
CircAGFG1modulates autophagy and apoptosis of macrophages infected by Mycobacterium tuberculosis via the Notch signaling pathway
Introduction
Tuberculosis, an infectious disease with high mortality rates, kills 1.5 million people each year. Since 2015, the incidence of tuberculosis has seen a cumulative reduction of only 6.3% (1). Some study found that overweight is a risk of latent tuberculosis infection (2). However, the prevalence of overweight is high in children (3) and adults (4) in China. Although this shows that the overall mortality of tuberculosis is decreasing, it has not been eradicated. Tuberculosis occurs when Mycobacterium tuberculosis (Mtb), a species of bacteria, infects the lung. Bacillus Calmette Guérin (BCG), is the current vaccine in use for infants; however, it is not ideal for adults (2), and so far, there is no effective vaccine to protect against all strains of Mtb. Moreover, the current drug treatment faces the obstacle of drug resistance (5), and the current methods for diagnosing tuberculosis, such as bacteriological detection, sputum smear, and CT lack efficiency (6-8). With all of this considered, there is an urgent need to clarify the pathological mechanisms of Mtb so that effective therapeutic targets and diagnosis biomarkers can be identified.
Cell apoptosis and autophagy are two vital defense mechanisms against microbial invasion. Cell apoptosis facilitates the clearance of Mtb infection, to the benefit of the host. Different to cell apoptosis, autophagy, as a highly important physiological process in maintaining cellular homeostasis, promotes cell survival. Autophagy is usually considered as the critical defense mechanism against infection (9); however, in Mycobacterial infections, Mycobacteria exploit autophagy to ensure their intracellular survival (10). Therefore, it is of great significance to find the important regulator behind the autophagy and apoptosis processes seen in mycobacterium infection.
Circular RNAs (circRNAs) with a closed loop structure, a type of non-coding RNA, have been confirmed as the critical regulatory mechanism in many physiological processes, such as autophagy and apoptosis, and act as an important player in the occurrence and pathologic processes of many diseases. CircRNAs have been regarded as the rising star in the development of respiratory diseases including silicosis, lung cancer, and pulmonary tuberculosis (11). Previous studies identified 170 circRNAs that were aberrantly expressed in patients with tuberculosis, which hints at the potential role of circRNAs in diagnosing tuberculosis (12). Specifically, circAGFG1, a new-found circRNA, is reported to contribute to non-small cell lung cancer (13), although its effect in tuberculosis are not fully understood. Therefore, this study investigated the effect of circAGFG1 in tuberculosis.
Non-coding RNAs play a critical role in regulating the function of a variety of signaling pathways, resulting in the promotion or inhibition of many diseases (14-16). The Notch signaling pathway has been reported as a crucial signaling pathway in tuberculosis. After Mtb infection, the Notch signaling pathway is activated (17). Inhibition of the Notch signaling pathway reverses the effect of Th1/Th2 imbalance, which is pivotal in the immune response of tuberculosis patients (18). As reported by many previous studies, the Notch signaling pathway is confirmed to be an important regulator of autophagy. The relationship between the Notch signaling pathway and circAGFG1 in tuberculosis was also explored in this study.
As macrophages are the cells targeted during Mycobacterium invasion, Mycobacterium-infected macrophages served as the cell model in our study.
Methods
Cell acquisition, isolation, culture and transfection
Alveolar macrophages in bronchoalveolar lavage were obtained from active tuberculosis donors and health donors via fiber-optic bronchoscopy, according to the previous literature (19,20). None of the donors had any other underlying diseases. Signed informed consent was obtained from all of the patients before participation in any part of the study.
Cell isolation and culture
The isolation was performed as described before in previous the literature (20,21). To establish the cell model of tuberculosis, the alveolar macrophages from healthy controls were cultured in RPMI 1640 medium. Subsequently, the cells were infected with different concentrations of Mycobacterium (1, 10, or 100 nm) for a period of 4 h. All cells were preserved in serum-free freezing medium (Teyebio, Shanghai, China).
