
Identification of a six-gene signature associated with tumor mutation burden for predicting prognosis in patients with invasive breast carcinoma
Introduction
Breast cancer (BC) is one of the major malignancies among females, and mortality remains the second main reason for cancer deaths worldwide (1). In recent years, the diagnosis of BC mainly depends on pathological tests, imaging examinations, and assessments of tumor markers (2). Because of a high recurrence rate of BC, the age of BC onset has gradually become younger. Therefore, the early detection and diagnosis of BC play an essential role in preventing the genesis and development of BC (3). It is thus necessary to identify possible molecular biomarkers for the diagnosis, prognosis, and survival risk prediction for BC.
Tumor mutation burden (TMB) is a new biomarker for immune therapy and is defined as the number of somatic, coding, base substitution, and indel mutations per megabase (4). It is widely acknowledged that the TMB plays a vital role in the occurrence and development of cancer (5). In one study, cancer patients with high levels of TMB showed stronger immunotherapeutic responses than those with low levels of TMB (6). Exploring gene expressions that are closely correlated with TMB levels can, therefore, be of great importance, and TMB has also been linked to cancer prognosis (7). Thus, there is an urgent need to identify potential TMB-related gene signature for BC prognosis.
In the current study, we firstly identified DEGs based on TMB by RNA sequencing data from The Cancer Genome Atlas (TCGA). Then, functional enrichment analyses were further employed to analyze the biological roles of these DEGs. Finally, we constructed a six-gene signature for BC by using TCGA datasets and bioinformatics analysis and verified that the prognostic value of this signature was associated with some clinical factors.
Methods
Data source
We obtained the RNA-seq datasets with the clinic information of BC patients comprising 474 tumor samples and 481 normal samples from the The Cancer Genome Atlas Breast Invasive Carcinoma (TCGA-BRCA) datasets and analyzed them using the limma package (8) with the false discovery rate (FDR) <0.05 and |log(fold change) >1|. Subsequently, we downloaded the masked somatic mutation data from TCGA database. Then, we classified BC patients into the low-TMB subtype and the high-TMB subtype by using the median of TMB data as the cutoff value and compared the expression levels of TMB-related genes between both subtypes.
Functional enrichment analysis and protein-protein interaction (PPI) network analysis of differentially expressed genes (DEGs)
To investigate potential biological processes (BP), cellular components (CC), molecular functions (MF), and signaling pathways related to the DEGs, we applied the Gene Ontology (GO) and Kyoto encyclopedia of genes and genomes (KEGG) pathway analyses by clusterProfiler package (9) in R. The potential connections between the DEGs were then visualised using the STRING database (10).
Cox proportional hazards model construction and model validation
By combining these prognostic gene expression markers with the regression coefficient (β) from the multivariate Cox proportional hazards regression analysis, a risk score model was established based on the results of multivariate Cox regression analysis. Then, the formula of risk score was calculated as follows: expression of gene1 × β1gene1 + expression of gene2 × β2gene2 + expression of genen × βngenen. According to the median risk score, all patients were assigned into the high-risk and low-risk groups. Proportional assumptions for Cox proportional hazard model were examined by Kaplan-Meier analysis. Receiver operating characteristic (ROC) and the area under the curve (AUC) were plotted by using R package survival ROC. In addition, the distribution of risk score and survival status, along with the gene expression levels of each patient, was analyzed by using the R software.
Correlation analysis of immune infiltration analysis
The CIBERSORT method (11) was used to estimate immunocyte population fractions, which could sensitively and accurately distinguish 22 phenotypes of immune cells. Then, in subsequent review, those with the CIBERSORT P<0.05 were considered eligible. Moreover, we employed 29 immune-related gene sets, which represent different immune cell types, functions, and pathways. We performed the single-sample gene set enrichment analysis (ssGSEA) to calculate the activity or enrichment levels of immune cells, functions, or pathways in tumor samples.
