Tumor mutation burden and JARID2 gene alteration are associated with short disease-free survival in locally advanced triple-negative breast cancer
Introduction
Triple-negative breast cancer (TNBC) is the most invasive subtype with high recurrence and metastasis rates, accounting for 15–20% of total breast cancer population (1). Lacking of effective targeted bioagents, endocrine therapy, and immunotherapy, treatment of TNBC mainly depends on chemotherapy. The currently recommended schemes for TNBC chemotherapy exhibit relatively limited efficacy and obvious side-effects, which lead to the poor clinical outcomes (2).
Even now, locally advanced breast cancer (LABC) still accounts for a large proportion of TNBC patients in China. The risk of local recurrence and distant metastasis in LABC is significantly higher than that of the disease in early stages (3). The National Comprehensive Cancer Network (NCCN) guideline recommends neoadjuvant chemotherapy (NAC) as the standard therapy for LABC. Currently, the efficacy evaluation of NAC mainly depends on pathologic complete response (pCR) (4). NAC has boosted the pCR rate up to 50–60% in early TNBC, significantly improved the survival period. However, due to the heavy tumor load, the pCR rate in LABC is lower than 20%. Thus, the majority of LABC after NAC are non-pCR patients, which brings a considerable challenge to clinical practice (5).
Cancer cells genetic alteration play a key role in chemoresistance and tumor metastasis, which directly causes the failure of TNBC therapy. TP53 as well as the retinoblastoma tumor suppressor gene (RB1), are the two critical tumor suppressors and important in cell proliferation regulation (6). The dysregulation of TP53 and RB1 genes is the typical status of tremendous tumor cells (7). Unfortunately, no efficient therapeutic strategy focus on TP53 has been developed until now. Although the CDK4/6 inhibitors targeting RB1 pathway showed successful clinical utility in metastatic breast cancer, the features of patients who would get benefit remains poorly understood (8).
The jumonji AT-rich interactive domain 2 (JARID2) encodes a Jumonji and AT-rich interaction domain (ARID)-domain-containing protein, which is a DNA-binding protein that functions as a transcriptional repressor (9). JARID2 was considered to be deregulated in many kinds of cancers and promoted their progression. For example, JARID2 was highly expressed in hepatocellular carcinoma (HCC) tissues and promoted epithelial and mesenchymal transition (10). Knockdown of JARID2 impaired the invasive and sphere-forming abilities of bladder cancer cells, as well as reduced the population of tumor initiating cells (11), suggesting that JARID2 plays an important role in bladder cancer progression. Nevertheless, the role of JARID2 in TNBC remains unknown.
Hyper-activation of phosphorylation cascades, which is initiated by protein tyrosine kinases (PTK), is a typical biological characteristic of TNBC. Protein tyrosine phosphatases (PTPs) serves as the antagonists of PTK, and PTP non-receptor type 13 (PTPN13) is the highest molecular weight (270 kDa) PTPs (12). Expression of PTPN13 gene was proved an independent prognostic marker for overall survival (OS) in breast cancer (13). PTPN13 was identified as one of the three most frequently mutated PTPs in colorectal carcinoma. However, little is known about PTPN13 gene alteration in TNBC (14).
In addition, fibroblasts and immune cells interacting with cancer cells in tumor microenvironment (TME) also play an important role in tumor cells chemoresistance and invasion. Through genetic and phenotypic changes, cancer cells also educate focal fibroblasts and immune cells to promote the TME transform towards an immunosuppressive status (15). Currently, the immune checkpoint blockades represents the most promising approach in cancer immunotherapy and highlights the treatment of various cancers. At present, anti-programmed cell death protein 1 (PD-1) and anti-programmed death-ligand 1 (PD-L1) have improved the prognosis of patients with advanced metastatic TNBC (16). However, patients acquire anti-tumor effect by anti-PD-1/PD-L1 therapy have been limited in certain patient subgroups. Researchers has found that expression of PD-1/PD-L1 in tumor tissues is indicative of immune checkpoint blockades response, but its predictive value is still insufficient. Recently, tumor mutation burden (TMB) has emerged as a potential biomarker for immune checkpoint blockade therapy (17). Meanwhile, tumor mutation characteristic of TNBC is complicated, with significant differences in chemosensitivity, local recurrence, and metastasis rates (18). Most recently, next-generation sequencing (NGS) emerges an effective technology for analyzing tumor heterogeneity and predicting immunotherapy efficacy (19). Here, we enrolled 14 cases of locally advanced TNBC; all were non-pCR patients after NAC, 7 developed disease progression within 12 months after operation [short disease-free survival (short-DFS)], the others developed disease progression after 1 year (long-DFS). The post-operation tumor tissues were collected. NGS targeting 422 common tumor genes and in vitro experiments were performed aiming to explore prognostic biomarkers and potential therapeutic molecular targets for non-pCR LABC patients.