Cell transfection
The cells were then transfected with pLO-circAGFG1, miRNA-1257 mimic, and siRNA-1257 (GenePharma Co. Ltd., Shanghai, China) by using Lipofectamine 2000 (Invitrogen, Carlsbad, USA), according to the manufacturer’s instructions.
Western blotting
The cells were lysed in lysis buffer to extract total protein. Protein concentration was measured using Bio-Rad Protein Assay Kit (Bio-Rad Laboratories, USA). Subsequently, the proteins were resolved with 15% SDS-PAGE and then transferred onto PVDF membranes. After blocking with 5% skim milk, the membranes were incubated with the primary antibodies at 4 °C overnight. The membranes were then incubated along with the secondary antibody. An ECL chemiluminescence kit (Teyebio, Shanghai, China) was used to obtain the bands.
CCK-8 assay
The cells of the study groups were cultured in 96-well plates at a density of 1×105. The cells were then measured using the Cell Counting Kit-8 kit (Teyebio, Shanghai, China), according to the manufacturer’s instructions. The viability of the cells was detected by measuring the absorbance at 450 nm using a microplate reader.
Flow cytometry
Annexin V-FITC/propidium iodide (PI) Apoptosis Detection Kit (BD Biosciences, USA), was used to measure cell apoptosis, according to the instructions of the manufacturer. Briefly, after suspension in binding buffer, the cells were stained using PI and Annexin V in darkness. Flow cytometer (BD Biosciences, USA) was used to analyze the stained cells.
Reverse transcription polymerase chain reaction (RT-PCR) assay
TRIzol reagent (Invitrogen, Carlsbad, CA, USA) was used to resolve total RNA, and SuperScriptIII® kit (Invitrogen) was used for the reverse-transcription of RNA into cDNA. PRISM 7500 Sequence Detection System (Life Technologies, Grand Island, NY, USA) was applied for PCR assay. RT-PCR was conducted under the following conditions: 95 °C for 5 min, then 40 circles at 95 °C for 10 s and 65 °C for 30 s. The 2−ΔΔCt method was used to calculate gene expression level.
Luciferase reporter assay
TargetScan was applied to predict the targets of circAGFG1 and miR-1257. As predicted by TargetScan, circAGFG1 targeted miR-1257, which in turn targeted NOTCH2. To verify the target of circAGFG1, 293T cells were co-transfected with miR-1257 mimics or mimic-control together with circAGFG1 wild type (WT) or circAGFG1 mutant (MUT). To verify the target of miR-1257, 293T cells were co-transfected with miR-1257 mimics or mimic-control together with NOTCH2 WT or NOTCH2 MUT. Lipofectamine 2000 (Invitrogen) was used for transfection according to the instructions of the manufacturer. Dual Luciferase Reporter Assay System (Promega) was applied to measure luciferase activity.
Statistical analysis
SPSS 17.0 (SPSS Inc, Chicago, IL, USA) software was applied for data analysis. Data were expressed as mean ± standard deviation (SD). Significant differences between groups were evaluated by one-way ANOVA or Student’s t-test. Statistical significance was deemed to exist when P<0.05.
Results
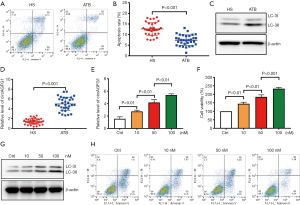
The levels of autography and circAGFG1 were increased and cell apoptosis was decreased in Mtb-infected macrophages
After Mtb infection, cell apoptosis for immune escape was inhibited and autophagy was activated (22,23). Consistent with previous literature, autophagy and cell survival were enhanced and apoptosis was inhibited in the macrophages from active tuberculosis (ATB) patients (Figure 1A,B). To further explore the role of circAGFG1 on autophagy and apoptosis in macrophages, PCR and Western blot assays were conducted. Our data demonstrated that the circAGFG1 level was elevated in macrophages from active Mtb patients when compared with those from healthy controls (Figure 1C,D). We further evaluated the influence of Mtb on cell apoptosis, cell survival, autophagy, and circAGFG1 level in vitro, and the results were consistent with those in Mtb-infected patients, which was in dose-dependent manner (Figure 1E,F,G). These findings indicate that circAGFG1 and autophagy, as well as cell apoptosis, which were all changed by Mtb, may play a critical role in tuberculosis.
