
A CT-derived deep neural network predicts for programmed death ligand-1 expression status in advanced lung adenocarcinomas
Introduction
Programmed cell death 1 receptor (PD-1) and its ligand programmed death ligand-1 (PD-L1) expression on activated B and T cells of tumor allows them to escape host immune system (1,2). PD-L1 expression is associated with poor prognosis (3-5), and is related to good therapeutic response to PD-L1 inhibitor therapy for patients with advanced non-small cell lung cancer (NSCLC). Anti-PD-L1 antibodies, targeting PD-L1 pathway, have yielded improved therapeutic efficacy and superior toxicity profiles compared with chemotherapy and brought favorable patient outcome (6-8). PD-L1 expression remains a crucial predictor in selecting patients for immunotherapy (9). However, reasonable use of these agents has been restricted by the lack of decisive predictive biomarkers (10). Identification of effective predictive biomarkers can enable safe and valid use of these agents. Nowadays, the detection of PD-L1 expression mainly depends on immunohistochemistry method of tissue sample. However, in clinical practice, the amount of tumor tissues obtained by fine needle aspiration (FNA) is rather limited. In some cases, they can only meet pathology diagnosis and gene test. However, further PD-L1 detection is also essential for some patients to guide therapy. At this time, in order to get PD-L1 expression status, FNA may be needed to be operated again, causing risk of human-made tumor metastasis and more economic burden to patients. Therefore, it is necessary to explore other substitutable and noninvasive tools to predict molecular expression status for advanced lung cancer patients confirmed by histological pathology. The search of more efficient and convenient biomarkers to predict PD-L1 expression before immune checkpoint inhibitor therapy is essential and can help physicians judge who would benefit from such treatments.
Despite a few clinical predictors have been discovered to predict PD-L1 expression, such as inflammatory markers (11) [C-reactive protein (CRP), neutrophil-lymphocyte ratio (NLR), lymphocyte-monocyte ratio (LMR) and platelet-lymphocyte ratio (PLR), etc.] and demographic characteristics[age, gender, smoking status, driver mutation(s), etc.] (12,13), the results are still controversial (14). Refinement of established biomarkers and discovery of novel ones are necessary. To the best of our knowledge, the role of imaging, as an important means of lung cancer diagnosis and therapeutic efficacy evaluation tool, is largely unknown in this aspect.
It is reported that computed tomography (CT) imaging features were correlated with driver gene mutation status of lung cancer (15-18). For molecular aspect, till now, there are only limited numbers of reports regarding correlations between CT imaging features and PD-L1 expression (19,20) in lung cancer patients. And they mainly focused on early stage lung adenocarcinomas (LUAD) which is usually treated by surgical resection while immunotherapy is applied more widely in patients with advanced stage. Therefore, it is necessary to study the correlation between imaging features and PD-L1 expression in advanced lung cancer patients. DL, emerging as a promising and prospective imaging tool, its predictive efficacy in PD-L1 expression has not been reported yet.
In the present study, we aimed to explore the relationship between PD-L1 expression status and CT features including morphologic characteristics and DL, in order to noninvasively predict PD-L1 expression in patients with advanced LUAD. We present the following article in accordance with the STARD reporting checklist (available at http://dx.doi.org/10.21037/atm-19-4690).
Methods
Patients and clinical characteristics
Patients diagnosed with unresectable and advanced LUAD (AJCC stage III and IV) by FNA biopsy and immunohistochemistry of the original tumor tissue, from lung cancer database of the First Affiliated Hospital of Sun Yat-sen University from June 1, 2018 to March 1, 2019 were included. This study was conducted in accordance with the Declaration of Helsinki (as revised in 2013) and was approved by institutional ethics board of the First Affiliated Hospital of Sun Yat-sen University {No. [2018] 010} and informed consents were waived.
