
A novel machine learning algorithm to predict disease free survival after resection of hepatocellular carcinoma
Introduction
Hepatocellular carcinoma (HCC) is the most common primary liver neoplasm and the second leading cause of cancer death in the world (1-3). Hepatocarcinogenesis occurs mostly in precancerous cirrhotic livers. The cirrhosis does not only foster hepatocarcinogesis but also limits treatment options and outcome in patients suffering from HCC. Liver transplantation (LT) is the most radical form of tumor resection and additionally can reconstitute liver function (4). However, because of the scarcity of donor organs transplantation can only be deployed subsidiary to other curative treatments (5). Therefore, a dynamic stepwise algorithm to allocate the best therapy is desired. Historically, clinical decision algorithms however have excluded patients from potentially curative surgery in favor of early HCCs with the longest predicted survival after transplantation (6). Another possible clinical treatment algorithm could be to define an oncologically satisfying outcome and treat patients with the most accessible treatment (7). This would trade the longest predicted survival as goal for the highest net benefit of the individual. However, to reallocate patients based on these principles, survival prediction after treatment is key. It should be readily available and as accurate as possible (8).
Several variables have been reported to accurately predict survival after LR in HCC. The proposed predictors have mostly been used to either describe tumor biology and immunology (9-12) or underlying liver disease (10,12). Although promising, unfortunately, these predictors have not been adopted into clinical routine nor are they recommended by the national associations for the Study of the Liver (3,4). This might be explained by either the high experimental expenditure in some cases or a lack of reproducibility of the retrospectively obtained clinical models. Additionally, other models are only available post hoc and therefore are not truly predictive (13).
The use of bioinformatics allows for complex multidimensional analyses of survival predictors. Mathematical algorithms and packages are designed to calculate sustainable and generalizable models, that prevent overfitting and accurately predict independent data sets. One of the most promising algorithms is Random Forest (RF) machine learning (ML) (14). It creates a multitude of decision trees based on both regression and classification variables. By majority vote the obtained model calculates the most likely prediction (14). With this ML, and specifically RF, could improve outcome prediction and guide complex multilayered treatment decisions.
The aim of this study was to use a ML algorithm based on easily accessible variables to accurately predict disease free survival (DFS) of HCC patients after LR.
Methods
Study groups and predictive variables
Patients scheduled for resection because of suspected HCC from 1st January 2007 until 1st April 2018 were included in the prospectively maintained data base. Inclusion was truncated at this date to ensure a minimum follow-up of 1 year. Ethical approval was obtained from the institutional review board (EK 19-395) at the Ludwig-Maximilian University in Munich. The need for an informed consent was waived by the institutional review board. This trial complies with the TRIPOD Statement (15).
Clinical indication for surgical resection was based on the recommendation of our multidisciplinary specialized tumor board, which is attended by experienced liver surgeons to evaluate the resectability of the patients (16). HCC was diagnosed based on pathognomonic magnet resonance imaging, computed tomography or biopsy. In general LR as a destination therapy was offered to all patients with preserved liver function and no cirrhosis or compensated cirrhosis. This was defined as: Child-Turcotte-Pugh-Score (CTP) A or B+ without clinical (esophageal varices) and radiographic signs of portal hypertension (enlarged spleen) and a serum bilirubin of ≤2 mg/dL. Portal hypertension is a negative predictor for early postoperative death through liver failure. In some guidelines it is named as a relative contraindication for liver surgery (17-19). Major resections were defined as resections of 4 or more segments. All other resections were defined as minor resections (20).
Twenty-six preoperatively readily available variables were used for this analysis. Clinical variables, such as sex, age, presence of cirrhosis and ascites, CTP and its underlying disease were noted. Radiographic parameters consisted of number, size of the largest lesion and whether the tumors were deemed inside Milan-Criteria. With these information patients were staged according to Barcelona Clinics Liver Cancer (BCLC) staging system. Additionally, the following laboratory values were acquired: Bilirubin (BILI; mg/dL), Albumin (ALB; g/L), Aspartate Transferase (AST; U/L), Alanine Transferase (ALT, U/L), alpha-Fetoprotein (AFP; ng/mL), (aPTT; s), International Normalized Ratio (INR), Creatinine (CREA; mg/dL), C-Reactive Protein (CRP; mg/L), Leukococytes (WBC, 106/L), Platelets (PLT; 106/L). Finally, with these values and clinical data additional scores were calculated. The formulas for the Model of Endstage Liver Disease (MELD), Kings-Score (KS) and modified Glascow Prognostic Score (mGPS) can be found in our supplementary material (http://fp.amegroups.cn/cms/11f739089d6cadd46ac0f4cbfa36a50f/atm.2020.04.16-1.pdf, p1) (10,21,22).
