
Mitotically associated long non-coding RNA is a tumor promoter in anaplastic thyroid cancer
Introduction
Anaplastic thyroid cancer (ATC), one of the deadliest of all cancers, is responsible for as much as 14–39% of mortality caused by thyroid cancer (1,2). Approximately 50% of ATC patients present with distant metastasis, and the disease has a median survival of a mere 5 months (3,4). ATC responds poorly to traditional therapies; however, dabrafenib and trametinib is the first targeted regimen to receive approval by FDA and achieved an overall response rate of 69% in a phase II clinical trial,, significantly improving the survival of ATC patients with BRAF mutation (5). Thus, understanding the molecular mechanisms underlying the initiation and progression of ATC is important and would help us to identify potential molecular targets for treating the disease.
Owing to the rarity of ATC and its extensive infiltration by macrophages, studying the genetic landscape of ATC presents a significant challenge (6). Several studies based primarily on genomic analyses have reported multiple mutations in oncogenes and tumor suppressors in ATC (7,8). Recent studies have shown that the TERT promoter mutation was one of the most frequently identified mutations in ATC along with the BRAFV600E and RAS mutations (9). Nevertheless, only 3% of RNAs are protein-coding messenger RNAs and the rest 97% RAN molecules are non-coding RNAs (ncRNAs) (10).
Long non-coding RNAs (lncRNAs) are a class of ncRNAs that exceed 200 nt in length. LncRNAs are widely expressed in human tissues and are strongly associated with cancer progression (11). HATAIR, for instance, which is a lncRNA that is overexpressed in multiple cancers, promotes cancer invasiveness and metastasis via interaction with PRC2 and silencing of anti-metastatic genes (12). However, the role of lncRANs in ATC tumorigenesis is unclear. Recently, a small number of lncRNAs have been linked with ATC. One study identified a low expression of the lncRNA PTCSC3 in ATC tissues and cells. Meanwhile, the overexpression of PTCSC3 inhibited the drug resistance of ATC to doxorubicin via the STAT3/INO80 pathway (13). Another study identified 19 upregulated and 28 downregulated lncRNAs in 9 ATC samples compared with normal tissue. Of these lncRNAs, PAR5 exerted a tumor-suppressive effect in ATC by impairing the oncogenic activity of EZH2 (14).
In the current study, we performed whole transcriptome sequencing of three ATC and two normal thyroid (NT) tissue samples and characterized the lncRNA expression profile of ATC. We subsequently defined the role of a lncRNA, mitotically associated long non-coding RNA (MANCR), in ATC. We believe that this study will reinforce our understanding of lncRNAs in the initiation and progression of ATC.
We present the following article in accordance with the MDAR reporting checklist (available at http://dx.doi.org/10.21037/atm-20-4530).
Methods
Tissue samples and whole transcriptome sequencing
Three ATC and two NT tissue samples were sequenced. Total RNA from the samples using TRIzol reagent (Invitrogen Inc., CA, USA). Ribosomal RNA was removed from total RNA with Ribo-Zero Gold (Illumina, San Diego, CA, USA) before the construction of RNA-seq libraries. Comparisons of transcriptome library sequencing data and human reference genomes were performed with TopHat (http://tophat.cbcb.umd.edu/), and gene expression was quantified with HTSeq (https://pypi.org/project/HTSeq/). The sequencing data was mapped with TopHat2 software, and the expression counts of each lncRNA were determined by HTSeq. DEseq2 was used to analyze the differential expression profiles of the lncRNAs based on the gene counts.
Cell culture and transfection
Five ATC cell lines (SW1736, C643, CAL-62, BTH101, and 8305C), three papillary thyroid cancer (PTC) cell lines (TPC1, K1, and BCPAP), and one NT cell line (Nthy-ori3-1) were obtained for analysis. SW1736, 8305C, TPC1, and K1 were obtained from Cell Bank of University of Colorado Cancer Center; C643, CAL-62, BTH101, and BCPAP were obtained from the Typical Culture Preservation Commission Cell Bank, Chinese Academy of Sciences (Shanghai, China); and Nthy-ori 3-1 was purchased from Sigma-Aldrich, Inc. (MO, USA).