All the cases were first-visit patients without any treatment and underwent enhanced chest CT examination after admission. Metastasis were evaluated by contemporaneous CT/MRI/PET-CT scans or biopsy. Patient characteristics for analysis included age, gender, smoking status, AJCC stage, pathologic tumor-node-metastasis (TNM) stage, driver gene mutation status, and PD-L1 expression. Definition: smoking status (non-smokers were defined as having smoked <100 cigarettes/life, former and current smokers were regarded as smokers), AJCC stage (AJCC Cancer Staging Handbook, 7th ed. New York, NY: Springer, 2009), TNM stage (7th Edition TNM classification of lung cancer staging system). In the current study, driver gene mutation status indicated wild-type and mutation type, the latter containing EGFR, ALK, KRAS, TP53, BRAF, HER2, MET, RET and ROS1.
Immunohistochemical detection of PD-L1 protein expression
Tumor tissue were deparaffinized and hydrated for immunohistochemical analysis by using primary rabbit monoclonal antibody against human PD-L1 (SP263; 1:2,000; Roche Ventana, Tucson, AZ, USA). The immunohistochemistry results for PD-L1 expression depended on the intensity of cell membrane staining, the proportion of PD-L1-positive cells showing membranous staining were independently evaluated by three investigators, and the consensus was reached when existing controversy and was regarded as the final results. Membrane PD-L1 expression on tumor cells was defined by tumor proportion scores (TPS), PD-L1 TPS ≥1% was defined as positive expression and PD-L1 TPS <1% was defined as negative expression. In our study cohort, PD-L1 TPS <1% were observed in 81cases, 1–5% in 1 case, 5–10% in 1 case, 10–25% in 1 case, 25–50% in 5 cases, ≥50% in 38 cases respectively. TPS cutoff of 5% and 10% were not applied in the subgroup analysis due to the almost unchanged patient numbers compared to TPS cutoff of 1% subgroups.
Morphologic CT characteristics and scanning protocol
Enhanced chest CT images of the included patients were acquired within one week prior to biopsy. Imaging characteristics were assessed by two experienced chest radiologists (XY Yang and X Wu) through PACS reading workstation, with experience of 21 and 25 years, respectively. The consensus was reached when divergence of views existed. Both of them were blinded to PD-L1 expression status and the cases were mixed with other types of lung cancer cases such as squamous cell carcinoma and small cell lung cancer as well as stage I and II adenocarcinoma to avoid bias. Definition of Morphologic CT imaging characteristics: tumor size indicated maximum axial diameter. Lesion type was classified as nodule (maximum diameter <3 cm), mass (maximum diameter ≥3 cm), inflammatory type (patchy lesion with unclear boundary). Necrosis meant low density in CT plain scan and without enhancement in enhanced scan. The volume percentage of necrosis components was divided into 0–25%, 25–50%, 50–75%, 75–100%.
CT scan was performed with a 64-row multidetector CT scanner (Aquilion 64, Canon, Japan) and a 320-row multidetector CT scanner (Aquilion ONE ViSION, Canon, Japan), in the craniocaudal direction during inspiration. For 64-row multidetector CT scanner, the scan parameters were as follows: tube voltage 120 kV and tube current 200 mAs; beam pitch, 0.828; rotation time, 0.5 second; beam collimation, 64 mm × 0.5 mm. For 320-row multidetector CT scanner, the scan parameters were as follows: tube voltage 120 kV and automatic tube current; beam pitch, 0.813; rotation time, 0.5 second; beam collimation, 80 mm × 0.5 mm. Axial thin-section multidetector CT images of the whole lung were reconstructed with slice thickness and spacing of 1 and 0.8 mm by using a high-spatial frequency algorithm. Iopromide (300 mgI/mL, Schering Pharmaceutical Ltd.) was used as contrast agent for enhanced scanning, and 80–100 mL was injected at 3–4 mL/s flow rate.
DL-3D DenseNet model
Segmentation and data preprocessing
The image data extracted by manual segmentation labels using the annotation tool “ITK-SNAP” (www.itksnap.org) (21) contains the region of interest (ROI) of lung cancer tissue only which were placed by three experienced chest radiologists (XY Yang, X Wu and DD Chang) layer by layer in enhanced axial thin-section CT images. The dataset generated using labels of tumor contour in each slice of CT image were combined to from a 3D image for further DL analysis. 3D images for each lung cancer lesion were converted to a fixed cube with shape 128×128×64 to fit the input of the proposed 3D neural network. Cubic interpolation method and zero-padding of the resized image along the axis was performed to finally generate a uniform three-dimensional image.