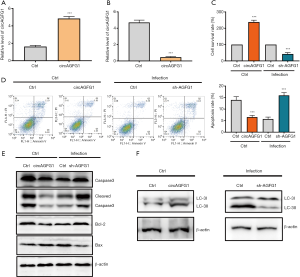
CircAGFG1 overexpression inhibited apoptosis and promoted autophagy in normal macrophages, while silencing circAGFG1 had the opposite effect on apoptosis and autophagy in Mtb-infected macrophages
Compared with corresponding controls, CircAGFG1 was upregulated in the circAGFG1 overexpression group and downregulated in the circAGFG1 knockdown group, suggesting that the overexpression and knockdown of circAGFG1 were successfully achieved (Figure 2A,B). In comparison with the controls, circAGFG1 overexpression enhanced cell viability (Figure 2C) and decreased cell apoptosis (Figure 2D) in normal cells, which indicated that both circAGFG1 and Mtb had a similar effect on autophagy and apoptosis. The effects of Mtb on macrophages was realized via the elevation of the level of circAGFG1. Moreover, silencing circAGFG1 in Mtb-infected macrophages had the opposite impact on autophagy and cell apoptosis (Figure 2C,D). Western blot analysis of apoptosis-related proteins, including cleavedcaspase-3, bcl-2, and bax were consistent with the results described above. The anti-apoptosis protein bcl-2 was increased by circAGFG1 overexpression. Meanwhile, pro-apoptosis proteins were inhibited by circAGFG1 overexpression in normal macrophages while knockdown of circAGFG1 in Mtb-infected macrophages had the opposite effect (Figure 2E). In comparison with the controls, circAGFG1 overexpression enhanced expression of LC-3II/LC-3I (Figure 2F) in normal cells. All these findings indicated that circAGFG1 induced by Mtb played a vital role in autophagy and cell apoptosis. Furthermore, silencing circAGFG1 counteracted the effects of Mtb on autophagy and cell apoptosis; a finding which could pave the way in providing a promising avenue in tuberculosis therapy.
miRNA1257 was the target of circAGFG1
As predicted by CircInteractome (https://circinteractome.nia.nih.gov/), circAGFG1 targeted miRNA1257 (Figure 3A). This was then verified by luciferase reporter assay. Among all of the groups, luciferase activity was lowest in the mimics group of WT type, further confirming miRNA1257 to be the target of circAGFG1 (Figure 3B). The miRNA1257 level in the healthy controls was elevated in comparison with the ATB group (Figure 3C), indicating its aberrant expression, and potential to serve as the biomarker in the diagnosis of tuberculosis. Moreover, the level of miRNA1257 was negatively related to the level of circAGFG1, suggesting that miRNA1257 was targeted and silenced by circAGFG1 (Figure 3D). To investigated this, we measured the level of miRNA1257 in the macrophages with circAGFG1 induction/knockdown, which showed the opposite results (Figure 3E). All these results indicated miRNA1257 to be the downstream molecule of circAGFG1, which may play a certain role in pulmonary tuberculosis.