Follow-up
According to the recommendation of the interdisciplinary tumor board HCC patients were structurally followed-up with screening cross sectional imaging every 3–6 moths in the first year. After an uneventful first year the intervals were changed to every 6 months. From 2 years on patients were screened every year. If patients did not choose to participate in our structured follow-up, periodical telephone interviews were conducted. Disease free survival (DFS) was defined as the time-period from the index operation to recurrence or death of the patient. Survival times for overall survival (OS) were calculated from the date of resection to the date of death. Last contact with the patients was censored.
Statistical analysis
All included variables were obtained before the index operation, making this an intention-to-treat analysis in a retrospective setting. All calculations were conducted with the RStudio software (Version 1.1.463, RStudio Inc., Boston, MA, USA). Loaded packages were caret, factoextra, FactoMineR, ggplot2, mice, pROC, randomForest, VIM and survival, survminer (23-29). Code for decision curve analysis was obtained from a publication by Zhang et al. (30). In general, normally distributed data is summarized with the mean and standard deviation (± SD) and compared using t-test. Classification variables were noted in a contingency table and compared using the Chi2-Test. A P value of <0.05 was considered statistically significant.
As detailed in Figure 1 we used the following workflow to analyze the data:
- Preprocessing
- Imputation of missing values
- Random data partitioning
- Variable Selection (training data)
- Recursive Feature Elimination (RFE)
- Clinical curation of selected variables (anti-classification)
- Modelling (training data)
- Resampling to balance data
- RF Modelling
- Prediction of independent test data (test data)
- Performance measurement (test data)
- Receiver operating characteristic (ROC) curve
- Area under the curve (AUC)
- Kaplan Meier Analysis
- Decision Curve Analysis (DCA)
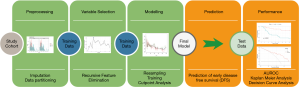
First, preprocessing was done by imputing missing values with the RF algorithm within the “mice” package for R. Observations with more than 50% missing data were excluded from the analysis. Visual evaluation of kernel density plots was performed for variables with more than 10% missing data. Additionally, to test for unwanted skewing of data through imputation, we recalculated the entire model with exclusion of important variables with more than 10% missing values as described by Sterne et al. (http://fp.amegroups.cn/cms/11f739089d6cadd46ac0f4cbfa36a50f/atm.2020.04.16-1.pdf, p2-3) (31).
After preprocessing, simple Kaplan Meier Analysis was performed for the entire cohort. Univariate and multivariate analyses of all variables were conducted to explore the data set for independently predictive variables. This analysis was separate from the ML modelling to show the predictive power of singular predictors obtained by classical retrospective analysis. This analysis was performed for 3 different time-points. First 90 days, which represents in-hospital mortality (32). Second, 24 months, which indicates early recurrence, and lastly the entire follow-up period was used as time-point to explore effects on long-term DFS. For univariate analysis we utilized standard Cox survival regression. For multivariate analysis we chose a stepwise forward and backward variable selection process published by Collet et al. (33). Categorical variables were only considered if more than 10 events occurred in each group. C-Statistics, Receiver Operating Curves (ROC) and their Area under the Curve (AUC) were calculated from variables that were shown to be independently predictive in the multivariate analysis.
The entire cohort was split randomly 70% to 30% in a training and a test data set using the createDataPartition formula from the caret ensemble package (24). Training data was used to develop the model and perform cross-validation (CV) for hyperparameter tuning. The test data was left untouched and used for testing the model after developing the model.