3D DenseNet model building
A 3D DenseNet (three-dimensional densely connected convolutional networks) model was selected as the base model in our experiment due to its obvious advantages of excellent over-fitting resistance when training data is relatively scarce and its high computational efficiency serving the purpose of the subsequent five-fold cross validation compared to the others such as ResNet (Deep Residual Learning for Image Recognition). The 3D model was formed by modifying the kernel of each convolutional and pooling layer of the typical DenseNet 2D images to 3D forms. Binary cross-entropy loss of our model was optimized by stochastic gradient descent (SGD) with mini-batch size of 16. We used cosine annealing to schedule our learning rate for total 900 steps by setting initial learning rate to 1e-6 and minimum learning to 1e-7. The architecture of our 3D DenseNet model as given in Figure 1. In our study, for the relatively small sample size, data augmentation was also applied to further alleviate overfitting. The data augmentation includes random image crop and flip, and randomly editing window width and center. During model training, transfer learning was adopted to speed up learning process as well as taking advantage of the pretrained model which already contained weights that could be used to extract general features. We transform the 2D pretrained model using ImageNet dataset to 3D by modifying the 2D kernels to 3D kernels.
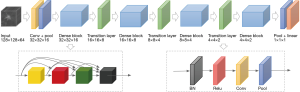
Cross-validation and evaluation metrics
A stratified five-fold cross-validation procedure was adopted to evaluate the prediction performance of DL model in which the data were divided up randomly into five groups without overlapping according to PD-L1 expression where each group has the same proportion of data. During training, each group in turn was chosen as validation set and the others were used as training set. The final evaluation result was based on the performance of the five models.
The prediction metrics of the DL model [including area under the receiver operating characteristic curves (AUC), accuracy (ACC), specificity (SP), sensitivity (SN)] was evaluated according to the performance of the five models which were trained and validated. The model was implemented under the MXNet (version 1.2.0, Apache Software Foundation, Forest Hill, MD USA) framework by using Python programming language (version 2.7.12) and was trained on NVIDIA GeForce GTX 1080 GPUs (NVIDIA, Beijing, China).
Statistical analysis
Clinical and image variables were grouped according to PD-L1 expression status. Statistical analyses were performed by using SPSS 22.0 (IBM, USA). Categorical variables were compared by χ2 test, Fisher’s exact test. Continuous variables were compared using Mann-Whitney U test. P<0.05 was considered as statistically significant. Note: variables of presence of pleural tag, air bronchogram, calcification, cavitation, margin, lobulation, spiculation, vessel convergence, vascular involvement showed were treated as binary variable (“yes” or “no”) for statistical analysis. Additionally, according to the reported (22,23), smoking status may corelate with PD-L1 expression in lung cancer patients, herein, variable of smoking status was treated as both continuous variable (pack year) and binary variables. In addition, for morphologic CT imaging characteristics, Multivariate logistic regression analysis was applied to explore independent predictors of PD-L1 expression, variables included in this analysis were age, gender, smoking status, driver-gene mutation status, vascular involvement, lung metastasis and lung metastasis type. The distinguishing efficacy of the statistically significant variable was presented by AUC.
Results
Correlations between clinicopathological characteristics and PD-L1 expression
The clinicopathological characteristics of the 127 LUAD patients included in the present study were showed in Table 1. Fifty-three (41.7%) patients were female, the median age of all patients was 63 years, 64 (50.4%) had never smoked, 29 (22.8%) were AJCC stage III and 98 (77.2%) were stage IV, 72 (56.7%%) were T3–T4 and 49 (38.6%) were N3, sensitive gene mutations were observed in 103 (81.1%) cases. PD-L1 was positive in 46 patients (36.2%) and negative in the remaining 81 (63.8%) cases, PD-L1-TPS <50% were observed in 89 (70.1%) patients and ≥50% were seen in the remaining 38 (29.9%) cases respectively. Statistical analysis showed no significant difference of all included clinical indicators both between PD-L1-TPS 1% subgroups and between PD-L1-TPS 50% subgroups. In contrast, EGFR mutation was associated with female gender (P=0.001) and non-smokers (P=0.000) as shown in Table S1.
