design and mechanisms of allosteric inhibitors for mitochondrial branched-chain α-ketoacid dehydrogenase kinase. Proc Natl Acad Sci U S A 2013;110:9728-33. [Crossref] [PubMed]
- Xue P, Zeng F, Duan Q, et al. BCKDK of BCAA catabolism cross-talking with the MAPK pathway promotes tumorigenesis of colorectal cancer. EBioMedicine 2017;20:50-60. [Crossref] [PubMed]
- Kusaczuk M, Bartoszewicz M, Cechowska-Pasko M. Phenylbutyric acid: simple structure - multiple effects. Curr Pharm Des 2015;21:2147-66. [Crossref] [PubMed]
- Al-Keilani MS, Al-Sawalha NA. Potential of phenylbutyrate as adjuvant chemotherapy: An overview of cellular and molecular anticancer mechanisms. Chem Res Toxicol 2017;30:1767-77. [Crossref] [PubMed]
- Meng M, Jiang JM, Liu H, et al. Effects of sodium phenylbutyrate on differentiation and induction of the P21WAF1/CIP1 anti-oncogene in human liver carcinoma cell lines. Chin J Dig Dis 2005;6:189-92. [Crossref] [PubMed]
- Wang CT, Meng M, Zhang JC, et al. Growth inhibition and gene induction in human hepatocellular carcinoma cell exposed to sodium 4-phenylbutanoate. Chin Med J (Engl) 2008;121:1707-11. [Crossref] [PubMed]
- Yu S, Zhao Z, Sun L, et al. Fermentation results in quantitative changes in milk- derived exosomes and different effects on cell growth and survival. J Agric Food Chem 2017;65:1220-8. [Crossref] [PubMed]
- García-Albiach R, Pozuelo de Felipe MJ, Angulo S, et al. Molecular analysis of yogurt containing Lactobacillus delbrueckii subsp. bulgaricus and Streptococcus thermophilus in human intestinal microbiota. Am J Clin Nutr 2008;87:91-6. Erratum in: Am J Clin Nutr 2008 Jun;87(6):1969. [Crossref] [PubMed]
- Pedersen HK, Gudmundsdottir V, Nielsen HB, et al. Human gut microbes impact host serum metabolome and insulin sensitivity. Nature 2016;535:376-81. [Crossref] [PubMed]
- Cummings NE, Williams EM, Kasza I, et al. Restoration of metabolic health by decreased consumption of branched-chain amino acids. J Physiol 2018;596:623-45. [Crossref] [PubMed]
- Fujita K, Iwama H, Miyoshi H, et al. Diabetes mellitus and metformin in hepatocellular carcinoma. World J Gastroenterol 2016;22:6100-13. [Crossref] [PubMed]
- Jo W, Yu ES, Chang M, et al. Metformin inhibits early stage diethylnitrosamine- induced hepatocarcinogenesis in rats. Mol Med Rep 2016;13:146-52. [Crossref] [PubMed]
- Das BK, Choukimath SM, Gadad PC. Asarone and metformin delays experimentally induced hepatocellular carcinoma in diabetic milieu. Life Sci 2019;230:10-8. [Crossref] [PubMed]
- Cunha V, Cotrim HP, Rocha R, et al. Metformin in the prevention of hepatocellular carcinoma in diabetic patients: A systematic review. Ann Hepatol 2020;19:232-7. [Crossref] [PubMed]
- Melnik BC, Schmitz G. Metformin: an inhibitor of mTORC1 signaling. J Endocrinol Diabetes Obes 2014;2:1029.
- Ling S, Song L, Fan N, et al. Combination of metformin and sorafenib suppresses proliferation and induces autophagy of hepatocellular carcinoma via targeting the mTOR pathway. Int J Oncol 2017;50:297-309. [Crossref] [PubMed]
- S Sonnet D. Metformin inhibits branched chain amino acid (BCAA) derived ketoacidosis and promotes metabolic homeostasis in MSUD. Sci Rep 2016;6:28775. [Crossref] [PubMed]
- Zemdegs J, Martin H, Pintana H, et al. Metformin promotes anxiolytic and antidepressant-like responses in insulin-resistant mice by decreasing circulating branched-chain amino acids. J Neurosci 2019;39:5935-48. [Crossref] [PubMed]
- Fuchs CJ, Hermans WJH, Holwerda AM, et al. Branched-chain amino acid and branched-chain ketoacid ingestion increases muscle protein synthesis rates in vivo in older adults: a double-blind, randomized trial. Am J Clin Nutr 2019;110:862-72. [Crossref] [PubMed]
Canada’s Dietary Guidelines - Zhang W, Xiang YB, Li HL, et al. Vegetable-based dietary pattern and liver cancer risk: results from the Shanghai women's and men's health studies. Cancer Sci 2013;104:1353-61. [Crossref] [PubMed]
- Koumbi L. Dietary factors can protect against liver cancer development. World J Hepatol 2017;9:119-25. [Crossref] [PubMed]
- Michaëlsson K, Wolk A, Langenskiöld S, et al. Milk intake and risk of mortality and fractures in women and men: cohort studies. BMJ 2014;349:g6015. [Crossref] [PubMed]
- Tognon G, Nilsson LM, Shungin D, et al. Nonfermented milk and other dairy products: associations with all-cause mortality. Am J Clin Nutr 2017;105:1502-11. [Crossref] [PubMed]
- Michaëlsson K, Byberg L. Mixing of apples and oranges in milk research: A cohort analysis of non-fermented milk intake and all-cause mortality. Nutrients 2020;12:1393. [Crossref] [PubMed]
- Fraser GE, Jaceldo-Siegl K, Orlich M, et al. Dairy, soy, and risk of breast cancer: those confounded milks. Int J Epidemiol 2020;49:1526-37. [Crossref] [PubMed]