Therefore secondly, RFE was used on the training data to select appropriate variables for the model. RFE itself is a ML algorithm implemented in the caret package. It allows for backwards selection of predictive variables. The least important predictors are eliminated sequentially thereby creating a more accurate model. As ML algorithms may discriminate based on socioeconomic factors, it may lead to the development of ethically non-acceptable patient selection (ML bias). Therefore, we have excluded gender and history of alcohol abuse from machine model training (anti-classification) (34).
Thirdly, modelling steps were done simultaneously using the train function within the caret package. We used a RF algorithm to construct the model. RF was proposed by Breiman in 2001 (14). It grows k (default: k=500) decision trees, which can be used with regression and classification variables. In contrast to other tree-based ML algorithms like “eXtreme Gradient Boosting”, these trees are grown at the same time not sequentially. This and repeated CV limits overfitting to the training data. Because of its robustness and the possibility for nested validation using CV we chose the RF approach. To account for the unbalanced data set, we tested down-, up-sampling and the SMOTE algorithm as well as outcome weighing within the RF training to prevent misclassification in case of a dominating majority-class. The sampling technique that created the most generizable model was chosen.
Lastly the tuned and cross validated final model was used to predict the test data (see above) which was untouched throughout the entire procedure. Performance on test data was measured using the ROC curve and AUC analysis. The obtained predictions from the RF model were added to the test data set. Based on the calculated probability patients were either classified as “High Risk” or “Low Risk” patients. Based on this classification we drew the Kaplan-Meier curve and calculated the survival difference. Additional to the aforementioned performance measures decision curve analysis was used to depict the clinical net benefit of our RF model (12,35). Relevant code from the workflow is made available in the supplementary Material (http://fp.amegroups.cn/cms/11f739089d6cadd46ac0f4cbfa36a50f/atm.2020.04.16-1.pdf, p 4-7).
Results
Study cohort
One hundred and eighty-one patients were scheduled by the interdisciplinary tumorboard for LR because of HCC. In the preprocessing analysis one observation showed itself to have more than 50% missing values, thus this patient was removed from the analysis. Therefore 180 patients were included into the analysis. As described above we analyzed the data imputation for stability. Detailed description of the analysis of imputed data can be found in the supplement document pp 5-6 including graphs (http://fp.amegroups.cn/cms/11f739089d6cadd46ac0f4cbfa36a50f/atm.2020.04.16-1.pdf, Figures 1,2) of the observed changes after imputing largest tumor measured and AFP.
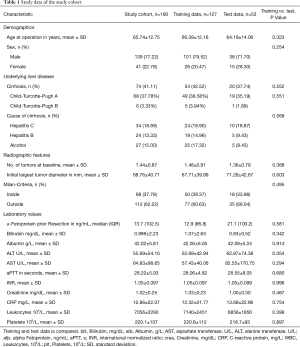
The demographic and clinical characteristics as well as mean laboratory values of the entire study cohort are listed in Table 1. Patients scheduled for operation because of HCC were 65.74±12.75 years old. 139 (77.22%) were male and 41 (22.78%) were female. 74 (41.11%) had signs of cirrhosis in the preoperative cross-sectional imaging or confirmed cirrhosis through biopsy. Of those patients with cirrhosis most patients were classified as CTP A (37.78%) and 3.33% showed a compensated CTP B+ cirrhosis. Underlying disease was evenly distributed between HCV, HBV, and alcoholic cirrhosis (Table 1). Most patients appeared to be outside Milan-Criteria on imaging at treatment decision 112 (62.22%). Overall Survival of all patients was 83.1% [76.5; 87.9], 63.4% [54.3; 71.1] and 56.6% [46.6; 65.4] at 1,3 and 5 years of follow-up, respectively. DFS of the entire study cohort at the same time-points was 61.6% [53.77; 68.5], 38.2% [30.0; 46.3] and 27.4% [19.1; 36.3] (Figure 2).