We present the following article in accordance with the STROBE reporting checklist (available at http://dx.doi.org/10.21037/atm-20-3773).
Methods
Ethical compliance
All procedures performed in studies involving human participants were in accordance with the ethical standards of the institutional and/or national research committee and with the Declaration of Helsinki (as revised in 2013). This study was approved by the institutional ethnics committee of the Fourth Hospital of Hebei Medical University, China. The authors had access to information that could identify individual participants during or after data collection.
Patient enrollment and sample preparation
A total of 14 patients were enrolled, who were diagnosed with stage IIb-IIIc primary invasive ductal TNBC and treated at the Fourth Hospital of Hebei Medical University between January 2014 and December 2016 were eligible in this study. All were adult females aged 35 years old or over who underwent radical breast surgery after 4–8 cycles of NAC, and had no previous exposure to radiotherapy, chemotherapy or immunotherapy for breast cancer.
Evaluation of tumor response was performed by independent evaluators, blinded to the arm assignment, and consisted of 2 oncologists and 2 pathologists. Tumor response status was defined according to the Response Evaluation Criteria in Solid Tumors Committee (RECIST). Specifically, complete response (CR) was defined as the disappearance of all the lesions both in the breast specimen and draining nodes. The primary endpoint objective response rate (ORR) was composed of tumor response classifications of CR and partial response (PR). Other analysis endpoints included pCR and disease-free survival (DFS).
The pCR was defined as a lack of histological evidence of invasive tumor cells in the surgical breast specimen and draining nodes. The presence of residual ductal carcinoma-in situ was not included in the pCR category after neoadjuvant treatment.
All patients were followed up after diagnosis until December 2019. Any diagnosis of metastasis and/or recurrence of breast cancer needed to be clinically demonstrated by either imaging studies of ultrasound, computed tomography CT, magnetic resonance imaging (MRI), bone scan, or other imaging modalities, and/or by history and physical exam.
In total, 14 post-operational formalin-fixed paraffin-embedded (FFPE) blocks and blood samples were collected. For FFPE samples, only those samples harboring above 20% tumor cell content were considered qualified and subsequently tested. Written consent from all patients was collected according to the ethics regulations of the Fourth Hospital of Hebei Medical University, China. Collected samples were sent to the core facility of Nanjing Shihe Geneseeq Biotechnology Inc. (Nanjing, China) for targeted NGS analysis.
DNA extraction
In total, 6–8 sections, 10 µm thickness, were collected from breast cancer FFPE samples per patient, which were de-paraffinized with xylene, and genomic DNA was extracted using QIAamp DNA FFPE Tissue Kit (QIAGEN, Hilden, Germany) following the manufacturer’s instructions. Genomic DNA from matched blood samples were prepared with DNeasy Blood & Tissue kit (QIAGEN) and sequenced for the purpose of identifying germline mutations. Purified genomic DNA was qualified by Nanodrop 2000 for A260/280 and A260/A230 ratios (Thermo Fisher Scientific). All DNA samples were quantified by Qubit 3.0 using the dsDNA HS Assay Kit (Life Technologies) according to the manufacturer’s recommendation.
Library preparation
Next, 500 ng–1 µg of extracted tumor genomic DNA was fragmented into 300–350 bp using Diagenode Bioruptor Plus (Bioruptor, Belgium). In brief, sequencing libraries were prepared with KAPA Hyper Prep kit (KAPA Biosystems) according to the optimized protocols. Fragmented DNA was selected using AxyPrep TM FragmentSelect-I Kit (Axygen), then proceeded with end-repairing, A-tailing, and adapter ligation with indexed adapters. Libraries were then subjected to polymerase chain reaction (PCR) amplification and purification before targeted enrichment.
Targeted next generation sequencing
A customized NGS panel targeting the whole exons and partial introns of 422 cancer-related genes, as listed in Figure S1, was used for hybridization enrichment. DNA libraries from different samples were marked with unique barcodes during library preparation and pooled together up to a total amount of 2.5 µg and subjected to probe-based hybridization using IDT xGen Lockdown reagents (IDT, Coralville, IA, USA) and Dynabeads M-270 (Thermo Fisher). Captured libraries were on-beads amplified with Illumina p5 and p7 primers in KAPA HiFi HotStart ReadyMix (KAPA Biosystems). The final library was quantified by KAPA Library Quantification kit (KAPA Biosystems) as the manufacturer’s instructions. Bioanalyzer 2100 (Agilent, Stanta Clara, CA, USA) was used to determine the fragment size distribution of the final library.