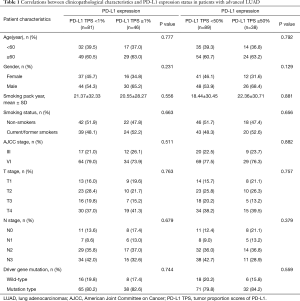
Full table
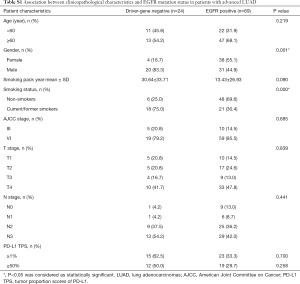
Full table
Associations between morphologic CT characteristics and PD-L1 expression
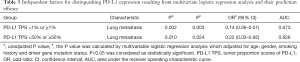
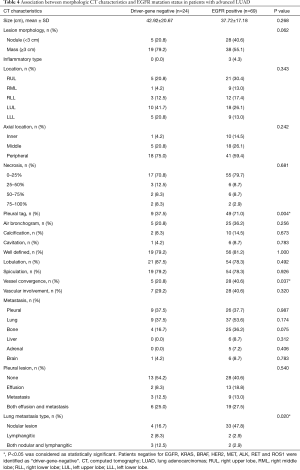
Among 127 patients and as given in Table 2, univariate analysis revealed that, among the morphologic CT characteristics, only vascular involvement (P=0.022) and lung metastasis (P=0.002) were associated with PD-L1-negative/positive expression status, they were more commonly (58% vs. 37%, 56.8% vs. 28.3%) presented in PD-L1-negative subgroup. Whilst, for PD-L1-TPS of 50% expression status, only lung metastasis (P=0.010) showed statistical significance, and was more commonly (53.9% vs. 28.9%) presented in PD-L1-TPS <50% expression group. Whilst, multivariate logistic regression analysis showed that only lung metastasis (P=0.003 for TPS 1%, P=0.024 for TPS 50%) was significantly associated with PD-L1 expression despite of different PD-L1 TPS cutoff values, given in Table 3. ROC curves of the independent morphologic CT characteristic for predicting PD-L1-TPS status of 1% (AUC 0.673) and 50% (AUC 0.626) was shown in Figure 2. In contrast, with regard to EGFR mutations, a univariate analysis revealed that EGFR-mutated adenocarcinomas were only statistically associated with lung metastasis type (P=0.020), as shown in Table 4.
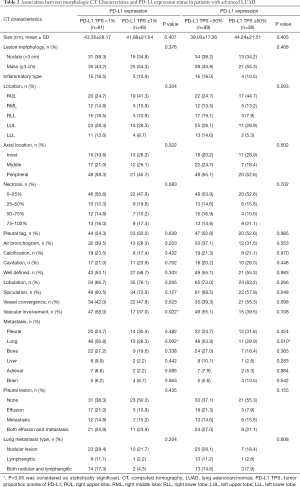
Full table
Full table
Full table
PD-L1 prediction efficiency evaluation by DL 3D DenseNet model
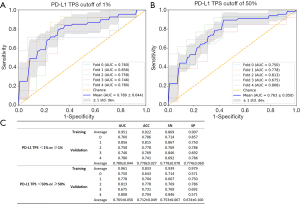
Five-fold cross-validation of the proposed DL-3D DenseNet model was applied to yield the results of prediction efficacy. The final result was computed by averaging the results of the five folds in respect to each metric. The results of the five rounds experiments and the final results were shown in Figure 3. In the validation dataset, when PD-L1-TPS cutoff value of 1% was used to group the patients, average AUC, accuracy, sensitivity and specificity were 0.784, 0.769, 0.773, 0.760. Whilst, when PD-L1-TPS cutoff value of 50% was used, average AUC, accuracy, sensitivity and specificity were 0.765, 0.712, 0.753, 0.674.