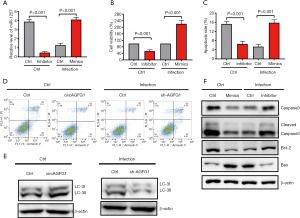
The effects of circAGFG1 on autophagy and cell apoptosis were realized by sponging miRNA1257
For further investigation, we explored if the role of circAGFG1 on autophagy and cell apoptosis was realized through its targeting of miRNA1257. Since miRNA1257 had already been confirmed as the target of circAGFG1, we evaluated the effects of either miRNA1257 overexpression or silencing on autophagy and cell apoptosis. As presented in the data, the overexpression and knockdown of miRNA1257 were successfully achieved (Figure 4A). By silencing miRNA1257, cell viability and LC3II/I level were increased, and cell apoptosis was inhibited in the cells without infection (Figure 4B,C,D,E,F), which was consistent with the effects of circAGFG1 overexpression. After miRNA1257 overexpression in the infected cells group, cell viability and LC3II/I were decreased and cell apoptosis was increased, which was consistent with the results of circAGFG1 knockdown (Figure 4D,E,F). Furthermore, the effect of miRNA1257 knockdown on apoptosis and autophagy were in line with the results of circAGFG1 overexpression. All these findings support the idea that the role of circAGFG1 was realized through its targeting of miRNA1257. The cell apoptosis-related proteins were also measured to further confirm the effect of miRNA1257 on apoptosis (Figure 4F). In non-infected normal cells, the anti-apoptosis protein, bcl-2, was increased by miRNA1257 knockdown while pro-apoptosis proteins, including bax and cleaved caspase 3, were decreased, which was in accordance with the anti-apoptosis effect seen with miRNA1257 knockdown. MiRNA1257 overexpression exerted the opposite effect on apoptosis-related proteins in infected cells, which presented as the evidence of the pro-apoptosis effect of miRNA1257 overexpression. The effects of miRNA1257 overexpression and circAGFG1 silencing on apoptosis were in accordance. Concurrently, the effects of silencing miRNA1257 and circAGFG1 overexpression were consistent. This further confirmed that the effects of circAGFG1 on apoptosis were realized via sponging miRNA1257.
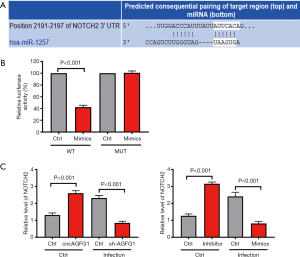
miRNA1257 functions by sponging Notch 2
For further exploring the downstream target molecule of circAGFG1/miRNA1257 that is involved in tuberculosis, we conducted a search of TargetScan to predict the target of miRNA1257. As revealed by results of our search, Notch 2 was the target of miRNA1257 (Figure 5A). This was further confirmed by luciferase reporter assay in which the luciferase activity was the lowest in the miRNA1257 mimics and the Notch 2 co-transfection wild-type group (Figure 5B). In the uninfected cells, both circAGFG1 overexpression and miRNA1257 inhibition increased the Notch 2 level, this corresponded with the pattern in the regulatory axis of circAGFG1/MiRNA1257/Notch 2 (Figure 5C). In the infected cells, the Notch 2 level was decreased either by sh-circAGFG1 or MiRNA1257 mimic, reflecting the regulatory role of the circAGFG1/MiRNA1257/Notch 2 axis. These findings indicated that the role of circAGFG1 was realized via the targeting of miRNA1257 to regulate the Notch signaling.
Discussion
Mtb has surpassed human immunodeficiency virus to become the infectious disease with the highest mortality (24). In vivo, Mtb has a long latent period, which presents a challenge for its diagnosis and treatment. Timely diagnosis and treatment are of great significance for curbing the spread of the disease (25). An increasing number of studies have demonstrated that Mtb can change the expression of non-coding RNAs, resulting in immune escape (26,27). In the current study, we found circAGFG1 to be aberrantly expressed in patients with active tuberculosis, which hints that it may play a critical role in pulmonary tuberculosis. After a comprehensive investigation, we found that circAGFG1 modulated autophagy and apoptosis of Mtb-infected macrophages via the Notch signaling pathway.
Macrophages, as the major cell targeted by Mtb, were investigated next. Our data revealed that circAGFG1 was upregulated in the macrophages from patients with pulmonary tuberculosis, indicating that circAGFG1 may be the functional gene that is changed by Mtb.