FFPE samples were sequenced with an average sequencing depth of at least 600× on Illumina Hiseq 4000 platform (Illumina, San Diego, CA, USA). The sequencing data were first demultiplexed by bcl2fastq v2.16.0.10 (Illumina, Inc.) and then subjected to Trimmomatic (8) for FASTQ file quality control (QC). Leading/trailing low quality (base Phred score below 15) or N bases were removed. Qualified reads were mapped to the reference human genome hg19 using Burrows-Wheeler Aligner (BWA-MEM, v.0.7.12) (9) with BWA-MEM algorithm and default parameters to create SAM files. Picard 1.119 (http://picard.sourceforge.net/) was used to convert SAM files to compressed BAM files which were then sorted according to chromosome coordinates. The Genome Analysis Toolkit (GATK, version 3.4-0) was modified and used to locally realign the BAMs files at intervals with indel mismatches and recalibrate base quality scores of reads in BAM files.
Variant filtering and annotation
Single nucleotide polymorphisms (SNPs) and indels were called by VarScan2 and HaplotypeCaller/UnifiedGenotyper in GATK, and common variants were filtered out by using dbSNP and the database from the 1,000 Genomes project. Whole blood control was used for identifying germline mutations. Structural abnormalities, such as gene fusions were identified by FACTERA, and copy number variations (CNVs) were analyzed with ADTEx.
Cell culture and transfection
The human breast cancer cell lines, including MCF-7, BT-474, SK-BR-3, and MD-MBA-231, along with skin fibroblast cells and esophageal squamous cell carcinoma cell lines, KYSE170 and TE,1 were cultured in Dulbecco’s Modified Eagle Medium (DMEM) or RPMI 1640 (Gibco, USA) medium supplemented with 10% fetal bovine serum (FBS) (Gibco, USA), 50 units of penicillin, and 50 µg/mL of streptomycin. All cells were cultured under the standard conditions of 5% CO2 at 37 °C. For transfection, cells were transfected with the indicated combinations of JARID2-silencing RNA [human JARID2 small interfering RNA (siRNA)] using EndoFectinTM-Max transfection reagent (GeneCopoeia, Guangzhou, China) according to the manufacturer’s instructions. Briefly, 1 day before transfection, 1.5×105 cells were plated on a 6-well plate in 2 mL medium free of antibiotics. The next day, transfection reagents containing 0.1 nmol of siRNA and 5 µL of Lipofectamine RNAiMAX in a final volume of 260 µL with Opti-MEM I were added to each well and incubated for 24 hours prior to further assay. A non-targeting sequence was used as a negative control.
RNA extraction and reverse transcription polymerase chain (RT-PCR)
Cells were lysed at the indicated time points, total RNA was extracted from cells with the TRIzol reagent according to the manufacturer (Invitrogen, USA). The concentration and quality of RNA were determined for each sample with 260/280 nm ratio with a Nanodrop Spectrophotometer (ND-100, Thermo, Waltham, MA). cDNA was synthesized using All-in-One First-Strand cDNA Synthesis kit (GeneCopoeia, China). Reverse transcription polymerase chain reaction (RT-PCR) was performed by using 2× Es Taq MasterMix Dye (CWBIO, China). Products of RT-PCR were separated using 1.5% agarose gels and were examined by ultraviolet (UV) irradiation. RT-PCR-based amplification was performed using the primers in Table S1.

Full table
Western blot
Cells were lysed to extract proteins with radioimmunoprecipitation assay (RIPA) reagent at the indicated time. The extracted protein samples were analyzed using 8% or 10% SDS-polyacrylamide gel electrophoresis (SDS-PAGE), and the separated proteins were transferred onto polyvinylidene fluoride (PVDF) membranes (Millipore, Billerica, MA, USA). Each membrane was blocked with 5% nonfat dry milk in tris-buffered saline with Tween20 (TBS-T) for 1 h, followed by incubation with primary antibody at 4 °C overnight. The membrane was then incubated with horseradish peroxidase (HRP)-conjugated secondary antibody before it was examined with an enhanced chemiluminescence (ECL) kit. The antibodies for western blot are shown in Table S2.
Full table
Statistical analysis
Statistical analysis was performed by using the SPSS 21.0 and GraphPad Prism 5 software. Data are shown as mean ± standard deviation (SD) and examined by using student’s t-test or one-way analysis of variance (ANOVA). Chi-squared statistical tests were employed to analyze the results. OS was defined from the time of diagnosis to the time of death from any cause. DFS was defined from the time of diagnosis until progressive disease was confirmed or until death from any cause. DFS was estimated by the Kaplan-Meier method and compared with log-rank test. All experiments were performed in triplicate, and all tests were two-sided, with P values <0.05 being considered significant.