Discussion
Our present study aimed to non-invasively predict PD-L1 expression status through CT-derived morphologic features and DL in advanced LUAD. We found that, among the morphologic CT characteristics, only lung metastasis was significantly associated with PD-L1 expression status, and it was more commonly presented in PD-L1-negative and in PD-L1-TPS <50% LUAD compared to PD-L1-positive and PD-L1-TPS ≥50% LUAD. AUC of morphologic characteristics for predicting PD-L1 expression status were less than 0.700. For DL-3D DenseNet model, it improved the prediction efficacy with AUC more than 0.750. Nevertheless, the prediction value with AUC 0.750 is only moderate, and a further improvement is still needed to avoid PD-L1 analysis on the histological specimen. To our knowledge, this is the first study to demonstrate the association between PD-L1 expressing status and CT features in advanced LUAD, which may serve as an important alternative marker for clinical PD-L1 detection.
For patient characteristics, it was reported that smoking history is associated with higher PD-L1 tumour proportion score (2,6,22,23), high PD-L1 expression (≥50%) was correlated with current/former smoking history. In our study, we did not find this correlation regardless of whether smoking status was treated as continuous variable (pack year) or binary variable. This controversial finding needs further study and confirmation.
Imaging as a non-invasive method has great potency in predicting gene expression and molecular level for many kinds of tumors (24,25), and is likely to become an important alternative marker for treatment decision making. For lung cancer and as to PD-L1 expression prediction, to the best our knowledge, there are only limited studies (19,20) focused on predicting of PD-L1 expression via morphologic CT characteristics. They found that presence of convergence, cavitation, absence of surrounding GGO, air bronchogram, shape, pleural indentation, tumor mean CT value, the ratio of consolidation mass to tumor mass were significantly associated with PD-L1 expression. However, no similar findings were found as in our study cohort of LUAD in which only lung metastasis was significantly associated with PD-L1 expression status. The possible reason may be, in their study, a large number of the enrolled patients were stage I-II. GGO and air bronchogram which might represent lung cancer lesion with relatively early stage were common in their study, however, these imaging signs were relatively less common in our study cohort. On the contrary, lung metastasis which was not mentioned in their studies was a significant characteristic correlated with PD-L1 expression status despite of different PD-L1 TPS cutoff values.
Interestingly, it was reported that pulmonary metastasis is associated with EGFR mutation in LUAD (26-28), patients with EGFR-positive LUAD were more prone to lung metastasis than those with wild-type EGFR, and they tended to be diffuse and random, including miliary metastases. However, in these studies, they did not mention the status of PD-L1 expression, so the relationship between PD-L1 expression status and lung metastasis was unknown. In our study, to further confirm the correlation between morphologic CT features and PD-L1 expression, we further analyzed the correlation between CT characteristics and EGFR mutation, finding that pleural tag, vessel convergence and lung metastasis type (random nodular like lesion) were significantly associated with EGFR mutation which was in accordance with the reported (26,27,29). As to lung metastasis, we did not find its association with EGFR mutation. According to the results of our study, we may assume that lung metastasis is associated with PD-L1 expression status in advanced LUAD, and this finding deserves further study and validation.
Despite these encouraging findings, the predictive efficiency of morphologic characteristics (no more than 0.70) was still limited. To further explore the relationship between CT imaging and PD-L1 expression status, the emerging and prospective method of DL which is an approach called convolutional neural network (CNN) analysis was then applied. As a machine learning algorithm of artificial intelligence (AI), it enables automated imaging features extraction with high data throughput, rather than limited to a number of visually inspected image-based features observed by naked eyes (30-32). Hold a great promise of acquiring a more accurate imaging assessment and may be a potent tool to solve some tough problems. Furthermore, there has been articles reported that CT-based radiomics (another algorithm of machine learning) can be used to predict gene mutation status of LUAD (33-35), based on the theory that tumor phenotype measured quantitatively through radiomics should reflect tumor genotype. Additionally, AI can also predict cancer mutation status in other organ and for other kinds of mutation types (36-38). As for our present study and inspired by theirs, we believe that AI also have the potential to predict molecular expression status such as PD-L1. In fact, till now, some researches have tried to use machine learning algorithm to predict PD-L1 expression through pathological images (39), and we try to predict it through CT images.
