Methylation of the MAOA promoter is associated with schizophrenia
Introduction
Schizophrenia is a chronic and severe mental illness with many clinical manifestations, negatively affecting thought, perception, emotion, and behavior (1,2). Schizophrenia shows much heterogeneity in the type, symptoms, and course of the disease. Although lifetime morbidity is as low as 1%, it can still lead to serious and negative consequences, including disability (3). Patients with schizophrenia are more violent compared to mentally healthy people or those with other mental disorders (4). Additionally, the quality of life for patients with schizophrenia is degraded while simultaneously imposing a heavy burden on families and society (5). Therefore, there is an urgent need to understand better the mechanisms underlying the pathophysiology of schizophrenia.
The development of molecular biological techniques has aided the discovery of polymorphic sites associated with schizophrenia. Monoamine oxidase A (MAOA) plays a crucial role in the metabolism of biogenic monoamines, including serotonin, dopamine, and epinephrine (6). MAOA and neurotransmitter abnormalities are closely related to the development of schizophrenia (7,8). The gene encoding MAOA is located on the X chromosome with a variable nucleotide repeat (VNTR), located 1.2-kb (Xp11.4–p11.3) upstream of the MAOA coding sequence and consists of a 30-bp repeat sequence (9). Carrying 2 or 3 VNTRs is defined as having low MAOA activity, while greater than 3 is referred to as high MAOA activity (7). Some studies have revealed that individuals with low MAOA activity alleles have a higher risk of schizophrenia (10,11). However, another study showed that schizophrenia in women is associated with high MAOA activity (12). Therefore, the inconsistency of these results suggested the involvement of alternative molecular mechanisms that could influence gene expression and lead to the development of schizophrenia.
Epigenetic modifications, including methylation, are essential gene molecular regulatory mechanisms to allow genes to cope with environmental changes. DNA’s methylation refers to the selective addition of a methyl group to the CpG nucleotide of methyltransferase-catalyzed DNA to form 5-methylcytosine (13). Studies have shown that DNA methylation can regulate gene expression in mental disorders by altering chromatin structure, DNA conformation, DNA stability, and DNA-protein interactions (14,15). DNA’s methylation variation is an essential contributor to individual phenotypic, differences, and abnormalities in methylation that are closely related to schizophrenia, violent aggression, and other complex diseases. A DNA methylation analysis of 485,764 CpG sites across the genome revealed 234 sites displayed significantly different methylation in schizophrenia patients (16). This study supported the idea that DNA methylation could regulate the pathophysiological processes involved in schizophrenia. However, the relationship between DNA methylation of MAOA and the onset of schizophrenia has not been reported before.
In this study, we extracted the methylation status of the MAOA gene promoter in 151 patients with schizophrenia (104 males and 47 females) and 247 controls (204 males and 43 females). Our results found that methylation of MAOA-2 and MAOA-3 is associated with schizophrenia.
Methods
The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013). The study was approved by the Ethics Committee of Kunming Medical University (No: KY2019.57).
Samples selection criteria
We investigated methylation of the MAOA gene promoter in a total of 151 individuals with schizophrenia (104 males and 47 females) in the Yunnan Psychiatric Hospital, Yunnan, China. Criteria for admission of schizophrenia: (I) in line with DSM-V diagnostic criteria; (II) diagnosis was made by at least two psychiatrists independently; (III) Yunnan Han nationality, age 18–60. Exclusion criteria: (I) combined with brain organic diseases, brain trauma or serious physical diseases; (II) combined with nervous system diseases and other mental disorders; (III) pregnant or lactating women; (IV) other drug abuse history within 6 months. Accordingly, 247 unrelated subjects (204 males and 43 females) were also recruited as control subjects. Criteria for admission of control group: (I) the individuals whose gender, nationality and age match the above case group; (II) no genetic history; no history of major physical diseases. The control group also met the above exclusion criteria. Written informed consent was obtained from the patients themselves or their family members and guardians before enrollment.