The SW1736, C643, and BCPAP cells were grown in RPMI 1649 medium; the CAL-62, BTH101, K1, BCPAP, Nthy-ori3-1 cells were grown in DMEM medium; and the 8305C cells were grown in MEM medium. All cells were cultured with 10% heat-inactivated fetal bovine serum (FBS), 100 IU/mL penicillin, and 10 µg/mL streptomycin at 37 °C with 5% CO2. MANCR was knocked down in SW1736 cells with two ASOs designed by and purchased from Ribobio Co., Ltd (Guangzhou, China). (Table S1). MANCR overexpression construct was used to overexpress MANCR in SW1736 cells, and an empty vector was used as the negative control. After 24 h of transfection, all cells were harvested for further analysis.

Full table
qRT-PCR
Total RNA was extracted from cell lines using TRIzol reagent (Invitrogen, Inc.) following the manufacturer’s protocol. Next, a reverse transcription (RT) reaction was performed using PrimeScriptTM RT reagent kit (Takara Bio, Inc., Japan) to convert total RNA to cDNA, and cDNA templates were mixed with gene-specific primers for MANCR (Table S1) and internal control β-actin. Then, quantitative real time polymerase chain reaction (qPCR) was carried out using ABI7900 HT Fast Real time system (Thermo Fisher Scientific Inc., MA, USA) to quantitate MANCR expression. Finally, the comparative Ct (2−ΔΔCt) method was used to analyze the threshold cycle (Ct) values.
CCK-8 assay
Cell proliferation was analyzed by cell counting Kit-8 (CCK-8) assay (Sigma-Aldrich; Merck KGaA). After 24 h of transfection, SW1736 cells were seeded in 96-well plates. An aliquot of 10 µL of CCK-8 solution was added to each well and the plate was incubated at 37 °C for 4 h. The absorbance reading was performed at 450 nm using a spectrophotometer (Thermo Scientific TM NanoDropTM 8000; Thermo Fisher Scientific, Inc., MA, USA) every 24 h. Five replicates were measured for each experimental group.
Transwell assay
Transwell assay (Corning, Tewksbury, MA, USA) was performed to evaluate cell invasion ability. Cells were seeded into the upper chamber of a Matrigel-coated (Sigma-Aldrich) insert, and medium containing 10% FBS was placed into the lower chamber as chemoattractant. After 24 h of incubation at 37 °C, the invaded cells were fixed with methanol and then stained with 0.5% crystal violet. The images of the cell were obtained with an IX71 inverted microscope (Olympus, Tokyo, Japan).
Apoptosis
Flow cytometry was used to analyze cell apoptosis 24 h after transfection. The collected cells were double stained with FITC-annexin V and propidium iodide (PI), and then analyzed using a flow cytometer (FACScan; BD Biosciences, NJ, USA) equipped with CellQuest software (BD Biosciences, NJ, USA). Apoptotic cells were marked based on Annexin V expression (Annexin V+/PI− and Annexin V+/PI+). Data from independent experiments performed in triplicate were studied to compare the number of apoptotic cells.
The Cancer Genome Atlas (TCGA) and GEO datasets
TCGA dataset for thyroid cancer was downloaded (https://www.cbioportal.org) (15). The raw data of the GSE29265 (9 ATC samples, 20 PTC samples, and 20 NT samples) and GSE65144 (12 ATC samples and 13 NT samples) datasets were retrieved from the Gene Expression Omnibus (GEO) (http://www.ncbi.nlm.nih.gov/geo/) and analyzed using R software packages.
Statistical analysis
Data preparation and statistical analyses were performed using SPSS version 20.0 (Chicago, IL, USA), GraphPad Prism version 8.2.1 (La Jolla, CA, USA), gene set enrichment analyses (GSEA) version 4.0.3 (http://www.gsea-msigdb.org), and R version 3.6.2 (Vienna, Austria). One-way analysis of variance (ANOVA) and Student’s t-test were used for statistical comparisons. The Kaplan-Meier method and log-rank test were used for survival analysis. Two-tailed P values were adopted, with P<0.05 considered significant.