Results
Patient characteristics
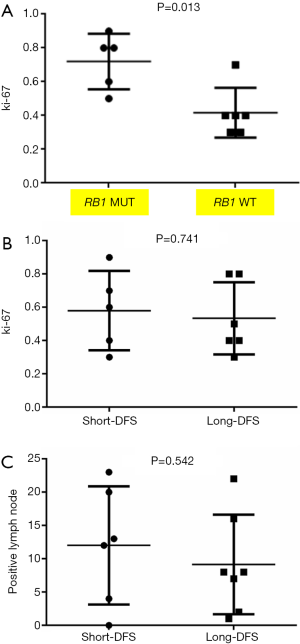
In this study, the data of 14 TNBC patients in stage IIb-IIIc were collected, and the proportion of patients in stage III was 78.6% (11/14). All patients received NAC with paclitaxel and anthracycline for 4–8 cycles. All were non-pCR TNBC proven by pathological examination after radical surgery. The median age of TNBC patients was 48 [37–71] years. Patients were followed up after operation: 7 patients developed disease progression within 12 months after operation and were assigned to the short-DFS group (n=7), while the others developed progression after 1 year and were assigned to the long-DFS group (n=7). No significant difference in age, tumor size, lymph node metastasis, clinical stage, and Ki-67 score was found between the 2 groups. The clinicopathological characteristics of patients are presented in Table 1.
Full table
Overall genetic mutation number and types identified by NGS
Four hundred and twenty-two cancer-related genes (see Figure S1 for gene list) were screened in tissue samples of TNBC patients by NGS. Figure 1A represents the gene mutation number of each patient, and Figure 1B shows the gene alteration map of the overall cohort. A total of 72 alterations were detected within 14 patients, ranging from 1–8 per patient with a median mutation number of 5. As shown in Figure 2A, the median genetic mutation in the short-DFS group was higher than that of the long-DFS group (6.0 vs. 4.3; P=0.094), indicating the more genetic mutations in the former group. The overall mutation types are displayed in Figure 2B. A total of 6 mutation types were detected, with the majority of missense mutation in 50.0% (36/72) patients. Figure 2C and Figure 2D show the mutation types of either group separately, with a higher missense mutation rate of 57.0% (24/42) in the short-DFS group compared with the 40.0% rate (12/30) of the long-DFS group. No correlation between mutation types and DFS was found.
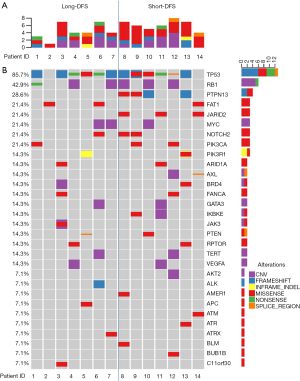
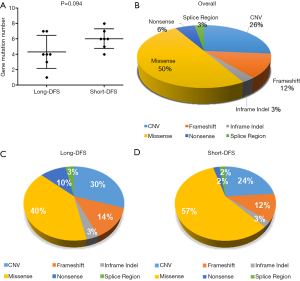
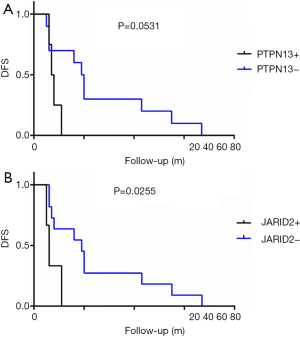
For the total cohort, the 30 mutation genes within the 422-gene panel are shown in Figure 1. The genes with the highest mutation frequency included TP53 (12/14, 85.7%), RB1 (6/14, 42.9%), PTPN13 (4/14, 28.6%), FAT1 (3/14, 21.4%), JARID2 (3/14, 21.4%), MYC (3/14, 21.4%), Notch2 (3/14, 21.4%), and PIK2CA (3/14, 21.4%). Among these genes, TP53 displayed the highest mutation frequency (12/14, 85.7%), while the most common mutation type was frameshift. No correlation between TP53 mutation and DFS was found in the entire cohort (P=0.512). There were equal TP53 mutations in the long-DFS and short-DFS groups (6/7 and 85.7%, respectively). The second highest gene was RB1 (6/14, 42.9%) and the main mutation type was CNV. No correlation between RB1 mutation and DFS (P=0.605) was found in total patients. Each group displayed the same RB1 mutation rates separately (3/7, 42.9%). Interestingly, patients with high Ki-67 expression were more likely to have RB1 mutations in the tumor tissues (P=0.013), as shown in Figure 3A. While neither Ki-67 status (Figure 3B) nor the number of metastatic lymph nodes (Figure 3C) showed significant difference between these two groups. The third highest gene was PTPN13 (28.6%, 4/14), with all of these mutation types occurring in patients with short-DFS. The PTPN13 mutation frequency showed significant difference between these two groups (P=0.018). However, no difference in DFS was found between the PTPN13 mutation and wild type patients (P=0.053) (Figure 4A).
