
Machine learning-based CT radiomics method for predicting hospital stay in patients with pneumonia associated with SARS-CoV-2 infection: a multicenter study
Introduction
The coronavirus disease 2019 (COVID-19) has become a global challenge since the December 2019 (1-3). Clinical characteristics of patients with severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) infection have been reported, and the median hospital stay of forty-seven discharged patients was 10 days (1). Studies showed that patients’ condition in Wuhan worsens on the 10th day after illness onset (2). However, patients with symptoms longer than 10 days outside Wuhan were less severe than those in Wuhan (3). Therefore, the hospital stay in patients with SARS-CoV-2 infection is one of the prognostic indicators, and its non-invasive predicting tool is important for assessing the patients’ clinical outcome.
Chest CT is recommended as a routine test in the diagnoses and monitoring of COVID-19 since ground-glass opacities and consolidation are the most relative imaging features in pneumonia associated with SARS-CoV-2 infection (4-6). On the basis of our previous work utilizing quantitative CT for COVID-19 (7), we hypothesized that high-throughput information hidden behind CT images (8) had potential in discriminating the hospital stay. The study aimed to develop and test machine learning-based CT radiomics models for predicting hospital stay in patients with pneumonia associated with SARS-CoV-2 infection. We present the following article in accordance with the STROBE reporting checklist (available at http://dx.doi.org/10.21037/atm-20-3026).
Methods
Study population
This retrospective, multicenter study was conducted according to principles of the Declaration of Helsinki (as revised in 2013) and approved by the ethics commissions of The First Hospital of Lanzhou University (LDYYLL2020-15). The need for written informed consent from the participants was waived. The patient’s personal data have been secured.
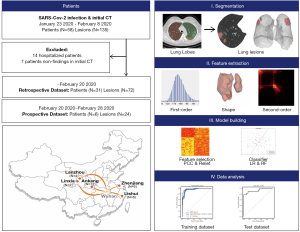
Patients with laboratory-confirmed SARS-CoV-2 infection and their initial CT images were enrolled from five designated hospitals between January 23, 2020 and February 8, 2020, with final follow-up on February 20, 2020 (Figure 1). Most patients received antiviral treatment with interferon inhalation, lopinavir and ritonavir, combined with probiotics. Patients were discharged once the results of two real-time fluorescence polymerase-chain-reaction tests taken 24 hours apart were negative for SARS-CoV-2 antigens. Patients without pneumonia findings or those remained in hospital were excluded. Sample size consideration was shown in Supplementary. In this study, the optimal cut-off value of hospital stay was determined to be 10 days based on previous studies (1-3), by which patients were classified into short-term hospital stay (≤10 days) and long-term hospital stay (>10 days).
CT radiomics features and model building
The pipeline of radiomics model was shown (Figure 1), and features extraction and model building were performed on lung lobe-level. CT examinations followed routine non-contrast chest CT scan protocols in each institution. Details of CT scan and images preprocessing was shown in Supplementary: Methods. Images containing lesions were segmented using Python (3.6, https://www.python.org) and 3Dslicer (version 4.10.0; https://www.slicer.org/) with two steps. First, lung lobes of each patient were segmented automatically using algorithms based on U-net (9), and results were checked and modified by one radiologist (QY). Next, lesions in each lung lobe were labeled semi-automatically using serval seeds placed within lesion region to generate the contours. Three experienced radiologists (SH, YW and XM) evaluated segments of each lesion and reached a consensus. All imaging processes were blinded to clinical data.
In total, 1,218 features were calculated per lesion patch. First-order, shape and second-order features were extracted from original images and wavelet filter applied images using pyradiomics (10). Two supervised learning algorithms, logistic regression (LR) and random forest (RF), were used to build the model and verify the robustness of features (Supplementary: Methods) (11). We applied 5-fold cross-validation on the training dataset to prove model performance.