We further explored the effects of circAGFG1 on Mtb-infected macrophages. The body’s defense mechanism is an essential study for pathogen invasion, and cell apoptosis is the vital defense mechanism against intracellular pathogens. Cell apoptosis can eliminate invading microorganisms as part of innate immune response and restrict the transmission of the infection (28,29). Autophagy was found to be another critical defense mechanism in many types of pathogen infection. Autophagy is set apart from cell apoptosis as it maintains homeostasis without inducing cell death; this is exploited by Mtb and facilitates its intracellular survival (30). Previous reports have demonstrated that Mtb exploits autophagy via its unique function, named “enhanced intracellular survival”, thus increasing its survival (31).
In the current study, in line with previous research, cell apoptosis was inhibited and autophagy was enhanced in the macrophages of patients with pulmonary tuberculosis (23,32). We also evaluated the relationship between circAGFG1 and Mtb. Mtb was observed to upregulate the level of circAGFG1 in a concentration-dependent manner, while apoptosis was inhibited and autophagy was enhanced. Based on our results, we subsequently wanted to confirm whether the defense mechanisms changed by Mtb were related to the aberrant expression of circAGFG1. Therefore, the effect of circAGFG1 overexpression on cell apoptosis and autophagy in the macrophages was investigated.
Remarkably, circAGFG1 overexpression also inhibited cell apoptosis and enhanced autophagy in the normal macrophages, which was consistent with the effect of Mtb on the infected cells. This finding confirmed that the effect of Mtb on cell apoptosis and autophagy was realized via the overexpression of circAGFG1, which suggests that circAGFG1 may offer a novel therapeutic target for pulmonary tuberculosis. It was further verified in the infected macrophages; after circAGFG1 knockdown, the effect of Mtb on cell apoptosis and autophagy was nullified. As such, circAGFG1 was revealed to be an excellent treatment target for pulmonary tuberculosis.
CircRNA functions by inhibiting certain miRNAs which plays a further role via regulating the expression of its downstream target gene (33-36). In the present research, miRNA-1257 was found to be the target gene of circAGFG1. miRNA-1257 was a newly discovered non-coding RNA, which has not been reported before in relation to pulmonary tuberculosis. Furthermore, the effect of circAGFG1 can be replicated by silencing miRNA-1257 in normal cells. From the results, we can see that Mtb, circAGFG1 overexpression, and silencing miRNA-1257 in normal macrophage exert the same effects on cell autophagy and apoptosis. This indicates that the circRNA/miRNA-1257 axis is the main target of Mtb.
The target gene in the downstream of miRNA-1257 was also explored. As revealed both by TargetScan and luciferase reporter assay, Notch2 was the target gene of miRNA-1257. Notch2 plays a vital role in the Notch signaling pathway and was found to be upregulated in patients with tuberculosis (17). Recent research has suggested that by blocking the Notch signaling pathway, the imbalance of Th1/Th2 in patients with tuberculosis can be addressed (18). The targeting of Notch2 by miRNA-1257 unveiled in this research may provide another promising target for tuberculosis treatment.
Conclusions
In this study, we found a new functional axis in Mtb-infected macrophages: CircAGFG1/miRNA-1257/Notch2. The changes in this functional axis induced by MT inhibit apoptosis and enhance autophagy. The findings in this research are vital and present a new therapeutic target for tuberculosis treatment.
Acknowledgments
Funding: None.