Results
LncRNA expression profiling in ATC
Whole transcriptome sequencing of three ATCs and two adjacent NT samples was conducted. In one case, the ATC had completely replaced the thyroid and therefore no adjacent NT was collected. The expression profiling of the lncRNAs is illustrated in Figure 1A,B. Of 11,085 sequenced lncRNAs, 182 lncRNAs were identified to be differentially expressed in ATC compared with NT. Among these lncRNAs, 67 were upregulated (P<0.05), and 115 were downregulated in ATC (P<0.05). Of the downregulated lncRNAs, PTCSC3 (P<0.001) was reported as a tumor suppressor in PTC (16), while the overexpression of PTCSC3 inhibited the drug resistance of ATC to doxorubicin (13). LINC00982 (P<0.001) was a divergent transcript of RDM16 and a biomarker for poor survival in lung adenocarcinoma and gastric cancer (17,18). Meanwhile, ADAMTS9-AS2 (P<0.001) was a tumor suppressor in multiple cancers and was associated with the PI3K/AKT pathway (19,20). Of the upregulated lncRNAs, mitotically associated long non-coding RNA (MANCR or LINC00704) (P=0.0016) was related to cell division in aggressive breast cancer (21).
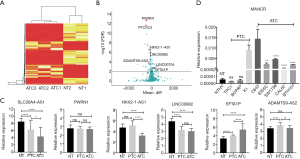
Next, the expressions of the differentially expressed lncRNAs were validated by combining the GSE29265 and GSE65144 datasets (21 ATC, 20 PTC and 33 NT). The expressions of SLC26A4-AS1, LINC00982, and NKX2-1-AS1 were significantly downregulated in ATC compared with NT (P<0.0001). The expression of ADAMTS9-AS2 was downregulated in ATC compared with PTC (P<0.01). These lncRNAs were tumor suppressors in ATC, as indicated by our sequencing data. SFTA1P was significantly overexpressed in ATC compared with PTC and NT, which indicated its role as an oncogene in ATC. The expressions of PWRN1 were not significantly different among the groups (Figure 1C).
Clinical validation of MANCR expression
The expressions of the differentially expressed lncRNAs were validated further using one NT, three PTC, and five ATC cell lines. Strikingly, the relative expression of MANCR was significantly increased in all 5 ATC cell lines compared with NT cell line (C643 P=0.0242; 8305C P=0.0096; SW1736 P<0.0001; CAL62 P<0.0001; and BTH101 P<0.0001). MANCR was also overexpressed in PTC cell line K1 (P=0.0332). However, there was no significant difference between MANCR expression in PTC cell line TPC1 (P=0.1229) and BCPAP (P=0.4444) and that in the NT cell line (Figure 1D).
Since MANCR was not an annotated lncRNA in the GEO datasets, we used the TCGA 495 PTC cohort for clinical validation. MANCR overexpression was correlated with extrathyroidal extension (ETE, P=0.0029) and AJCC T stage IV (P=0.0009); however, it had no association with lymph node metastasis, distant metastasis, or disease-free survival (Figure 2A).
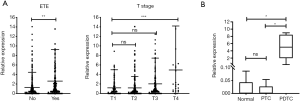
Next, MANCR expression was validated using the sequencing data of poorly differentiated thyroid cancer (PDTC), PTC, and adjacent NT samples. This cohort comprised five PDTC tissues, five PTC tissues, and five NT tissues, as was reported previously (22). Interestingly, the expression level of MANCR was similar in the NT and PTC samples (P=0.7591), and its expression was significantly elevated in the PDTC samples compared with the NT (P=0.0107) and PTC (P=0.0106) samples (Figure 2B). These results indicated that MANCR was an oncogene in ATC/PDTC.
MANCR is a tumor promoter in ATC
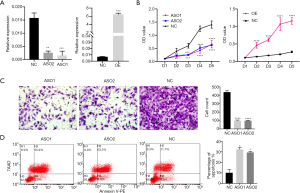
In order to define the biological role of MANCR in ATC, MANCR was transiently knocked down and overexpressed in SW1736 cells using ASOs and MANCR overexpression construct (Figure 3A). After 24 h of transfection, CCK8 assays were performed. Following knockdown, MANCR was shown to significantly inhibit the proliferation of SW1736 cells. Similarly, MANCR overexpression could increase cell proliferation in SW1736 cells (Figure 3B). Moreover, the results of the Transwell assays demonstrated that the knockdown of MANCR with both ASO1 and ASO2 in SW1736 cells decreased their invasive ability compared with the negative controls (Figure 3C). Finally, flow cytometry analyses were performed, showing that the knockdown of MANCR could induce cell apoptosis in SW1736 cells (Figure 3D). These results demonstrated that MANCR is a tumor promoter in ATC, in which it increases cancer cell proliferation and invasion and decreases apoptosis.