JARID2 mutation was related to the poor DFS
The overall mutation rate of FAT1, JARID2, MYC, NOTCH2, and PIK2CA was 21.4% (3/14). Other common mutation genes such as PTEN (2/14, 14.3%) and VEGFA (2/14, 14.3%) were also detected and distributed in the 2 groups. Among these genes, JARID2 mutation was only detected in the short-DFS group. Furthermore, patients with JARID2 mutation presented significantly shorter DFS period (P=0.026) (Figure 4B). In addition, the mutation type of JARID2 was mainly missense mutation, suggesting the possibility of activating specific downstream signals and relevant adverse prognostic factors by JARID2 mutation.
Downregulation of the JARID2 gene expression increased epithelial-mesenchymal transition (EMT) activities in MD-MBA-231 cells
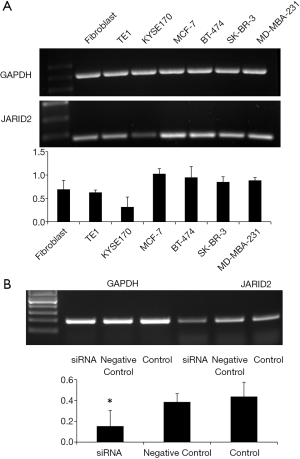
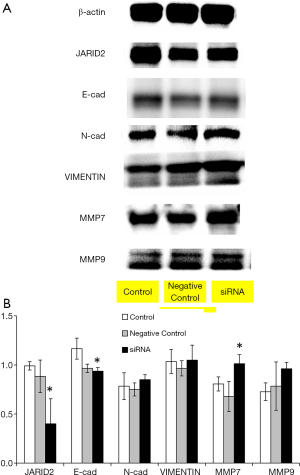
In order to further clarify the role of the JARID2 gene in TNBC, we performed relevant experiments in vitro. As shown in Figure 5A, we first detected the expression of JARID2 gene in various cell lines by RT-PCR. Breast cancer cell lines, including MCF-7, BT-474, SK-BR-3, and MD-MBA-231, presented a universal expression of JARID2. JARID2 gene expression was also detected in other control cell lines such as human skin fibroblasts (represented the cancer-associated fibroblasts in focal tumor or the stromal cells in TME) and human esophageal cancer cells, KYSE170 and TE1. Next, we knocked down JARID2 gene in MD-MBA-231 cells with siRNA (Figure 5B). We found that the E-cadherin expression decreased, while vimentin, MMP7, and MMP9 expression increased after downregulation of JARID2 protein levels (Figure 6). The activation of epithelial-mesenchymal transition (EMT) is one of the hallmarks of caner metastasis, which is characterized by E-cadherin downregulation and vimentin and MMP upregulation. Our data showed that knocking down JARID2 gene in MD-MBA-231 cells was capable of enhancing its EMT ability, which supported the results gathered by NGS.
Discussion
In this study, we enrolled 14 non-PCR patients with locally advanced TNBC treated by NAC and sequenced the post-operation tumor tissues by NGS. Both the mutation frequency and mutation types of short-DFS patients were higher than those of the long-DFS cases. The common mutation genes of breast cancer, including TP53, RB1, PIK3CA, and PTEN, were detected in both groups. The JARID2 gene mutation was only present in short-DFS patients and was significantly related to poor disease outcome. In vitro experiments demonstrated that downregulation of JARID2 could enhance EMT activity of TNBC cells, making JARID2 potential prognostic and therapeutic marker for non-PCR patients.
Immune checkpoint blockades are the focus of cancer immunotherapy in recent years, and TMB is recognized as a biomarker of immunotherapy. Cancer patients with high TMB are more likely to benefit from immunotherapy. Although whole exome sequencing (WES) is the standard method for TMB evaluation, the extremely high expense limits its clinical application. NGS, also known as high-throughput sequencing, is continuously making progress in breast cancer precision medicine (20). NGS can identify new treatment targets for individual breast cancer patients and predict the risk of recurrence and metastasis. At present, different sequencing platforms have formed their own related database and bioinformatics analysis systems to serve clinical practice (21). The platform we used is a commercialized targeted NGS of 422 cancer-related genes, including common cancer susceptibility genes and related signal pathway molecules, and is widely used in malignant tumor diagnosis and research (Figure S2). The genetic mutation types were in accordance with the data in The Cancer Genome Atlas (TCGA) (Figure S3).