DNA’s methylation
Blood was obtained from all individuals, and genomic DNA was extracted from Ethylenediaminetetraacetic acid (EDTA) blood using a QIAamp DNA Blood Mini Kit (QIAGEN company, German). Primer 3 (http://primer 3.ut.ee//) was used to design primers after bisulfite disposed. The Polymerase chain reaction (PCR) primers for MAOA-2 were: Forward—5'-AAGTYGGGGGTATAATTGTTTAGGTT-3'; Reverse—5'-CTAAAACCCCRAAAACCACTCT-3'. The PCR primers for MAOA-3 were: Forward—5'-GGGGGAGTYGGGTATTGTG-3'; Reverse—5'-ACCCCCACCTCAATACCTAAC-3'.
Gel electrophoresis and NanoDrop analysis
Agarose gel electrophoresis detected genomic DNA. The electrophoresis bands were visible, with no clear degradation and no RNA contamination. Genomic DNA quality was quantified using a Nanodrop 2000 (NanoDrop Technologies, USA). Samples with concentrations greater than or equal to 20 ng/µL or a total of 1 µg with an OD260/OD280 DA ≥1.8 were used (this amount can be used in 10 PCR panels).
Design of primers and optimization of single-locus PCR conditions
Primers were designed and provided by Genesky Company (Shanghai, China). The human genome treated with bisulfite was selected and amplified as primer-template to obtain a clear single band for later experiments.
The optimization of multiple PCR primers
The primers optimized in the last step were mixed into a panel of multiple PCR primers. On the particular method of capillary electrophoresis, we determined whether each pair of primers in multiple systems could be amplified efficiently and specifically, and then adjusted to optimize the composition and concentration of primers for the multiple PCR panels. The optimized multiple PCR primer panel was used to conduct multiple PCR amplification with the transformed sample genome as the template. The Genesky Company conducted multiple PCR amplification (Shanghai, China).
Treatment of DNA with bisulfite
The samples were processed by EZ DNA Methylation-God Kit (ZYMO company, USA), to convert cytosine C without methylation of genomic DNA into uracil U (Bisulfite Conversion Efficiency is shown in Figure S1).
Specific tag sequences were added to the samples
Specific tag sequences compatible with the Illumina platform (Illumina, USA) were introduced to the end of the library by PCR amplification using primers with Index sequences. The PCR procedure with 11 cycles was used to reduce the nonspecific amplification as much as possible.
Quantitative and computer sequencing
The final methyl target sequencing library was obtained by mixing Index PCR products of all samples. The fragment length of the library was verified by an Agilent 2100 Bioanalyzer (Agilent Technologies, USA) (see Figure S2).
Results
The total level of MAOA methylation between schizophrenia patients and healthy control subjects is similar
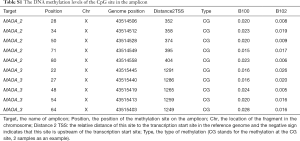
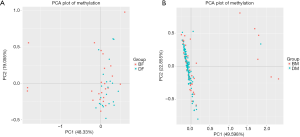
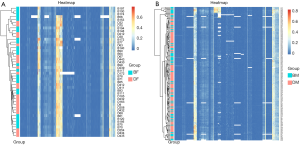
By using methylation sequencing of target regions (Table 1), we investigated the total DNA methylation profile of MAOA in 151 patients with schizophrenia and 247 controls (Table S1). Principal component analysis (PCA; Figure 1) and hierarchical clustering (Figure 2) were employed to visualize the DNA methylation profile of MAOA in all samples. Neither PCA nor hierarchical clustering analysis could separate the schizophrenia patients and the control subjects as expected when investigating all MAOA methylation sites.