MANCR is related to cell cycle
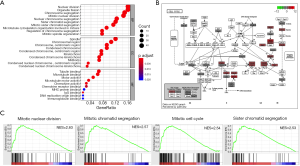
The whole transcriptome sequencing data was further analyzed, and 4,454 genes correlated with MANCR expression were identified (P<0.05), among which 3,216 were upregulated (logFC ≥1) and 66 were downregulated (logFC ≤−1) in ATC. A total of 451 genes were differentially expressed in ATC compared with NT (P<0.05). These genes included 335 protein-coding genes, 57 lncRNAs, 26 pseudogenes, and 33 other transcripts. Gene ontology (GO) analysis showed that the most relevant biological processes of these genes were nuclear division, organelle fission, chromosome segregation, and mitotic nuclear division, all of which are related to cell cycle (Figure 4A,B). Also, in gene set enrichment analysis (GSEA), 1,227 and 148 upregulated gene sets were identified in ATC and NT, respectively, using the molecular signatures database (MSigDB) GO gene sets. The top upregulated pathways in ATC included mitotic nuclear division [normalized enrichment scores (NES) =2.63, P<0.001], mitotic sister chromatic segregation (NES =2.5663998, P<0.001), mitotic cell cycle (NES = 2.5384307, P<0.001), and cell division (NES =2.5367835, P<0.001) (Figure 4C and Table S2). Interestingly, all the 11 pathways involving mitosis and 29 out of 34 pathways involving cell cycle were upregulated in the ATC samples.
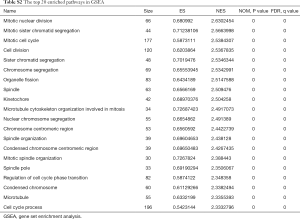
Full table
Discussion
In the current study, whole transcriptome sequencing was used to analyze the lncRNA expression profile of ATC. A unique expression profile of lncRNAs in ATC was identified, providing a unique perspective of ATC carcinogenesis.
Only a small number of studies have reported the genomic and transcriptome characteristics of ATC. Kunstman et al. analyzed 22 cases of ATC as well as 4 established ATC cell lines using whole exome sequencing. They observed new ATC-related mTOR, NF1, NF2, MLH1, MLH3, MSH5, MSH6, ERBB2, EIF1AX, and USH2A mutations, and identified established thyroid cancer genes mutations such as BRAF, TP53, and RAS-family genes (23). Landa et al. analyzed the transcriptome profiles of 37 tumors (17 PDTCs and 20 ATCs). Their data supported a model of tumorigenesis by which PDTC and ATC arise from well-differentiated tumors through the accumulation of key additional genetic abnormalities such as mutations in TP53 and TERT promoter, and PI3K/AKT/mTOR pathway effectors (7). Yoo et al. recently performed RNA sequencing in 13 ATCs and 12 advanced DTCs. They identified a fourth molecular subtype of thyroid cancer, ATC-like despite BRAF-like, RAS-like, and NBNR. They also promoted the activation of the JAK-STAT signaling pathway as a potential target in RAS-positive ATC (9). However, their analyses were not focused on lncRNAs.
Our study, on the other hand, analyzed lncRNA expression profiles in ATC. Among the identified lncRNAs, MANCR was reported as an oncogene in ATC for the first time, through clinical validation and in vitro experiments. MANCR is a lncRNA that was originally identified in the transcriptome profiling of a series of breast cancer cell lines (24). With the exception of the spleen, MANCR expression is very low in most normal tissues (21). Tracy et al. reported that MANCR was overexpressed in triple-negative breast cancer cell lines and patient samples. Similar to our findings, a striking reduction was seen in cell growth, and cell death was induced upon MANCR knockdown in breast cancer cells (21).
A recent study also identified MANCR as an oncogene in PTC by genome-wide analyses using datasets from TCGA and the GEO (25). The authors performed loss-of-function assays and observed that downregulation of MANCR could significantly impair the proliferation, colony formation, cell-cycle progression, and cell invasion of PTC cells via epithelial mesenchymal transition. Unlike their results, we observed no MANCR overexpression in the majority of PTC cell lines or patient samples compared with NT. However, MANCR was upregulated in ATC and in locally advanced PTC (ETE and T4 stage). These results indicated that MANCR might serve as a promoter in the anaplastic change of PTC.