Full table
Multivariate analysis of predictors for DFS
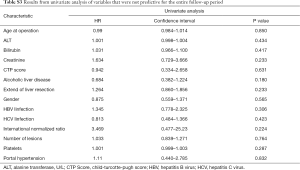
As mentioned above variable selection for the multivariate analysis was conducted according to the stepwise procedure published by Collet et al. DFS 90 days after resection was independently predicted by the serum creatinine and the presence of an HBV infection (Table S1). At 24 months predictive variables changed to only serum AFP (Table S2). Lastly, mGPS, BCLC, AFP, AST and the MELD Score showed themselves to be independently predictive of DFS over the entire follow-up period (Table 2, Table S3). Next, we analyzed the c-statistics of all independent predictors. The c-index for mGPS, BCLC, AFP, AST, HBV and MELD Score were 0.579, 0.571, 0.505, 0.595, 0.523 and 0.543 indicating a poor predictive power of the individual variables. ROC curves and AUC for continuous variables are depicted in Figure 3.

Full table

Full table

Full table
Full table

Creation of the training and test data sets
Before training and validating (Cross Validation) the algorithm, we split the study cohort 70/30. The training data set included n=127 patients. The test data set included 53 patients and remained untouched throughout the analysis and was only used for testing the final model.
Variables were compared between the training and the test data sets. None of the variables differed significantly between both cohorts. Detailed information can be found in Table 1.
RF Model training and validation
We used RFE to select the variables based on the accuracy they contribute to the final model. Seven variables (mGPS, aPTT, CRP, largest tumor size, number of lesions, age at time of operation and history of alcohol abuse) remained as predictors for the RF model (Figure 4).
As shown in Figure 4 the variables were chosen by the highest accuracy in the cross-validation cohort. The chosen variables included history of alcohol abuse, which we removed after curation to prevent machine bias. Lastly, mGPS, aPTT, CRP, largest tumor size, number of lesions and age at time of operation remained to calculate the RF model.
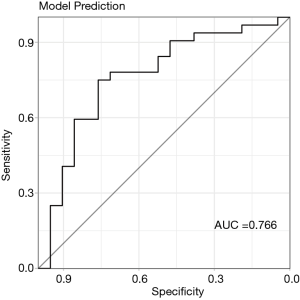
In case of unbalanced data, optimization for predictive accuracy is prone to favor the majority class in predicting outcome. As described in the method section the outcome classes were weighed to improve the prediction power of the RF model. “ALIVE” was weighed 0.01 and “EVENT” was weighed 0.0065. Additionally, up-, down-sampling and the SMOTE algorithm were used to improve prediction of the majority and minority outcome classes. Finally, based down-sampling was identified to be the most accurate and generizable method for predicting the independent test data. With RFE and down-sampling the RF model based on above mentioned variables reached an AUC of 0.766 (0.627–0.904) in predicting the test data (Figure 5) during the entire follow-up period.
RF Model validation for early DFS
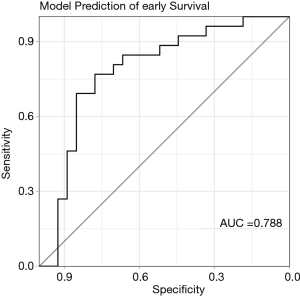
To predict early recurrence the above trained and validated RF model was used to predict early recurrence in the test data set. The RF model showed a strong predictive power for early recurrence with an AUC of 0.788 (0.658–0.919) (Figure 6). After dividing the patients into “High-Risk” (HR) and “Low-Risk” (LR), survival analysis was performed.
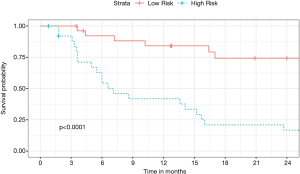
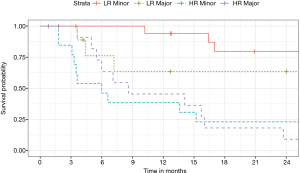
Twenty-seven patients were predicted to survive (LR) and 26 patients were predicted to have an event (HR). In the LR group survival was 84.1% (63.1, 93.7) at 1 year and 74.2% (51.0, 87.6) at 2 years. In the HR group survival was lower [1 year: 41.8% (22.4, 60.2), 2 years: 16.7% (5.2, 33.8)] (P<0.001). The survival curve is depicted in Figure 7. After dividing the LR and HR patients into major and minor resection no difference based on the extend of resection could be identified (Figure 8).