Full table
Full table
There are several differentially methylated sites in male and female schizophrenia patients
Although there was no significant difference between the total DNA methylation level of MAOA between schizophrenia patients and controls, we explored whether there was a difference in methylation at specific methylation sites on MAOA. Interestingly, we found several DNA methylation sites (Table 2) in the MAOA promoter differently methylated in schizophrenia when compared with healthy control samples.
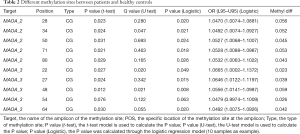
Full table
There are several differentially methylated segments in male and female schizophrenia patients
Next, we investigated whether there were some DNA segments, which were differentially methylated between the schizophrenia patients and healthy controls. By calculating the mean methylation level of all CpG sites on MAOA, we found that the methylation of MAOA-2 and MAOA-3 was significantly altered between schizophrenics and controls in men (P<0.005; Table 3; Figure 3). Similarly, there was a significant difference in MAOA-2 methylation between schizophrenics and controls in women (Table 4; Figure 4).

Full table
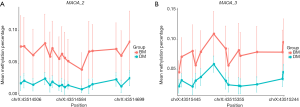

Full table
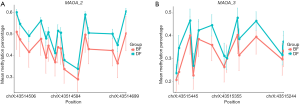
Differentially methylated haplotype identification
Furthermore, we conducted a methylation haplotype analysis to find differentially methylated haplotypes between schizophrenia and control patients. Finally, we discovered that some methylation haplotypes were significantly different between schizophrenia and control patients (Table 5).
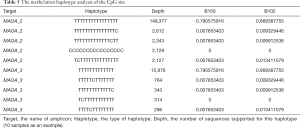
Full table
Discussion
In recent years, the relationship between DNA methylation and schizophrenia has been extensively explored (17-19). Schizophrenia is a complicated disease, which might be influenced by epigenetic modifications, including methylation, and is vulnerable to environmental factors (20). This present study reveals that methylation of the MAOA gene promoter at the MAOA-2 and MAOA-3 methylation sites is linked to susceptibility to schizophrenia in both males and females.
It is reported that DNA methylation—especially MAOA methylation perhaps is in response to environmental influences, may also be a factor concerning these mental disorders. The presently observed trend is towards CpG-specific MAOA hypomethylation-due to increased gene expression and decreased serotonin or norepinephrine availability. MAOA methylation has been linked to many psychiatric disorders. Studies reveal that in panic disorder patients, patients exhibit lower MAOA methylation compared to controls, and there is a negative correlation between baseline psychiatric disorder severity and MAOA methylation (21).
In addition to schizophrenia, DNA methylation of MAOA is also associated with other mental disorders and is gender-dependent. These results may be because the MAOA gene is found on the X chromosome. MAOA methylation status has been significantly associated with lifetime nicotine dependence and alcohol dependence in women, but not men (22). In a sample of female patients with acrophobia, MAOA methylation was significantly reduced in patients compared to controls, and treatment for this disorder could significantly enhance MAOA methylation (15). In male patients, hypermethylation of exon 1 and intron 1 of MAOA was associated with post traumatic stress disorder (PTSD) when compared to controls (22). Hypomethylation of the MAOA promoter region has also been significantly associated with smoking behavior in women (23). Our results also suggest a difference in MAOA methylation in male and female schizophrenia patients.
DNA’s methylation of MAOA contributes to mental disorders, including schizophrenia, through the regulation of serotonin. In a pharmacokinetic study investigating 61 female patients with major depression, decreased methylation at two individual CpG sites in the MAOA promoter region was linked to a slower response to treatment with serotonin reuptake inhibitors over 6 weeks (24). Abnormalities in DNA methylation at the MAOA promoter may be associated with schizophrenia in males (25). Males with antisocial personality disorder (ASPD) showed hypermethylation in the MAOA promoter compared to healthy men. These findings were associated with whole-blood serotonin levels and reduced transcriptional activity in vitro (26). A prospective examination of associations between epigenome-wide methylation patterns in cord blood at birth and propensity to develop conduct disorders between the ages of 4–13 years identified a subthreshold association for increased methylation in the vicinity of the MAOA gene (27).