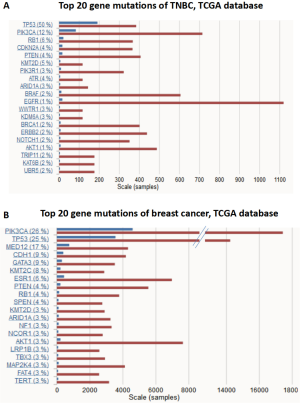
In our data, TP53 displayed the highest mutation frequency among the 422 cancer-related factors and the mutation types include missense, inactivation, insertion, and deletion mutations. TP53 is a common tumor suppressor in humans, whose expressed protein plays an important role in cell growth, proliferation, and DNA damage repair. Previous studies have also reported TP53 mutation occurred frequently in breast cancer (75%), which is likely related to cancer development and outcomes (22). Together with epidermal growth factor receptor (EGFR) and cytokeratin, TP53 has been regarded as an important marker for TNBC subtype biomarker. The high mutation rate of TP53 in TNBC was also shown in the COSMIC cancer database (50%) (Figure S3). Although numerous TP53 mutations have been detected across among multiple tumor types, no applicable therapeutic agent targeting TP53 is available for clinical practice (23).
NAC was originally used as the induced chemotherapy or primary chemotherapy for patients with inoperable breast cancer. Nowadays, NAC is the primary therapeutic strategy for LABC patients. Numerous clinical studies have confirmed that NAC is critical to improving the living quality and survival period (24). Although the chemosensitivity and pCR rate of TNBC are relatively high, compared with early breast cancer, the majority of locally advanced TNBC were non-pCR after NAC. More importantly, TNBC is a high-heterogeneity subtype of breast cancer and still lacks effective evaluation parameters (25). In this study, 14 patients with locally advanced TNBC who received NAC were retrospectively analyzed by NGS. The JARID2 mutation was only detected in the short-DFS group, which was significantly related to increased disease recurrence and metastasis; thus, the JARID2 mutation may be a potential predictive biomarker for these patients.
The Jumonji AT-rich interactive domain 2 (JARID2) encoded protein can modify chromatin structure and inhibit target gene transcription through DNA binding (26). At present, JARID2 is classified as a histone demethylase by virtue of its epigenetic regulation ability. Knocking down JARID2 in mouse endothelium decreased methylation on the histone H3 (H3K9) of the Notch1 gene locus, confirming that JARID2 is a transcriptional inhibitor of Notch1 and maintains the stem cell characteristics (27). However, the studies focus on JARID2 in malignant tumors are contradictory. Declining JARID2 gene expression was reported in B-chronic lymphoblastic leukemia and acute monocytic leukemia (28), while knockout of JARID2 gene was found to promote the proliferation of leukemia cells by accelerating G1/S transformation. Further analysis showed JARID2 negatively regulated the expression of CCND1 through increasing the methylation of H3K27 on its promoter, thus negatively regulating leukemia cell proliferation (29). However, downregulation of JARID2 significantly inhibited the proliferation, migration, and invasion of human ovarian cancer cells by reducing the expression of phosphorylated inositol 3 kinase (PI3K) and protein kinase B (Akt) (30). It has also been reported that overexpression of JARID2 increased the metastasis ability of lung cancer cells by promoting EMT process (31). Researchers have proven that JARID2 gene deletion is common in chronic myeloid malignancies, but no reports on the somatic mutation of JARID2 in breast cancer is available (32). Our data identified the mutation of JARID2 in TNBC only in short-DFS patients, which thus makes it a potential marker for prognosis. Data in vitro showed that inhibition of JARID2 gene expression in TNBC cell lines could modulate the EMT-related marker expression, which involved the decrease of E-cadherin and the increase of vimentin, MMP7, and MMP9 expression. EMT is an important biological process in cancer metastasis, in which epithelial cells are transformed into stromal cells. These data were consistent with the mutations detected in clinical specimens of short DFS by NGS.