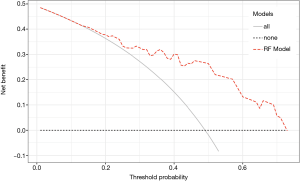
Additionally, DCA was conducted to examine the clinical benefit of the model. Figure 9 shows that our RF model added a relevant net benefit across a range of risk thresholds up to 0.72.
Discussion
Key findings
The aim of this study was to develop a ML model to accurately predict early DFS after LR of HCC.
The collected clinical and laboratory values were chosen based on previous publications identifying variables for prediction of HCC outcomes (5,7,21,36-42). By using bioinformatics, we identified relevant variables for the model. Although not all variables by themselves were independently predictive in the multivariate analysis, the combination of these within the RF ML added to the predictiveness of the final model. We tested our RF model in a separate test data set and calculated its performance.
With our RF model we were able to distinguish between patients at high or low risk for recurrence or death early after LR. Only 16.7% of HR patients had disease recurrence or died during the follow-up. In contrast 74.2% LR patients survived without disease recurrence. This difference was highly significant. The prediction of test data based on the model reached an AUC of 0.788 [0.658-0.919]. Additionally, the extend of resection had no influence on survival. DCA showed a clinical net benefit for using the RF model.
Up to now, no study has attempted to combine these routine variables with complex ML algorithms to predict survival after HCC resection.
Workflow
The presented workflow was designed to be used for a range of diseases and treatments. The first obstacle when analyzing clinical data is that not all variables will be available to researchers. In this scenario, either observations (patients) are excluded or missing data is imputed. Exclusion of patients leads to a reduction of the power of the analysis (23). The imputation of means and median values can be used as dummy variables, however these values will add more noise to the data (23). Imputation algorithms allow for multiple estimations and cross-validation of missing values. However, with naïve unchecked imputations these algorithms are in danger of creating a self-fulfilling prophecy (31). Therefore, in light of multiple observations with missing values in our data set we reviewed the variables included in the final model. We opted to remove the observations with missing values and ran the entire workflow again as proposed by Sterne et al. (31). We could conclusively show that the imputation of missing values did not skew the data (http://fp.amegroups.cn/cms/11f739089d6cadd46ac0f4cbfa36a50f/atm.2020.04.16-1.pdf, pp 2-3)
Another pitfall for ML models might be unbalanced outcome in data sets (in this study: disease recurrence or death). The model is thereby incentivized to predict the majority class, whilst misclassifying the minority class. To combat this, up-, down-sampling and SMOTE were used to balance the data (43,44). In our analysis the down-sampling technique yielded the most generizable model.
ML algorithms are superior in pattern recognition particularly in large data sets but may result in decision-making which due to the method ignore ethical values embedded in our society (34). In medicine gender, race and other stigmatized patient characteristics (e.g., history of alcohol abuse) may come out as discriminative variables, which we have deliberately excluded (anti-classification) (45,46). Even though, anti-classification is criticized because it might lead to intentional biases in the entire cohort, we advocate for a practical approach until algorithms for “Fair Machine Learning” are available (34).
Interpretation
Prediction of outcome after HCC resection has been published before. Especially, most variables that were included in our model have already been identified by classical retrospective multivariate analysis. Kinoshita and colleagues calculated multiple inflammation-based scores including mGPS. For 24 months of follow-up mGPS reached an AUC of 0.695 (47). These results could not be replicated in our study (AUC =0.579). CRP has also been tested individually in multiple studies. In a recently published study CRP could not reach AUC values above 0.6 for recurrence free survival (12). The same is also true for largest tumor size and number of lesions. Their individual predictive power is disputed in the literature and while independently predictive in validation cohorts in some studies, tumor size alone was not predictive in ours. However, BCLC A stadium was a positive prognostic indicator. The differing results in the literature might be based on the fact that cohorts are often not comparable. Especially western cohorts have a lower proportion of hepatitis patients (12,35). Additionally, analyses are often based on single center retrospective cohorts and only analyzed with multivariate analyses without appropriate test cohorts. In our multivariate analysis (separate from the ML algorithm), which we performed for 90 days and 24 months after operation as well as for the entire available follow-up, we could show that the predictiveness of variables changes. Predictive variables for early in-hospital DFS were creatinine and presence of HBV infection. At 24 months predictive variable shifted to AFP as a surrogate of tumor biology. The RF model designed in this work incorporated variables representing liver function and possibly tumor biology. These were not chosen by multivariate analysis but with a complex algorithm called RFE (24). Compared to multivariate analysis it is superior because it incorporates the complex relationships between variables within the multitude of trees (classification models) that are created by the Random Forest algorithm.