Limitation
In the process of sample collection, the number of female patients is significantly less than that of male patients, so that the number of female samples in this study is insufficient. This phenomenon has also attracted our attention, but the real reason for this phenomenon cannot be reasonably explained. We are considering whether the difference in the prevalence of schizophrenia between male and female patients is worthy of further study. Due to the limited number of female clinical samples used in the experiment, the general applicability of the results of this study is limited to a certain extent, so it is necessary to expand the sample size in future research.
Conclusions
Our study showed that the pathophysiology of schizophrenia might be mediated by DNA methylation of the MAOA promoter. However, the role of DNA methylation in the progression of schizophrenia and its underlying molecular mechanisms are still unclear. In future studies, we will investigate the methylation of other genes related to the pathogenesis of schizophrenia and investigate the changes induced by MAOA methylation in greater detail.
Acknowledgments
We thank the National Natural Science Foundation of China and each author.
Funding: The present study was supported by the National Natural Science Foundation of China (grant No. 31100906).
Footnote
Data Sharing Statement: Available at http://dx.doi.org/10.21037/atm-20-4481
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/atm-20-4481). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013). The study was approved by the Ethics Committee of Kunming Medical University (No: KY2019.57). Written informed consent was obtained from all subjects in our study.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Kelly DL, Rowland LM, Patchan KM, et al. Schizophrenia clinical symptom differences in women vs. men with and without a history of childhood physical abuse. Child Adolesc Psychiatry Ment Health 2016;10:5. [Crossref] [PubMed]
- Peuskens J. Clinical effectiveness in adults with chronic schizophrenia. Eur Neuropsychopharmacol 2004;14:S453-9. [Crossref] [PubMed]
- Aronow WS, Shamliyan TA. Effects of atypical antipsychotic drugs on QT interval in patients with mental disorders. Ann Transl Med 2018;6:147. [Crossref] [PubMed]
- Hettige NC, Kennedy JL, Vincenzo DL. Does a history of suicide attempt predict higher antipsychotic dosage in schizophrenia? Psychopharmacology (Berl) 2014;231:2507-13. [PubMed]
- Boyer L, Baumstarck K, Boucekine M, et al. Measuring quality of life in patients with schizophrenia:an overview. Expert Rev Pharmacoecon Outcomes Res 2013;13:343-9. [Crossref] [PubMed]
- Huang SY, Lin MT, Shy MJ, et al. Neither single-marker nor haplotype analyses support an association between monoamine oxidase A gene and bipolar disorder. Eur Arch Psychiatry Clin Neurosci 2008;258:350-6. [Crossref] [PubMed]
- Bendre M, Comasco E, Checknita D, et al. Associations between MAOA-uVNTR genotype, maltreatment, MAOA methylation and alcohol consumption in young adult males. Alcohol Clin Exp Res 2018;42:508-19. [Crossref] [PubMed]
- Zhang Y, Ming QS, Yi JY, et al. Gene-Gene-Environment Interactions of Serotonin Transporter, Monoamine Oxidase A and Childhood Maltreatment Predict Aggressive Behavior in Chinese Adolescents. Front Behav Neurosci 2017;11:17. [Crossref] [PubMed]
- Zhang J, Chen Y, Zhang K, et al. A cis-phase interaction study of genetic variants within the MAOA gene in major depressive disorder. Biol Psychiatry 2010;68:795-800. [Crossref] [PubMed]