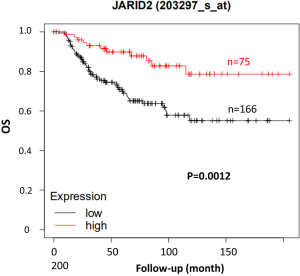
One limitation of this study is that no BRCA1/2 gene mutation was found in our patients. It has been reported that 5.3% and 11.2% TNBC patients have BRCA1/2 mutations in China, respectively (33). The frequency of BRCA1/2 gene mutation in Caucasians is as high as 40–50% in familial breast cancer patients, while that in Chinese is less than 20% (34). The failure to detect this mutation may be related to the small sample size of our study. The 14 patients should be followed up continuously to determine the effect of the mutation genes on OS. In addition, due to the high heterogeneity of TNBC, the application of these molecular markers is still in the exploratory stage. In order to better solve the therapeutic problem of locally advanced TNBC, the biomarkers should be further screened in the non-pCR TNBC population to effectively transform the new targets into clinical applications. Another limitation of this study is the small sample size, only 14 cases of TNBC were enrolled in our data. Including in vitro and in vivo experiments, more work should be conducted to further demonstrate JARID2 function. OS between short-DFS and long-DFS groups should be analyzed to explore in-depth value of JARID2 in clinical practice. Meanwhile, we used an online survival analysis tool KM Plotter (www.kmplot.com) to assess JARID2 mRNA expression with OS on breast cancer. As shown in Figure S4, TNBC patients with high gene expression of JARID2 have a significantly better prognosis compared with low expression ones (P=0.0012), which confirm our data from another perspective.
Acknowledgments
The authors would like to thank Ms. Huichai Yan and Ms. Jiangxin Wang for their excellent technical assistance, and all the investigators for participating in this study.
Funding: This work was supported by the China Anti-Cancer Association Research Grant.
Footnote
Reporting Checklist: The authors have completed the STROBE reporting checklist. Available at http://dx.doi.org/10.21037/atm-20-3773
Data Sharing Statement: Available at http://dx.doi.org/10.21037/atm-20-3773
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/atm-20-3773). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. All procedures performed in studies involving human participants were in accordance with the ethical standards of the institutional and/or national research committee and with the Declaration of Helsinki (as revised in 2013). This study was approved by the institutional ethnics committee of the Fourth Hospital of Hebei Medical University, China (No. 201107209). Informed consent was obtained from all individual participants included in the study.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Miller KD, Nogueira L, Mariotto AB, et al. Cancer treatment and survivorship statistics, 2019. CA Cancer J Clin 2019;69:363-85. [Crossref] [PubMed]
- Wang RX, Xu XE, Huang L, et al. eEF2 kinase mediated autophagy as a potential therapeutic target for paclitaxel-resistant triple-negative breast cancer. Ann Transl Med 2019;7:783. [Crossref] [PubMed]
- Tran WT, Childs C, Probst H, et al. Imaging Biomarkers for Precision Medicine in Locally Advanced Breast Cancer. J Med Imaging Radiat Sci 2018;49:342-51. [Crossref] [PubMed]
- Cortazar P, Zhang L, Untch M, et al. Pathological complete response and long-term clinical benefit in breast cancer: the CTNeoBC pooled analysis. Lancet 2014;384:164-72. [Crossref] [PubMed]
- Choi M, Park YH, Ahn JS, et al. Assessment of pathologic response and long-term outcome in locally advanced breast cancers after neoadjuvant chemotherapy: comparison of pathologic classification systems. Breast Cancer Res Treat 2016;160:475-89. [Crossref] [PubMed]
- Knudsen ES, Nambiar R, Rosario SR, et al. Pan-cancer molecular analysis of the RB tumor suppressor pathway. Commun Biol 2020;3:158. [Crossref] [PubMed]
- Wang DY, Jiang Z, Ben-David Y, et al. Molecular stratification within triple-negative breast cancer subtypes. Sci Rep 2019;9:19107. [Crossref] [PubMed]
- Wang M, Hu Y, Hou L, et al. A clinical study on the use of Huaier granules in post-surgical treatment of triple-negative breast cancer. Gland Surg 2019;8:758-65. [Crossref] [PubMed]
- Jung J, Mysliwiec MR, Lee Y. Roles of JUMONJI in mouse embryonic development. Dev Dyn 2005;232:21-32. [Crossref] [PubMed]
- Lei X, Xu JF, Chang RM, et al. JARID2 promotes invasion and metastasis of hepatocellular carcinoma by facilitating epithelial-mesenchymal transition through PTEN/AKT signaling. Oncotarget 2016;7:40266-84. [Crossref] [PubMed]