Identification of small molecular drugs
We compared the six-gene signature with those in the Connectivity Map (cMap) database (12) to identify candidate small molecular drugs associated with BC. We then divided DEGs into the upregulated group and downregulated group. Next, we used these genes to query the cMap database. A connectivity score from -1 to 1 indicated the stimulation of the compound, while a negative score indicated the inhibition of the compound.
Statistical analysis
All the statistical analyses were performed using the R package. Survival curves were established by the Kaplan-Meier method and the log-rank test. Univariate and multivariate Cox proportional-hazard models were used to evaluate the hazard ratios of prognostic factors. We applied the ‘RMS’ package in R to plot the nomogram and calibration analysis. A P value <0.05 was viewed as statistically significant.
Results
Identification of DEGs between two TMB groups
A total of 343 DEGs were screened between low/high-TMB groups from the TCGA-BRCA dataset according to the screening criteria, including 168 down-regulated and 175 up-regulated genes (Figure 1A). Then, GO and KEGG analyses were applied to investigate the biological roles of the DEGs. The functional enrichment analysis revealed that these DEGs were mostly enriched in epidermis development, extracellular matrix, and receptor-ligand activity among BP, CC, and MF, respectively (Figure 1B). KEGG analysis demonstrated that these DEGs primarily participated in protein digestion and absorption (Figure 1C).
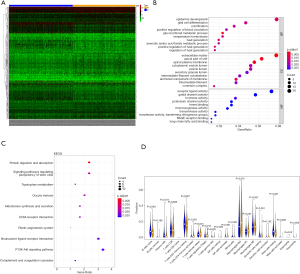
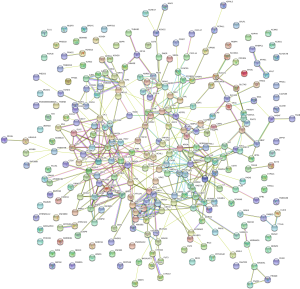
To further explore the relationship between high and low levels of TMB and infiltration of immune cells, The CIBERSORT algorithm was used to measure the relative proportions of immune cells in low/high TMB groups. It revealed that the abundance levels of naive B cells, resting CD4 memory T cells, resting dendritic cells, and resting mast cells were significantly lower in the high-TMB group than the low-TMB group. Conversely, T cells follicular helper, Tregs, and macrophage subsets (M0/M1) were significantly higher in the high-TMB group (Figure 1D, P<0.05), indicating that TMB levels can affect the cancer progression. Moreover, the PPI network of the DEGs was further constructed by the STRING database with 273 nodes and 448 edges (Figure 2).
Construction of the Cox regression model and survival analysis
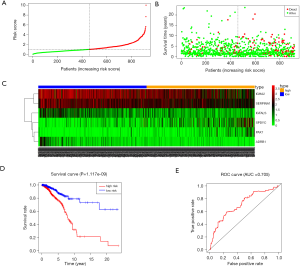
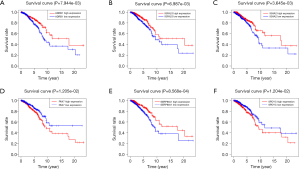
Multivariable Cox proportional hazards stepwise regression with backward selection was used to build a prognostic model. Then, we calculated the risk score for each patient in the datasets. Hence, we established a six-gene signature as follows: risk score = (‒0.2224 × IGFALS) + (0.2232 × PAX7) + (0.1243 × SPDYC) + (‒0.1224 × IGHA2) + (‒0.1172 × SERPINA1) + (‒0.3558 × ADRB1). Then, the distribution of risk score, survival status, and the corresponding heatmap of the gene expression levels of each patient in the signature was analyzed and plotted (Figure 3A,B,C). In addition, the survival risk curve showed that patients in the high-risk group had a significantly shorter overall survival time compared with patients in the low-risk group (Figure 3D). ROC analysis revealed reliable performance in the survival prediction of the model, and the AUC was 0.705 (Figure 3E). Moreover, survival analysis using Kaplan-Meier showed that these 6 genes were closely associated with the overall survival of BC patients (Figure 4, P<0.05).