- Gallardo-Pujol D, Andréspueyo A, Maydeuolivares A. MAOA genotype, social exclusion and aggression: An experimental test of a gene-environment interaction. Genes Brain Behav 2013;12:140-5. [Crossref] [PubMed]
- Edwards AC, Dodge KA, Latendresse SJ, et al. MAOA-uVNTR and early physical discipline interact to influence delinquent behavior. J Child Psychol Psychiatry 2010;51:679-87. [Crossref] [PubMed]
- Verhoeven FE, Booij L, Kruijt AW, et al. The effects of MAOA genotype, childhood trauma, and sex on trait and state-dependent aggression. Brain Behav 2012;2:806-13. [Crossref] [PubMed]
- Robertson KD, Jones PA. DNA methylation: past, present and future directions. Carcinogenesis 2000;21:461-7. [Crossref] [PubMed]
- Chen D, Meng L, Pei F, et al. A review of DNA methylation in depression. J Clin Neurosci 2017;43:39-46. [Crossref] [PubMed]
- Schiele MA, Ziegler C, Kollert L, et al. Plasticity of Functional MAOA Gene Methylation in Acrophobia. Int J Neuropsychopharmacol 2018;21:822-7. [Crossref] [PubMed]
- Kinoshita M, Numata S, Tajima A, et al. DNA Methylation Signatures of Peripheral Leukocytes in Schizophrenia. Neuromolecular Med 2013;15:95-101. [Crossref] [PubMed]
- Hass J, Walton E, Wright C, et al. Associations between DNA methylation and schizophrenia-related intermediate phenotypes - a gene set enrichment analysis. Prog Neuropsychopharmacol Biol Psychiatry 2015;59:31-9. [Crossref] [PubMed]
- Iwamoto K, Bundo M, Yamada K, et al. DNA methylation status of SOX10 correlates with its downregulation and oligodendrocyte dysfunction in schizophrenia. J Neurosci 2005;25:5376-81. [Crossref] [PubMed]
- Walton E, Liu J, Hass J, et al. MB-COMT promoter DNA methylation is associated with working-memory processing in schizophrenia patients and healthy controls. Epigenetics 2014;9:1101-7. [Crossref] [PubMed]
- Philibert RA, Bohle P, Secrest D, et al. The association of the HOPA12bp polymorphism with schizophrenia in the NIMH genetics initiative for schizophrenia sample. Am J Med Genet B Neuropsychiatr Genet 2007;144B:743-7. [Crossref] [PubMed]
- Ziegler C, Richter J, Mahr M, et al. MAOA gene hypomethylation in panic disorder-reversibility of an epigenetic risk pattern by psychotherapy. Transl Psychiatry 2016;6:e773. [Crossref] [PubMed]
- Philibert RA, Gunter TD, Beach SRH, et al. MAOA methylation is associated with nicotine and alcohol dependence in women. Am J Med Genet B Neuropsychiatr Genet 2008;147B:565-70. [Crossref] [PubMed]
- Tiili EM, Mitiushkina NV, Sukhovskaya OA, et al. The genotypes and methylation of MAO genes as factors behind smoking behavior. Pharmacogenetics & Genomics 2017;27:394-401. [Crossref] [PubMed]
- Howe AS, Buttenschã HN, Bani-Fatemi A, et al. Candidate genes in panic disorder: meta-analyses of 23 common variants in major anxiogenic pathways. Mol Psychiatry 2016;21:665-79. [Crossref] [PubMed]
- Chen Y, Zhang J, Zhang L, et al. Effects of MAOA promoter methylation on susceptibility to paranoid schizophrenia. Hum Genet 2012;131:1081-7. [Crossref] [PubMed]
- Checknita D, Maussion G, Comai S, et al. Monoamine oxidase A gene promoter methylation and transcriptional downregulation in an offender population with antisocial personality disorder. Br J Psychiatry 2015;206:216-22. [Crossref] [PubMed]
- Cecil CAM, Walton E, Jaffee SR, et al. Neonatal DNA methylation and early-onset conduct problems: A genome-wide, prospective study. Dev Psychopathol 2018;30:383-7. [Crossref] [PubMed]
(English Language Editor: J. Chapnick)