- Zhu XX, Yan YW, Ai CZ, et al. Jarid2 is essential for the maintenance of tumor initiating cells in bladder cancer. Oncotarget 2017;8:24483-90. [Crossref] [PubMed]
- Abaan OD, Toretsky JA. PTPL1: a large phosphatase with a split personality. Cancer Metastasis Rev 2008;27:205-14. [Crossref] [PubMed]
- Wang Z, Shen D, Parsons DW, et al. Mutational analysis of the tyrosine phosphatome in colorectal cancers. Science 2004;304:1164-6. [Crossref] [PubMed]
- Révillion F, Puech C, Rabenoelina F, et al. Expression of the putative tumor suppressor gene PTPN13/PTPL1 is an independent prognostic marker for overall survival in breast cancer. Int J Cancer 2009;124:638-43. [Crossref] [PubMed]
- El-Kenawi A, Hänggi K, Ruffell B. The Immune Microenvironment and Cancer Metastasis. Cold Spring Harb Perspect Med 2020;10:a037424. [Crossref] [PubMed]
- Kwa MJ, Adams S. Checkpoint inhibitors in triple-negative breast cancer (TNBC): Where to go from here. Cancer 2018;124:2086-103. [Crossref] [PubMed]
- Chan TA, Yarchoan M, Jaffee E, et al. Development of tumor mutation burden as an immunotherapy biomarker: utility for the oncology clinic. Ann Oncol 2019;30:44-56. [Crossref] [PubMed]
- Voutsadakis IA. High Tumor Mutation Burden and Other Immunotherapy Response Predictors in Breast Cancers: Associations and Therapeutic Opportunities. Target Oncol 2020;15:127-38. [Crossref] [PubMed]
- Kuo FC, Mar BG, Lindsley RC, et al. The relative utilities of genome-wide, gene panel, and individual gene sequencing in clinical practice. Blood 2017;130:433-9. [Crossref] [PubMed]
- Barroso-Sousa R, Jain E, Cohen O, et al. Prevalence and mutational determinants of high tumor mutation burden in breast cancer. Ann Oncol 2020;31:387-94. [Crossref] [PubMed]
- Curtis C, Shah SP, Chin SF, et al. The genomic and transcriptomic architecture of 2,000 breast tumours reveals novel subgroups. Nature 2012;486:346-52. [Crossref] [PubMed]
- Walerych D, Napoli M, Collavin L, et al. The rebel angel: mutant p53 as the driving oncogene in breast cancer. Carcinogenesis 2012;33:2007-17. [Crossref] [PubMed]
- Synnott NC, Murray A, McGowan PM, et al. Mutant p53: a novel target for the treatment of patients with triple-negative breast cancer? Int J Cancer 2017;140:234-46. [Crossref] [PubMed]
- Early Breast Cancer Trialists’ Collaborative Group (EBCTCG). Long-term outcomes for neoadjuvant versus adjuvant chemotherapy in early breast cancer: meta-analysis of individual patient data from ten andomized trials. Lancet Oncol 2018;19:27-39. [Crossref] [PubMed]
- Harbeck N, Gluz O. Neoadjuvant therapy for triple negative and HER2-positive early breast cancer. Breast 2017;34:S99-103. [Crossref] [PubMed]
- Kinkel SA, Galeev R, Flensburg C, et al. Jarid2 regulates hematopoietic stem cell function by acting with polycomb repressive complex 2. Blood 2015;125:1890-900. [Crossref] [PubMed]
- Mysliwiec MR, Bresnick EH, Lee Y. Endothelial Jarid2/Jumonji is required for normal cardiac development and proper Notch1 expression. J Biol Chem 2011;286:17193-204. [Crossref] [PubMed]
- Mayer RL, Schwarzmeier JD, Gerner MC, et al. Proteomics and metabolomics identify molecular mechanisms of aging potentially predisposing for chronic lymphocytic leukemia. Mol Cell Proteomics 2018;17:290-303. [Crossref] [PubMed]
- Qu Y, Yang Q, Liu J, et al. c-Myc is Required for BRAFV600E-Induced Epigenetic Silencing by H3K27me3 in Tumorigenesis. Theranostics 2017;7:2092-107. [Crossref] [PubMed]
- Cao J, Li H, Liu G, et al. Knockdown of JARID2 inhibits the proliferation and invasion of ovarian cancer through the PI3K/Akt signaling pathway. Mol Med Rep 2017;16:3600-5. [Crossref] [PubMed]
- Tange S, Oktyabri D, Terashima M, et al. JARID2 is involved in transforming growth factor-beta-induced epithelial-mesenchymal transition of lung and colon cancer cell lines. PloS One 2014;9:e115684. [Crossref] [PubMed]
- Puda A, Milosevic JD, Berg T, et al. Frequent deletions of JARID2 in leukemic transformation of chronic myeloid malignancies. Am J Hematol 2012;87:245-50. [Crossref] [PubMed]
- Armstrong N, Ryder S, Forbes C, et al. A systematic review of the international prevalence of BRCA mutation in breast cancer. Clin Epidemiol 2019;11:543-61. [Crossref] [PubMed]
- Lee JM, Ledermann JA, Kohn EC. PARP Inhibitors for BRCA1/2 mutation-associated and BRCA-like malignancies. Ann Oncol 2014;25:32-40. [Crossref] [PubMed]