Model performance
The cutoff point was defined on receiver operating characteristic (ROC) curves of training data by maximizing the sum of sensitivity and specificity. The model performance was evaluated using test dataset on lung lobe-level. Areas under the ROC curve (AUC), sensitivities, specificities, positive predictive value (PPV), and negative predictive value (NPV) were recorded. On patient-level, one was defined as long-term hospital stay once more than one lesion of lung lobe was labeled as long-term stay lesion, if not, as short-term hospital stay.
Statistical analysis
Categorical data were expressed in numbers (percentages), and continuous variables as median (interquartile range). Demographic and clinical characteristics of patients were assessed using Chi square test (Fisher’s exact test as appropriate) for categorical variables, and Mann-Whitney test for continuous variables in SPSS (version 22.0. IBM Crop. Armonk, NY, USA). Feature selection and model building were implemented with FeAture Explorer (FAE, v0.2.5, https://github.com/salan668/FAE) on Python (3.6). Test values like AUCs [95% confidence interval (95% CI)], sensitivity, specificity was calculated in SPSS and Python. A P value <0.05 was considered statistically significant.
Results
Patient characteristics
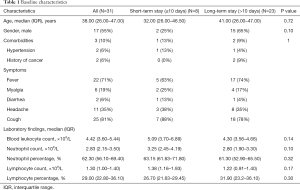
A total of 52 patients with laboratory-confirmed SARS-CoV-2 infection and initial CT images were enrolled from five designated hospitals in Ankang, Lishui, Zhenjiang, Lanzhou, and Linxia, China. As of February 20, 14 patients were still hospitalized, and 7 patients had non-findings in CT images. Therefore, 31 patients with 72 lesion segments were included in the final analysis. The training and inter-validation cohort comprised 26 patients (12 from Ankang, 8 from Lishui, 4 from Lanzhou, and 2 from Linxia) with 59 lesion segments, and test cohort comprised 5 patients from Zhenjiang with 13 lesion segments. The median age was 38.00 (interquartile range, 26.00–47.00) years and 17 (55%) were male. Comorbidities, symptoms and laboratory findings at admission were summarized in Table 1.
Full table
Performance of CT radiomics model
The CT radiomics model, based on 6 features (Table S1), showed the highest AUC on the training and inter-validation dataset. The performance of modeling using LR and RF methods was shown in Figure 2. On lung lobe-level, models using LR method significantly distinguished short- and long-term hospital stay [cut-off value 0.31, in test dataset, AUC 0.97 (95% CI, 0.83–1.0), sensitivity 1.0, specificity 0.89, NPV 1.0, and PPV 0.8]. Besides, models using RF method obtained satisfied results [cut-off value 0.68, in test dataset, AUC 0.92 (95% CI, 0.67–1.0), sensitivity 0.75, specificity 1.0, NPV 0.9, and PPV 1.0].

Full table
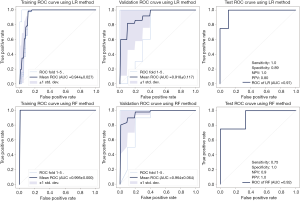
On patients-level, in training and inter-validation datasets, 6 of 6 patients were correctly classified as short-term stay by both models, and 20 of 20, 16 of 20 patients were correctly identified as long-term stay by RF and LR models, respectively. In test dataset, one of two patients were correctly classified as short-term stay, and three of three were correctly identified as long-term stay by RF and LR models.
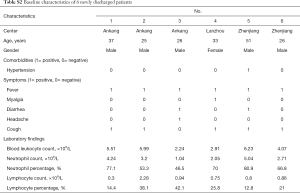
As of February 28, we followed up for a prospective dataset of six newly discharged patients with 24 lesions from designated hospitals (comorbidities, symptoms and laboratory findings were described in Table S2). All patients were correctly recognized as long-term stay using both RF and LR models developed on raw 52 patients.