Up to now, publications investigating ML prediction for outcomes after surgery are scarce. Recently, Kim et al. published a ML model based on radiomics and clinicopathological variables. With this they reached an AUC of 0.716 predicting the early DFS (2 years) after HCC resection. We concur with the authors, that using modern ML algorithms combined with clinical values and possibly experimental/computer generated variables could help to better predict survival after liver resection (48).
Limitations
The main limitation is the retrospective nature of this study. Even though we tested the model on an untouched set of patient data, more patients would make this model more stable and generizable. Also, the inclusion of more Hepatitis patients would make our approach more suitable for patients from e.g., Asia. Based on these encouraging results we plan to launch a prospective observational multinational study to further validate and improve the model. Because the complexity of a RF model could potentially impede deployment to study centers and clinical practice, we have developed an easy to use browser based online app (tiny.cc/hcc_model). With this, researchers are free to explore the model in other cohorts or to join our efforts to improve the RF model.
Another limitation of our study is that the RFE algorithm inside the caret package does not allow for nested resampling inside the feature selection. With this feature selection could be tuned to adjust for unbalanced data. We hope that a new version of the caret package will allow for resampling when selecting variables using RFE.
Conclusions
With the use of our workflow we were able to develop and test a RF model based on standard clinical and laboratory variables to accurately predict early DFS after liver resection in case of HCC. ML modelling could change future treatment allocation to offer LR to low risk patients and to list high risk patients for LT.
Acknowledgement
The authors thank Philipp Pöschke from the Institute for Informatics at the Ludwigs-Maximilian-University in Munich for his valuable help in coding the workflow and developing the online app.
Funding: None.
Footnote
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/atm.2020.04.16). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. The research was conducted ethically in accordance with the World Medical Association Declaration of Helsinki, ethical approval was obtained from the institutional review board (EK 19-395) at the Ludwig-Maximilian University in Munich.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Price TR, Perkins SM, Sandrasegaran K, et al. Evaluation of response after stereotactic body radiotherapy for hepatocellular carcinoma. Cancer 2012;118:3191-8. [Crossref] [PubMed]
- Malek NP, Schmidt S, Huber P, et al. The diagnosis and treatment of hepatocellular carcinoma. Dtsch Arztebl Int 2014;111:101-6. [PubMed]
- Marrero JA, Kulik LM, Sirlin CB, et al. Diagnosis, Staging, and Management of Hepatocellular Carcinoma: 2018 Practice Guidance by the American Association for the Study of Liver Diseases. Hepatology 2018;68:723-50. [Crossref] [PubMed]
- European Association for the Study of the Liver. EASL Clinical Practice Guidelines: Management of hepatocellular carcinoma. J Hepatol 2018;69:182-236. [Crossref] [PubMed]
- Schoenberg MB, Bucher JN, Vater A, et al. Resection or Transplant in Early Hepatocellular Carcinoma. Dtsch Arztebl Int 2017;114:519-26. [PubMed]
- Vitale A, Farinati F, Pawlik TM, et al. The concept of therapeutic hierarchy for patients with hepatocellular carcinoma: A multicenter cohort study. Liver Int 2019;39:1478-89. [Crossref] [PubMed]
- Schoenberg MB, Anger HJW, Hao J, et al. Development of novel biological resection criteria for safe and oncologically satisfying resection of hepatocellular carcinoma. Surg Oncol 2018;27:663-73. [Crossref] [PubMed]
- Steyerberg EW, Vickers AJ, Cook NR, et al. Assessing the performance of prediction models: a framework for traditional and novel measures. Epidemiology 2010;21:128-38. [Crossref] [PubMed]
- Sun XD, Shi XJ, Chen YG, et al. Elevated Preoperative Neutrophil-Lymphocyte Ratio Is Associated with Poor Prognosis in Hepatocellular Carcinoma Patients Treated with Liver Transplantation: A Meta-Analysis. Gastroenterol Res Pract 2016;2016:4743808.