In fact, multiple studies support that PDTC/ATCs arise from pre-existing PTCs which undergo important genetic alterations (7,26,27). Mutations in TERT promoter, which actives TERT transcription, display a stepwise increase in frequency along the spectrum of disease progression, which is consistent with the constitutive activation of MAPK signaling (7). P53 and β-catenin mutations, which were found with increasing incidence in PDTC and ATC but not in PTC, may also serve as a direct molecular trigger of tumor dedifferentiation (27).
Although the mechanism of MANCR in carcinogenesis has not been fully elucidated, our GSEA results indicated that its role was primarily related to mitosis and cell cycle. These results are consistent with the in vitro experiments in a previous study. Tracy et al. (21) revealed that in breast cancer, interphase cells had the lowest expression of MANCR, while early mitotic cells had the highest expression. They also demonstrated defective cytokinesis in MANCR knockdown cells.
Conclusions
In conclusion, our observations indicate a unique expression profile of lncRNA in ATC. MANCR, one of the most significantly overexpressed lncRNAs, is upregulated in PDTC/ATC. MANCR is a tumor promoter in ATC; it increases cancer cell proliferation and invasion and decreases apoptosis. The role of MANCR in the carcinogenesis of ATC is possibly associated with mitosis and cell cycle. Since MANCR expression is minimal in most normal tissues, it may serve as a potential target in the treatment of ATC.
Acknowledgments
We would like to thank Dr. Tian Liao, Dr. Ben Ma, Dr. Wan-Lin Liu, Dr. Li-Tao Han, and Dr. Wei-Bo Xu for technical help with this work.
Funding: This work was supported by the National Natural Science Foundation of China [81772851 and 81972501 to YLW, 81902721 to NSH], Shanghai Sailing Program [19YF1409300 to NSH], and Shanghai Anticancer Association EYAS PROJECT [SACA-CY1A04 to NSH].
Footnote
Reporting Checklist: The authors have completed the MDAR reporting checklist. Available at http://dx.doi.org/10.21037/atm-20-4530
Data Sharing Statement: Available at http://dx.doi.org/10.21037/atm-20-4530
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/atm-20-4530). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. The protocol for the research project has been approved by a suitably constituted Ethics Committee of Fudan University Shanghai Cancer Center (No. 050432-4-1805C), and it conforms to the provisions of the Helsinki Declaration as revised in 2013.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Kebebew E, Greenspan FS, Clark OH, et al. Anaplastic thyroid carcinoma. Treatment outcome and prognostic factors. Cancer 2005;103:1330-5. [Crossref] [PubMed]
- Kitamura Y, Shimizu K, Nagahama M, et al. Immediate causes of death in thyroid carcinoma: clinicopathological analysis of 161 fatal cases. J Clin Endocrinol Metab 1999;84:4043-9. [Crossref] [PubMed]
- Smallridge RC, Copland JA. Anaplastic thyroid carcinoma: pathogenesis and emerging therapies. Clin Oncol (R Coll Radiol) 2010;22:486-97. [Crossref] [PubMed]
- Wendler J, Kroiss M, Gast K, et al. Clinical presentation, treatment and outcome of anaplastic thyroid carcinoma: results of a multicenter study in Germany. Eur J Endocrinol 2016;175:521-9. [Crossref] [PubMed]
- Subbiah V, Kreitman RJ, Wainberg ZA, et al. Dabrafenib and Trametinib Treatment in Patients With Locally Advanced or Metastatic BRAF V600-Mutant Anaplastic Thyroid Cancer. J Clin Oncol 2018;36:7-13. [Crossref] [PubMed]
- Ryder M, Ghossein RA, Ricarte-Filho JC, et al. Increased density of tumor-associated macrophages is associated with decreased survival in advanced thyroid cancer. Endocr Relat Cancer 2008;15:1069-74. [Crossref] [PubMed]
- Landa I, Ibrahimpasic T, Boucai L, et al. Genomic and transcriptomic hallmarks of poorly differentiated and anaplastic thyroid cancers. J Clin Invest 2016;126:1052-66. [Crossref] [PubMed]