Full table
Discussion
In this study, machine learning-based CT radiomics models were developed and tested for predicting hospital stay in patients with pneumonia associated with SARS-CoV-2 infection. CT radiomics features hidden within lesions, ground-glass opacities and consolidation, were extracted, and demonstrated robust performance in building machine learning models using multicenter cohorts for training, and independent cohorts for test.
To the best of our knowledge, the present study is the first to investigate the correlation between high throughput CT features and prognosis of patients with pneumonia associated with SARS-CoV-2 infection, by developing and testing machine-learning models in an independent test cohort and a prospective cohort. The present study demonstrated the potential of radiomics features from CT as an imaging biomarker in predicting patient’s prognosis. We believe that CT features and models in this study could predict hospital stay in patients with COVID-19 pneumonia using initial CT scan and patients identified as long-term hospital stay should be paid more attention to avoid worse condition in course.
Though there were differences in CT scan parameters among centers, key features included in models were second-order, and focused on distribution, correlation and variance in gray level intensities, which described the relationship between voxels and hold quantitative information on the spatial heterogeneity of pneumonia lesions (11,12). Compared with first-order features, second-order features were not sensitive to absolute value and thus more robust. Moreover, the models showed satisfied AUCs more than 90% on both training and independent and prospective test process, which indicated that the models could be applied in a general situation. Similarity in AUCs, sensitivity and specificity for RF and LR models also demonstrated the robustness, according to prior study that classification method showed most dominant in variability of model (11).
The study was limited by small sample size. The percentage of short-term hospital stay is low in our multicenter cohorts, and semi-automated lesion segmentation might result in selection bias. A larger prospective multicenter cohort is needed to tune and test the machine learning-based CT radiomics models.
Conclusions
The machine learning-based CT radiomics features and methods showed feasibility and accuracy for predicting hospital stay in patients with pneumonia associated with SARS-CoV-2 infection.
Supplementary
Methods
Sample size consideration
The training and inter-validation cohort comprised 26 patients (12 from Ankang, 8 from Lishui, 4 from Lanzhou, and 2 from Linxia) with 59 lesion segments, and test cohort comprised 5 patients from Zhenjiang with 13 lesion segments. The test dataset was not used during model development.
In training, to balance the case-to-noncase ratio as 1:1, we used SMOTE method to up-sample the short-term hospital stay cases, which is a re-sampling technique commonly used in datasets. We applied 5-fold cross-validation on training dataset to prove model performance. We applied normalization on the feature matrix, and each vector had a zero center and unit standard deviation. Pearson correlation coefficient (PCC) of the feature pair was used to reduce feature space dimensionality. Relief was used to select the most outcome-related features. Six features were selected after PCC and relief, which making an event-per-predictor ratio >15 (95 cases after smote vs. 6 features) (13). Therefore, we believed that there was no big concern on the overfitting issue of our model at this sample size.
Image preprocessing
All CT followed routine chest CT scan protocols in each institution. Patients from Ankang were imaged with 1.25 mm slice thickness/120 kV CT from GE Medical Systems, Milwaukee, WI; patients from Lanzhou were imaged with 1 mm slice thickness/120 kV CT from Siemens Healthineers, Erlangen, Germany; patients from Zhenjiang were imaged with 1.5 mm slice thickness/120 kV CT from Siemens Healthineers, Erlangen, Germany; patients from Lishui were imaged with 3 mm slice thickness/120 kV CT from Siemens Healthineers, Erlangen, Germany; patients from Linxia were imaged with 2 mm slice thickness/120 kV CT from Siemens Healthineers, Erlangen, Germany.
Before feature extraction, all images would be resampled into isotropic voxels of unit dimension to ensure comparability, where 1 voxel corresponds to 1 mm. Image normalization was performed by centering it at the mean with standard deviation and remapping the histogram to fit within µ ± 3σ (µ: mean gray-level; σ: gray-level standard deviation).
RF and LR classifier
Logistic regression is a linear classifier that combines all the features. A hyper-plane was searched in the high dimension to separate the samples.