Association between the risk scores of the six-gene signature and clinicopathological features
For observing the association between the risk scores and each clinicopathological factors, we separately calculated the distribution of risk scores. We examined the expression of the 6 genes in high/low-TMB and high/low-risk patients in the TCGA-BRCA dataset (Figure 5A). We observed significant differences between the high- and low-risk groups concerning tumor grade (T, P<0.001) and lymph node (N, P<0.05). Furthermore, we found risk scores were significantly different between patients stratified by T grade (P<0.05), age (P<0.001), and status (P<0.001) (Figure 5B). Moreover, univariate and multivariate Cox analysis was applied to determine whether the six-gene signature can be employed to independently predict the survival of BC patients. The results showed that the six-gene risk score could be used to independently predict the overall survival of the patient (P<0.001, Figure 5C,D, Table 1). Then, we built a nomogram that integrated the clinicopathological factors with the six-gene signature to predict the survival probability of BC patients (Figure 5E). Meanwhile, calibration plots for the probability of 3- and 5-year survival were consistent between predicted values by the nomogram and measured values (Figure 5F,G).

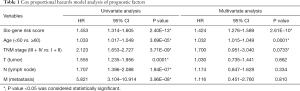
Full table
To further explore the relationship between the 6 TMB-related genes and the infiltration of immune cells, we conducted the ssGSEA algorithm to estimate the coefficient of the association of 6 genes in immune cell subsets. It was found that these 6 genes were closely associated with tumor-infiltrating immune cell subset, indicating that these genes mainly participate in immune response (Figure 6, P<0.05). Furthermore, we applied the cMap database to identify potential therapeutic drugs for BC, and the top 20 most distinct small molecule drugs with their enrichment scores were identified (Table 2). Nadolol was related to highly significant negative scores (‒0.949). The potential small molecules could reverse the expression levels of the 6 TMB-related genes induced by BC, and represent new directions for research in BC treatment.
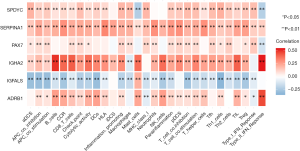
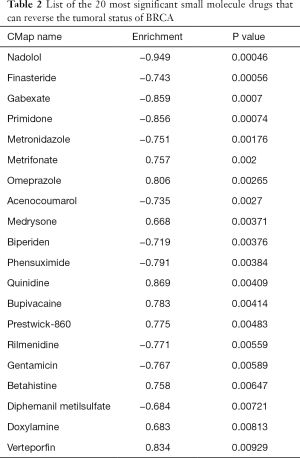
Full table
Discussion
BC is a heterogeneous disease with different biological properties caused by a variety of factors involving the accumulation of gene changes (13). Despite the improved diagnosis and therapy of BC, the prognosis is still poor (14). Finding accurate biomarkers for early diagnosis and a more accurate prognosis of BC will help to improve the efficiency of therapies for BC and provide molecular markers for targeted treatment (15). Thus, it is urgent to develop more sensitive and specific biomarkers for the prognosis of BC patients.
Currently, many studies are aimed at finding predictive biomarkers of immune responses. (16). TMB has been used to reveal the process of mutation accumulation in tumors and has proven to be effective in predicting the immune response in various cancers, including BC (17). Previous studies have reported that TMB is closely associated with the prognosis of cancer patients (18). In this study, a total of 343 TMB-related DEGs also showed a considerable difference between the 2 groups with different TMB levels, which may be of great significance. To demonstrate the molecular function of these DEGs, we applied the GO and KEGG analyses. The results indicated that DEGs were mainly enriched in epidermis development, extracellular matrix, and receptor-ligand activity, in addition to protein digestion and absorption. Moreover, when we compared the differential abundance of immune cells between the low and high TMB groups via CIBERSORT algorithm, the results revealed that the abundance levels of naive B cells, resting CD4 memory T cells, resting dendritic cells, and resting mast cells were significantly lower in the high-TMB group compared to those in the low-TMB group. Similarly, a previous study showed that increased overall survival was related to relatively larger fractions of resting CD4 memory T cells (19).