- Cillo U, Giuliani T, Polacco M, et al. Prediction of hepatocellular carcinoma biological behavior in patient selection for liver transplantation. World J Gastroenterol 2016;22:232-52. [Crossref] [PubMed]
- Morgul MH, Klunk S, Anastasiadou Z, et al. Diagnosis of HCC for patients with cirrhosis using miRNA profiles of the tumor-surrounding tissue - A statistical model based on stepwise penalized logistic regression. Exp Mol Pathol 2016;101:165-71. [Crossref] [PubMed]
- Gan W, Yi Y, Fu Y, et al. Fibrinogen and C-reactive protein score is a prognostic index for patients with hepatocellular carcinoma undergoing curative resection: a prognostic nomogram study. J Cancer 2018;9:148-56. [Crossref] [PubMed]
- Brunner SM, Rubner C, Kesselring R, et al. Tumor-infiltrating, interleukin-33-producing effector-memory CD8(+) T cells in resected hepatocellular carcinoma prolong patient survival. Hepatology 2015;61:1957-67. [Crossref] [PubMed]
- Breiman L. Random Forests. Machine Learning 2001;45:5-32. [Crossref]
- Collins GS, Reitsma JB, Altman DG, et al. Transparent Reporting of a multivariable prediction model for Individual Prognosis Or Diagnosis (TRIPOD): The TRIPOD Statement. Eur Urol 2015;67:1142-51. [Crossref] [PubMed]
- Jones RP, Vauthey JN, Adam R, et al. Effect of specialist decision-making on treatment strategies for colorectal liver metastases. Br J Surg 2012;99:1263-9. [Crossref] [PubMed]
- Bruix J, Reig M, Sherman M. Evidence-Based Diagnosis, Staging, and Treatment of Patients With Hepatocellular Carcinoma. Gastroenterology 2016;150:835-53. [Crossref] [PubMed]
- Forner A, Gilabert M, Bruix J, et al. Treatment of intermediate-stage hepatocellular carcinoma. Nat Rev Clin Oncol 2014;11:525-35. [Crossref] [PubMed]
- Heimbach JK, Kulik LM, Finn RS, et al. AASLD guidelines for the treatment of hepatocellular carcinoma. Hepatology 2018;67:358-80. [Crossref] [PubMed]
- Reddy SK, Barbas AS, Turley RS, et al. A standard definition of major hepatectomy: resection of four or more liver segments. HPB (Oxford) 2011;13:494-502. [Crossref] [PubMed]
- Cross TJ, Rizzi P, Berry PA, et al. King's Score: an accurate marker of cirrhosis in chronic hepatitis C. Eur J Gastroenterol Hepatol 2009;21:730-8. [Crossref] [PubMed]
- Kamath PS, Wiesner RH, Malinchoc M, et al. A model to predict survival in patients with end-stage liver disease. Hepatology 2001;33:464-70. [Crossref] [PubMed]
- van Buuren S, Groothuis-Oudshoorn K. mice: Multivariate Imputation by Chained Equations in R. J Stat Softw 2011. [Crossref]
- Kuhn M. Building Predictive Models in R Using the caret Package. J Stat Softw 2008. [Crossref]
- Kassambara A. Machine Learning Essentials. Practical Guide in R. STHDA; 2018.
- Robin X, Turck N, Hainard A, et al. pROC: an open-source package for R and S+ to analyze and compare ROC curves. BMC Bioinformatics 2011;12:77. [Crossref] [PubMed]
- Liaw A, Wiener M. Classification and regression by randomForest. R news 2002;2:18-22.
- Kowarik A, Templ M. Imputation with the R Package VIM. J Stat Softw 2016. [Crossref]
- Wickham H. ggplot2: Elegant Graphics for Data Analysis. New York: Springer-Verlag, 2016.