Compared to other CNN models, the densely connected convolutional networks (DenseNet) has better performance in classification tasks. Based on this, we further improved it by using 3D multimodal medical images, which was also applied in other organs (40). Moreover, as for relatively small sample size, a series of algorithms such as data augmentation were applied to avoid over fitting, and transfer learning was also adopted to speed up learning process as well as taking advantage of the pretrained model. In this study, we firstly assessed the distinguishing ability of DL-3D DenseNet for PD-L1 expression in LUAD patients. Promisingly, according to the results of our study, the 3D-DenseNet model achieved better performance in predicting PD-L1 expression in advanced LUAD compared with the morphologic characteristics, it may enhance our confidence in predicting PD-L1 expression status via CT image, thus it may be an alternative marker in immunotherapy decision making.
Our study has several limitations. First, since all the cases were advanced lung cancer patients, it was impossible to obtain whole tumor specimens surgically, thus tumor heterogeneity could not be avoided by FNA biopsy. Moreover, our promising findings deserve further validation with expanded samples. Due to the limited patient numbers, this study lacks external validation. Third, our study focused on LUAD and did not address other histologic subtypes.
Conclusions
In conclusion, compared to morphologic CT characteristics, CT-derived Deep Neural Network improves the prediction efficacy, it has the potential to predict PD-L1 expression status in advanced LUAD and may serve as an important alternative marker for clinical PD-L1 detection, thus help to select the best responders to immunotherapy.
Acknowledgments
Funding: This work was supported by the National Natural Science Foundation of China (No. 81771908, No. 81571750 and No. 81600065).
Footnote
Reporting Checklist: The authors have completed the STARD reporting checklist. Available at http://dx.doi.org/10.21037/atm-19-4690
Data Sharing Statement: Available at http://dx.doi.org/10.21037/atm-19-4690
Peer Review File: Available at http://dx.doi.org/10.21037/atm-19-4690
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/atm-19-4690). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. This study was conducted in accordance with the Declaration of Helsinki (as revised in 2013) and was approved by institutional ethics board of the First Affiliated Hospital of Sun Yat-sen University {No. [2018] 010} and informed consents were waived.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Pardoll DM. The blockade of immune checkpoints in cancer immunotherapy. Nat Rev Cancer 2012;12:252-64. [Crossref] [PubMed]
- Borghaei H, Paz-Ares L, Horn L, et al. Nivolumab versus Docetaxel in Advanced Nonsquamous Non-Small-Cell Lung Cancer. N Engl J Med 2015;373:1627-39. [Crossref] [PubMed]
- Chang YL, Yang CY, Huang YL, et al. High PD-L1 expression is associated with stage IV disease and poorer overall survival in 186 cases of small cell lung cancers. Oncotarget 2017;8:18021-30. [Crossref] [PubMed]
- Yeo MK, Choi SY, Seong IO, et al. Association of PD-L1 expression and PD-L1 gene polymorphism with poor prognosis in lung adenocarcinoma and squamous cell carcinoma. Hum Pathol 2017;68:103-11. [Crossref] [PubMed]
- Sun JM, Zhou W, Choi YL, et al. Prognostic Significance of PD-L1 in Patients with Non-Small Cell Lung Cancer: A Large Cohort Study of Surgically Resected Cases. J Thorac Oncol 2016;11:1003-11. [Crossref] [PubMed]