- Pozdeyev N, Gay LM, Sokol ES, et al. Genetic analysis of 779 advanced differentiated and anaplastic thyroid cancers. Clin Cancer Res 2018;24:3059-68. [Crossref] [PubMed]
- Yoo SK, Song YS, Lee EK, et al. Integrative analysis of genomic and transcriptomic characteristics associated with progression of aggressive thyroid cancer. Nat Commun 2019;10:2764. [Crossref] [PubMed]
- Mattick JS. The genetic signatures of noncoding RNAs. PLoS Genet 2009;5:e1000459. [Crossref] [PubMed]
- Teng M, Liu LY, Hua JT, et al. Orphan noncoding RNAs: novel regulators and cancer biomarkers. Ann Transl Med 2019;7:S21. [Crossref] [PubMed]
- Gupta RA, Shah N, Wang KC, et al. Long non-coding RNA HOTAIR reprograms chromatin state to promote cancer metastasis. Nature 2010;464:1071-6. [Crossref] [PubMed]
- Wang XM, Liu Y, Fan YX, et al. LncRNA PTCSC3 affects drug resistance of anaplastic thyroid cancer through STAT3/INO80 pathway. Cancer Biol Ther 2018;19:590-7. [Crossref] [PubMed]
- Pellecchia S, Sepe R, Decaussin-Petrucci M, et al. The Long Non-Coding RNA Prader Willi/Angelman Region RNA5 (PAR5) Is Downregulated in Anaplastic Thyroid Carcinomas Where It Acts as a Tumor Suppressor by Reducing EZH2 Activity. Cancers (Basel) 2020;12:E235. [Crossref] [PubMed]
- Gao J, Aksoy BA, Dogrusoz U, et al. Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal. Sci Signal 2013;6:pl1. [Crossref] [PubMed]
- Jendrzejewski J, He H, Radomska HS, et al. The polymorphism rs944289 predisposes to papillary thyroid carcinoma through a large intergenic noncoding RNA gene of tumor suppressor type. Proc Natl Acad Sci U S A 2012;109:8646-51. [Crossref] [PubMed]
- Lv W, Yu X, Li W, et al. Low expression of LINC00982 and PRDM16 is associated with altered gene expression, damaged pathways and poor survival in lung adenocarcinoma. Oncol Rep 2018;40:2698-709. [Crossref] [PubMed]
- Fei ZH, Yu XJ, Zhou M, et al. Upregulated expression of long non-coding RNA LINC00982 regulates cell proliferation and its clinical relevance in patients with gastric cancer. Tumour Biol 2016;37:1983-93. [Crossref] [PubMed]
- Xie S, Yu X, Li Y, et al. Upregulation of lncRNA ADAMTS9-AS2 Promotes Salivary Adenoid Cystic Carcinoma Metastasis via PI3K/Akt and MEK/Erk Signaling. Mol Ther 2018;26:2766-78. [Crossref] [PubMed]
- Cao B, Liu C, Yang G. Down-regulation of lncRNA ADAMTS9-AS2 contributes to gastric cancer development via activation of PI3K/Akt pathway. Biomed Pharmacother 2018;107:185-93. [Crossref] [PubMed]
- Tracy KM, Tye CE, Ghule PN, et al. Mitotically-associated lncRNA (MANCR) Affects Genomic Stability and Cell Division in Aggressive Breast Cancer. Mol Cancer Res 2018;16:587-98. [Crossref] [PubMed]
- Ma B, Jiang H, Wen D, et al. Transcriptome Analyses Identify a Metabolic Gene Signature Indicative of Dedifferentiation of Papillary Thyroid Cancer. J Clin Endocrinol Metab 2019;104:3713-25. [Crossref] [PubMed]
- Kunstman JW, Juhlin CC, Goh G, et al. Characterization of the mutational landscape of anaplastic thyroid cancer via whole-exome sequencing. Hum Mol Genet 2015;24:2318-29. [Crossref] [PubMed]
- Tracy KM, Tye CE, Page NA, et al. Selective expression of long non-coding RNAs in a breast cancer cell progression model. J Cell Physiol 2018;233:1291-9. [Crossref] [PubMed]
- Lu W, Xu Y, Xu J, et al. Identification of differential expressed lncRNAs in human thyroid cancer by a genome-wide analyses. Cancer Med 2018;7:3935-44. [Crossref] [PubMed]
- Nikiforova MN, Kimura ET, Gandhi M, et al. BRAF mutations in thyroid tumors are restricted to papillary carcinomas and anaplastic or poorly differentiated carcinomas arising from papillary carcinomas. J Clin Endocrinol Metab 2003;88:5399-404. [Crossref] [PubMed]
- Nikiforov YE. Genetic alterations involved in the transition from well-differentiated to poorly differentiated and anaplastic thyroid carcinomas. Endocr Pathol 2004;15:319-27. [Crossref] [PubMed]
(English Language Editor: J. Reynolds)