- Zhang Z, Rousson V, Lee WC, et al. Decision curve analysis: a technical note. Ann Transl Med 2018;6:308. [Crossref] [PubMed]
- Sterne JAC, White IQR, Carlin JB, et al. Multiple imputation for missing data in epidemiological and clinical research: potential and pitfalls. BMJ 2009;338:b2393. [Crossref] [PubMed]
- Schiergens TS, Dorsch M, Mittermeier L, et al. Thirty-day mortality leads to underestimation of postoperative death after liver resection: A novel method to define the acute postoperative period. Surgery 2015;158:1530-7. [Crossref] [PubMed]
- Collet D. Modelling Survival Data in Medical Research, Third Edition. Chapman & Hall/CRC Texts in Statistical Science. Chapman & Hall; 2014.
- Corbett-Davies S, Goel S. The measure and mismeasure of fairness: A critical review of fair machine learning. arXiv preprint arXiv:1808.00023, 2018.
- Huang JL, Fu YP, Jing CY, et al. A novel and validated prognostic nomogram based on liver fibrosis and tumor burden for patients with hepatocellular carcinoma after curative resection. J Surg Oncol 2018;117:625-33. [Crossref] [PubMed]
- A new prognostic system for hepatocellular carcinoma: a retrospective study of 435 patients: the Cancer of the Liver Italian Program (CLIP) investigators. Hepatology 1998;28:751-5. [Crossref] [PubMed]
- Andreou A, Gul S, Pascher A, et al. Patient and tumour biology predict survival beyond the Milan criteria in liver transplantation for hepatocellular carcinoma. HPB (Oxford) 2015;17:168-75. [Crossref] [PubMed]
- Bigourdan JM, Jaeck D, Meyer N, et al. Small hepatocellular carcinoma in Child A cirrhotic patients: hepatic resection versus transplantation. Liver Transpl 2003;9:513-20. [Crossref] [PubMed]
- Cha CH, Ruo L, Fong Y, et al. Resection of hepatocellular carcinoma in patients otherwise eligible for transplantation. Ann Surg 2003;238:315-21; discussion 321-3. [PubMed]
- Gwiasda J, Schulte A, Kaltenborn A, et al. Identification of the resection severity index as a significant independent prognostic factor for early mortality and observed survival >5 and >10 years after liver resection for hepatocellular carcinoma. Surg Oncol 2017;26:178-87. [Crossref] [PubMed]
- Kim JH, Han DS, Bang HY, et al. Preoperative neutrophil-to-lymphocyte ratio is a prognostic factor for overall survival in patients with gastric cancer. Ann Surg Treat Res 2015;89:81-6. [Crossref] [PubMed]
- Ni XC, Yi Y, Fu YP, et al. Prognostic Value of the Modified Glasgow Prognostic Score in Patients Undergoing Radical Surgery for Hepatocellular Carcinoma. Medicine (Baltimore) 2015;94:e1486. [Crossref] [PubMed]
- Kuhn M. Model training and tuning. In: The caret Package. Accessed 07/23 2019.Available online: https://topepo.github.io/caret/model-training-and-tuning.html
- Chawla NV, Bowyer KW, Hall LO, et al. SMOTE: synthetic minority over-sampling technique. J Artif Int Res 2002;16:321-57. [Crossref]
- Volkow ND, Fowler JS, Wang GJ, et al. Imaging dopamine's role in drug abuse and addiction. Neuropharmacology 2009;56 Suppl 1:3-8. [Crossref] [PubMed]
- Kendler KS, Gardner CO, Prescott CA. Toward a comprehensive developmental model for alcohol use disorders in men. Twin Res Hum Genet 2011;14:1-15. [Crossref] [PubMed]
- Kinoshita A, Onoda H, Imai N, et al. Comparison of the prognostic value of inflammation-based prognostic scores in patients with hepatocellular carcinoma. Br J Cancer 2012;107:988-93. [Crossref] [PubMed]
- Kim S, Shin J, Kim DY, et al. Radiomics on Gadoxetic Acid-Enhanced Magnetic Resonance Imaging for Prediction of Postoperative Early and Late Recurrence of Single Hepatocellular Carcinoma. Clin Cancer Res 2019;25:3847-55. [Crossref] [PubMed]