- Garon EB, Rizvi NA, Hui R, et al. Pembrolizumab for the treatment of non-small-cell lung cancer. N Engl J Med 2015;372:2018-28. [Crossref] [PubMed]
- Herbst RS, Soria JC, Kowanetz M, et al. Predictive correlates of response to the anti-PD-L1 antibody MPDL3280A in cancer patients. Nature 2014;515:563-7. [Crossref] [PubMed]
- Herbst RS, Baas P, Kim DW, et al. Pembrolizumab versus docetaxel for previously treated, PD-L1-positive, advanced non-small-cell lung cancer (KEYNOTE-010): a randomised controlled trial. Lancet 2016;387:1540-50. [Crossref] [PubMed]
- Hanna N, Johnson D, Temin S, et al. Systemic Therapy for Stage IV Non-Small-Cell Lung Cancer: American Society of Clinical Oncology Clinical Practice Guideline Update. J Clin Oncol 2017;35:3484-515. [Crossref] [PubMed]
- Sacher AG, Gandhi L. Biomarkers for the Clinical Use of PD-1/PD-L1 Inhibitors in Non-Small-Cell Lung Cancer: A Review. JAMA Oncol 2016;2:1217-22. [Crossref] [PubMed]
- Akamine T, Takada K, Toyokawa G, et al. Association of preoperative serum CRP with PD-L1 expression in 508 patients with non-small cell lung cancer: A comprehensive analysis of systemic inflammatory markers. Surg Oncol 2018;27:88-94. [Crossref] [PubMed]
- Lan B, Ma C, Zhang C, et al. Association between PD-L1 expression and driver gene status in non-small-cell lung cancer: a meta-analysis. Oncotarget 2018;9:7684-99. [Crossref] [PubMed]
- Tseng JS, Yang TY, Wu CY, et al. Characteristics and Predictive Value of PD-L1 Status in Real-World Non-Small Cell Lung Cancer Patients. J Immunother 2018;41:292-9. [Crossref] [PubMed]
- Lafuente-Sanchis A, Zuniga A, Estors M, et al. Association of PD-1, PD-L1, and CTLA-4 Gene Expression and Clinicopathologic Characteristics in Patients With Non-Small-Cell Lung Cancer. Clin Lung Cancer 2017;18:e109-16. [Crossref] [PubMed]
- Choi CM, Kim MY, Lee JC, et al. Advanced lung adenocarcinoma harboring a mutation of the epidermal growth factor receptor: CT findings after tyrosine kinase inhibitor therapy. Radiology 2014;270:574-82. [Crossref] [PubMed]
- Hsu JS, Huang MS, Chen CY, et al. Correlation between EGFR mutation status and computed tomography features in patients with advanced pulmonary adenocarcinoma. J Thorac Imaging 2014;29:357-63. [Crossref] [PubMed]
- Salvador-Coloma C, Lorente D, Palanca S, et al. Early radiological response as predictor of overall survival in non-small cell lung cancer (NSCLC) patients with epidermal growth factor receptor mutations. J Thorac Dis 2018;10:1386-93. [Crossref] [PubMed]
- Koo HJ, Kim MY, Park S, et al. Non-Small Cell Lung Cancer with Resistance to EGFR-TKI Therapy: CT Characteristics of T790M Mutation-positive Cancer. Radiology 2018;289:227-37. [Crossref] [PubMed]
- Toyokawa G, Takada K, Okamoto T, et al. Computed Tomography Features of Lung Adenocarcinomas With Programmed Death Ligand 1 Expression. Clin Lung Cancer 2017;18:e375-83. [Crossref] [PubMed]
- Wu T, Zhou F, Soodeen-Lalloo AK, et al. The Association Between Imaging Features of TSCT and the Expression of PD-L1 in Patients With Surgical Resection of Lung Adenocarcinoma. Clin Lung Cancer 2019;20:e195-207. [Crossref] [PubMed]
- Yushkevich PA, Piven J, Hazlett HC, et al. User-guided 3D active contour segmentation of anatomical structures: significantly improved efficiency and reliability. NeuroImage 2006;31:1116-28. [Crossref] [PubMed]