Random forest is an ensemble learning method which combining multiple decision trees at different subset of the training data set. Random forest is an effective method to avoid over-fitting.
Acknowledgments
Funding: This work was supported by Gansu Provincial COVID-19 Science and Technology Major Project; Shenyang Emergency Research Project for Prevention and Treatment of COVID-19 (grant number YJ2020-9-009); Guangxi Digestive Disease Clinical Medical Research Center Construction Project (grant number AD17129027); Shanxi Provincial Emergency Research Project for Chinese Medicine for Prevention and Treatment of COVID-19 (grant number 2020-YJ005); Zhenjiang Key Research and Development Plan for COVID-19 Emergency Project (grant number SH2020001).
Footnote
Reporting Checklist: The authors have completed the STROBE reporting checklist. Available at http://dx.doi.org/10.21037/atm-20-3026
Data Sharing Statement: Available at http://dx.doi.org/10.21037/atm-20-3026
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/atm-20-3026). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. This retrospective, multicenter study was conducted according to principles of the Declaration of Helsinki (as revised in 2013) and approved by the ethics commissions of The First Hospital of Lanzhou University (LDYYLL2020-15). The need for written informed consent from the participants was waived. The patient’s personal data have been secured.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Wang D, Hu B, Hu C, et al. Clinical Characteristics of 138 Hospitalized Patients With 2019 Novel Coronavirus-Infected Pneumonia in Wuhan, China. JAMA 2020;323:1061-9. [Crossref] [PubMed]
- Huang C, Wang Y, Li X, et al. Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China. Lancet 2020;395:497-506. [Crossref] [PubMed]
- Xu XW, Wu XX, Jiang XG, et al. Clinical findings in a group of patients infected with the 2019 novel coronavirus (SARS-Cov-2) outside of Wuhan, China: retrospective case series. BMJ 2020;368:m606. [Crossref] [PubMed]
- Zu ZY, Jiang MD, Xu PP, et al. Coronavirus Disease 2019 (COVID-19): A Perspective from China. Radiology 2020. [Epub ahead of print]. [Crossref] [PubMed]
- Pan F, Ye T, Sun P, et al. Time Course of Lung Changes at Chest CT during Recovery from Coronavirus Disease 2019 (COVID-19). Radiology 2020;295:715-21. [Crossref] [PubMed]
- Lei J, Li J, Li X, et al. CT Imaging of the 2019 Novel Coronavirus (2019-nCoV) Pneumonia. Radiology 2020;295:18. [Crossref] [PubMed]
- Qi X, Lei J, Yu Q, et al. CT imaging of coronavirus disease 2019 (COVID-19): from the qualitative to quantitative. Ann Transl Med 2020;8:256. [Crossref] [PubMed]
- Gillies RJ, Kinahan PE, Hricak H. Radiomics: Images Are More than Pictures, They Are Data. Radiology 2016;278:563-77. [Crossref] [PubMed]
- Hofmanninger J, Prayer F, Pan J, et al. Automatic lung segmentation in routine imaging is a data diversity problem, not a methodology problem. ArXiv200111767 Phys Stat. January 2020. Available online: http://arxiv.org/abs/2001.11767
- van Griethuysen JJM, Fedorov A, Parmar C, et al. Computational Radiomics System to Decode the Radiographic Phenotype. Cancer Res 2017;77:e104-e107. [Crossref] [PubMed]
- Parmar C, Grossmann P, Bussink J, et al. Machine Learning methods for Quantitative Radiomic Biomarkers. Sci Rep 2015;5:13087. [Crossref] [PubMed]
- Elshafeey N, Kotrotsou A, Hassan A, et al. Multicenter study demonstrates radiomic features derived from magnetic resonance perfusion images identify pseudoprogression in glioblastoma. Nat Commun 2019;10:3170. [Crossref] [PubMed]
- Chow S, Shao J, Wang H. Sample size calculations in clinical research. 2nd Ed. Chapman & Hall//CRC Biostatistics Series, 2008.