Footnote
Data Sharing Statement: Available at http://dx.doi.org/10.21037/atm-20-3048
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/atm-20-3048). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. The study was approved by the Ethics Committee of Third Hospital of Jilin University of Medicine and written informed consent was obtained from all patients.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Harding E. WHO global progress report on tuberculosis elimination. Lancet Respir Med 2020;8:19. [Crossref] [PubMed]
- Gu Y, Wu C, Yu F, et al. Application of endobronchial ultrasonography using a guide sheath and electromagnetic navigation bronchoscopy in the diagnosis of atypical bacteriologically-negative pulmonary tuberculosis. Ann Transl Med 2019;7:567. [Crossref] [PubMed]
- He L, Ren X, Chen Y, et al. Prevalence of overweight and obesity among primary school children aged 5 to 14 years in Wannan area, China. Nutr Hosp 2014;30:776-81. [PubMed]
- He L, Ren X, Qian Y, et al. Prevalence of overweight and obesity among a university faculty and staffs from 2004 to 2010 in Wuhu, China. Nutr Hosp 2014;29:1033-7. [PubMed]
- Islam MM, Hameed HMA, Mugweru J, et al. Drug resistance mechanisms and novel drug targets for tuberculosis therapy. J Genet Genomics 2017;44:21-37. [Crossref] [PubMed]
- Yoshiyama T, Kurosaki A, Ogata H, et al. Limited benefit of CT scans in tuberculosis contact tracing. J Infect Chemother 2019;25:764-8. [Crossref] [PubMed]
- Sander MS, Laah SN, Titahong CN, et al. Systematic screening for tuberculosis among hospital outpatients in Cameroon: The role of screening and testing algorithms to improve case detection. J Clin Tuberc Other Mycobact Dis 2019;15:100095. [Crossref] [PubMed]
- Ahmad M, Ibrahim WH, Sarafandi SA, et al. Diagnostic value of bronchoalveolar lavage in the subset of patients with negative sputum/smear and mycobacterial culture and a suspicion of pulmonary tuberculosis. Int J Infect Dis 2019;82:96-101. [Crossref] [PubMed]
- Yuk JM, Yoshimori T, Jo EK. Autophagy and bacterial infectious diseases. Exp Mol Med 2012;44:99-108. [Crossref] [PubMed]
- Yuan Q, Chen H, Yang Y, et al. miR-18a promotes Mycobacterial survival in macrophages via inhibiting autophagy by down-regulation of ATM. J Cell Mol Med 2020;24:2004-12. [Crossref] [PubMed]
- Wang J, Zhu M, Pan J, et al. Circular RNAs: a rising star in respiratory diseases. Respir Res 2019;20:3. [Crossref] [PubMed]
- Zhang X, Zhu M, Yang R, et al. Identification and comparison of novel circular RNAs with associated co-expression and competing endogenous RNA networks in pulmonary tuberculosis. Oncotarget 2017;8:113571-82. [Crossref] [PubMed]
- Xue YB, Ding MQ, Xue L, et al. CircAGFG1 sponges miR-203 to promote EMT and metastasis of non-small-cell lung cancer by upregulating ZNF281 expression. Thorac Cancer 2019;10:1692-701. [Crossref] [PubMed]
- Zhang M, Wang S, Tang L, et al. Downregulated circular RNA hsa_circ_0067301 regulates epithelial-mesenchymal transition in endometriosis via the miR-141/Notch signaling pathway. Biochem Biophys Res Commun 2019;514:71-7. [Crossref] [PubMed]
- Mao Y, Xu R. Circular RNA CDR1-AS contributes to pemetrexed and cisplatin chemoresistance through EGFR/PI3K signaling pathway in lung adenocarcinoma. Biomed Pharmacother 2020;123:109771. [Crossref] [PubMed]
- Peng S, Song C, Li H, et al. Circular RNA SNX29 Sponges miR-744 to Regulate Proliferation and Differentiation of Myoblasts by Activating the Wnt5a/Ca2+ Signaling Pathway. Mol Ther-Nucleic Acids 2019;16:481-93. [Crossref] [PubMed]
- Li QF, He XY, Xin T. Role of the Notch signaling pathway in children with tuberculosis. Zhongguo Dang Dai Er Ke Za Zhi 2019;21:1012-5. [PubMed]
- Li Q, Zhang H, Yu L, et al. Down-regulation of Notch signaling pathway reverses the Th1/Th2 imbalance in tuberculosis patients. Int Immunopharmacol 2018;54:24-32. [Crossref] [PubMed]
- Duque C, Arroyo L, Ortega H, et al. Different responses of human mononuclear phagocyte populations to Mycobacterium tuberculosis. Tuberculosis 2014;94:111-22. [Crossref] [PubMed]
- Lavalett L, Rodriguez H, Ortega H, et al. Alveolar macrophages from tuberculosis patients display an altered inflammatory gene expression profile. Tuberculosis 2017;107:156-67. [Crossref] [PubMed]
- Surewicz K, Aung H, Kanost RA, et al. The differential interaction of p38 MAP kinase and tumor necrosis factor-α in human alveolar macrophages and monocytes induced by Mycobacterium tuberculois. Cell Immunol 2004;228:34-41. [Crossref] [PubMed]
- Ke Z, Lu J, Zhu J, et al. Down-regulation of lincRNA-EPS regulates apoptosis and autophagy in BCG-infected RAW264.7 macrophages via JNK/MAPK signaling pathway. Infect Genet Evol 2020;77:104077. [Crossref] [PubMed]
- Li M, Cui J, Niu W, et al. Long non-coding PCED1B-AS1 regulates macrophage apoptosis and autophagy by sponging miR-155 in active tuberculosis. Biochem Biophys Res Commun 2019;509:803-9. [Crossref] [PubMed]
- Global tuberculosis report 2016. 2013;6.