- Gainor JF, Shaw AT, Sequist LV, et al. EGFR Mutations and ALK Rearrangements Are Associated with Low Response Rates to PD-1 Pathway Blockade in Non-Small Cell Lung Cancer: A Retrospective Analysis. Clin. Cancer Res 2016;22:4585-93. [Crossref] [PubMed]
- Rangachari D, VanderLaan PA, Shea M, et al. Correlation between Classic Driver Oncogene Mutations in EGFR, ALK, or ROS1 and 22C3-PD-L1 >/=50% Expression in Lung Adenocarcinoma. J Thorac Oncol 2017;12:878-83. [Crossref] [PubMed]
- Grossmann P, Stringfield O, El-Hachem N, et al. Defining the biological basis of radiomic phenotypes in lung cancer. Elife 2017;6:e23421. [Crossref] [PubMed]
- Podolsky MD, Barchuk AA, Kuznetcov VI, et al. Evaluation of Machine Learning Algorithm Utilization for Lung Cancer Classification Based on Gene Expression Levels. Asian Pac J Cancer Prev 2016;17:835-8. [Crossref] [PubMed]
- Hasegawa M, Sakai F, Ishikawa R, et al. CT Features of Epidermal Growth Factor Receptor-Mutated Adenocarcinoma of the Lung: Comparison with Nonmutated Adenocarcinoma. J Thorac Oncol 2016;11:819-26. [Crossref] [PubMed]
- Togashi Y, Masago K, Kubo T, et al. Association of diffuse, random pulmonary metastases, including miliary metastases, with epidermal growth factor receptor mutations in lung adenocarcinoma. Cancer 2011;117:819-25. [Crossref] [PubMed]
- Park K, Goto K. A review of the benefit-risk profile of gefitinib in Asian patients with advanced non-small-cell lung cancer. Curr Med Res Opin 2006;22:561-73. [Crossref] [PubMed]
- Liu Y, Kim J, Qu F, et al. CT Features Associated with Epidermal Growth Factor Receptor Mutation Status in Patients with Lung Adenocarcinoma. Radiology 2016;280:271-80. [Crossref] [PubMed]
- LeCun Y, Bengio Y, Hinton G. Deep learning. Nature 2015;521:436-44. [Crossref] [PubMed]
- Lakhani P, Prater AB, Hutson RK, et al. Machine Learning in Radiology: Applications Beyond Image Interpretation. J Am Coll Radiol 2018;15:350-9. [Crossref] [PubMed]
- McBee MP, Awan OA, Colucci AT, et al. Deep Learning in Radiology. Acad Radiol 2018;25:1472-80. [Crossref] [PubMed]
- Rios Velazquez E, Parmar C, Liu Y, et al. Somatic Mutations Drive Distinct Imaging Phenotypes in Lung Cancer. Cancer Res 2017;77:3922-30. [Crossref] [PubMed]
- Yang X, Dong X, Wang J, et al. Computed Tomography-Based Radiomics Signature: A Potential Indicator of Epidermal Growth Factor Receptor Mutation in Pulmonary Adenocarcinoma Appearing as a Subsolid Nodule. Oncologist 2019;24:e1156-64. [Crossref] [PubMed]
- Liu Y, Kim J, Balagurunathan Y, et al. Radiomic Features Are Associated With EGFR Mutation Status in Lung Adenocarcinomas. Clin Lung Cancer 2016;17:441-8.e6. [Crossref] [PubMed]
- Miles KA, Ganeshan B, Rodriguez-Justo M, et al. Multifunctional imaging signature for V-KI-RAS2 Kirsten rat sarcoma viral oncogene homolog (KRAS) mutations in colorectal cancer. J Nucl Med 2014;55:386-91. [Crossref] [PubMed]
- Yamamoto S, Korn RL, Oklu R, et al. ALK molecular phenotype in non-small cell lung cancer: CT radiogenomic characterization. Radiology 2014;272:568-76. [Crossref] [PubMed]
- Weiss GJ, Ganeshan B, Miles KA, et al. Noninvasive image texture analysis differentiates K-ras mutation from pan-wildtype NSCLC and is prognostic. PLoS One 2014;9:e100244. [Crossref] [PubMed]
- Sha L, Osinski BL, Ho IY, et al. Multi-Field-of-View Deep Learning Model Predicts Nonsmall Cell Lung Cancer Programmed Death-Ligand 1 Status from Whole-Slide Hematoxylin and Eosin Images. J Pathol Inform 2019;10:24. [Crossref] [PubMed]
- Liang S, Zhang R, Liang D, et al. Multimodal 3D DenseNet for IDH Genotype Prediction in Gliomas. Genes (Basel) 2018;9:382. [Crossref] [PubMed]