- Zumla A. Current trends and newer concepts on diagnosis, management and prevention of respiratory tract infections. Curr Opin Pulm Med 2013;19:189-91. [Crossref] [PubMed]
- Dutta RK, Kathania M, Raje M, et al. IL-6 inhibits IFN-gamma induced autophagy in Mycobacterium tuberculosis H37Rv infected macrophages. Int J Biochem Cell Biol 2012;44:942-54. [Crossref] [PubMed]
- Gu X, Gao Y, Mu D-G, et al. MiR-23a-5p modulates mycobacterial survival and autophagy during mycobacterium tuberculosis infection through TLR2/MyD88/NF-κB pathway by targeting TLR2. Exp Cell Res 2017;354:71-7. [Crossref] [PubMed]
- Mohareer K, Asalla S, Banerjee S. Cell death at the cross roads of host-pathogen interaction in Mycobacterium tuberculosis infection. Tuberculosis (Edinb) 2018;113:99-121. [Crossref] [PubMed]
- Krakauer T. Inflammasomes, Autophagy, and Cell Death: The Trinity of Innate Host Defense against Intracellular Bacteria. Mediators Inflamm 2019;2019:2471215. [Crossref] [PubMed]
- Jurkuvenaite A, Benavides GA, Komarova S, et al. Upregulation of autophagy decreases chlorine-induced mitochondrial injury and lung inflammation. Free Radic Biol Med 2015;85:83-94. [Crossref] [PubMed]
- Wei J, Dahl JL, Moulder JW, et al. Identification of a Mycobacterium tuberculosis gene that enhances mycobacterial survival in macrophages. J Bacteriol 2000;182:377-84. [Crossref] [PubMed]
- Zhou JS, Zhao Y, Zhou HB, et al. Autophagy plays an essential role in cigarette smoke-induced expression of MUC5AC in airway epithelium. Am J Physiol Lung Cell Mol Physiol 2016;310:L1042-52. [Crossref] [PubMed]
- Duffy FJ, Thompson E, Downing K, et al. A Serum Circulating miRNA Signature for Short-Term Risk of Progression to Active Tuberculosis Among Household Contacts. Front Immunol 2018;9:661. [Crossref] [PubMed]
- Lu C, Chen B, Chen C, et al. CircNr1h4 regulates the pathological process of renal injury in salt-sensitive hypertensive mice by targeting miR-155-5p. J Cell Mol Med 2020;24:1700-12. [Crossref] [PubMed]
- Mi B, Xiong Y, Chen L, et al. CircRNA AFF4 promotes osteoblast cells proliferation and inhibits apoptosis via the Mir-7223-5p/PIK3R1 axis. Aging (Albany NY) 2019;11:11988-2001. [Crossref] [PubMed]
- Wang L, Wang P, Su X, et al. Circ_0001658 promotes the proliferation and metastasis of osteosarcoma cells via regulating miR-382-5p/YB-1 axis. Cell Biochem Funct 2020;38:77-86. [Crossref] [PubMed]