Moreover, using univariate and stepwise multivariate Cox regression analyses, we established a prognostic signature for BC based on the expression of 6 genes, including IGFALS, PAX7, SPDYC, IGHA2, SERPINA1, and ADRB1. Among these, IGHA2 and SERPINA1 are considered risky prognostic genes in BC. Kang et al. reported that the differential expression of IGHA2 in the early and advanced stages of BC indicated that IGHA2 might be a novel marker for the progression of BC (20). Chan et al. showed that SERPINA1 expression is correlated with the outcome of ER+ and ER+/HER2+ BC (21). Moreover, Kang et al. reported that knockdown of ADRB can suppress tumor progression by inhibiting tumor growth, metastasis, and angiogenesis in BC, and that ADRB1 was abundantly expressed in several types of BC cells (22). As a member of the Spy1/RINGO family, SPDYC might also play a vital role in regulating both normal and abnormal growth processes in the breast (23).
Furthermore, some previous findings have shown that insulin-like growth factor binding protein (IGFALS) can act as a possible tumor-suppressor gene silenced by methylation in hepatocellular carcinoma (24). Also, He et al. showed that deregulated levels of PAX7 can contribute to muscle wasting in cancer cachexia (25). Previous studies also have suggested that SERPINA1 could inhibit caspase-3-mediated apoptosis and has been proven to predict the development of cutaneous squamous cell carcinoma (26). Finally, SERPINA1 has been shown to contribute to the development of metastatic features in epithelial ovarian cancer cells (27).
Through bioinformatics analyses, based on the six-gene signature, we found that BC patients with high-risk scores had a significantly decreased survival time compared with low-risk scores. ROC analysis demonstrated the relatively high sensitivity and specificity of the six-gene signature model. Similarly, in this study, the survival analysis of the six genes based on a prognostic signature all demonstrated a significant difference between the high and low expression groups. Then, we integrated the six-gene signature with other independent clinical variables to construct a comprehensive model for monitoring progression in BC, which suggested that the six-gene risk score could be used independently to estimate the overall survival of the patients. Based on the role of TMB in immune responses (28), ssGSEA analysis revealed that the 6 genes are closely associated with tumor-infiltrating immune cells. Nadolol was additionally identified as an important small molecular agent in BC’s development.
Conclusions
In this study, we integrated TMB with gene expression and built a prognostic prediction model that can improve survival prediction for BC patients and reveal the TMB-based prognostic signature which may be used as a prognostic biomarker for BC patients. Further experimental studies are still needed to confirm the established signature.
Acknowledgments
Funding: This work was supported by the Nantong Science and Technology Project of China (Grant No. MS12018060 and MS12019025).
Footnote
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/atm.2020.04.02). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. The study was approved by Ethics the Committee of Affiliated Hospital of Nantong University (ID: 2013-67). Informed consent was provided by all participants.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Siegel RL, Miller KD, Jemal A. Cancer statistics, 2019. CA Cancer J Clin 2019;69:7-34. [Crossref] [PubMed]
- McDonald ES, Clark AS, Tchou J, et al. Clinical Diagnosis and Management of Breast Cancer. J Nucl Med 2016;57:9S-16S. [Crossref] [PubMed]
- Elmore JG, Armstrong K, Lehman CD, et al. Screening for Breast Cancer. JAMA 2005;293:1245-56. [Crossref] [PubMed]
- Chalmers ZR, Connelly CF, Fabrizio D, et al. Analysis of 100,000 human cancer genomes reveals the landscape of tumor mutational burden. Genome Med 2017;9:34. [Crossref] [PubMed]
- Chan TA, Yarchoan M, Jaffee EM, et al. Development of tumor mutation burden as an immunotherapy biomarker: utility for the oncology clinic. Ann Oncol 2019;30:44-56. [Crossref] [PubMed]
- Endris V, Buchhalter I, Allgauer M, et al. Measurement of tumor mutational burden (TMB) in routine molecular diagnostics: in silico and real-life analysis of three larger gene panels. Int J Cancer 2019;144:2303-12. [PubMed]
- Wang X, Li M. Correlate tumor mutation burden with immune signatures in human cancers. BMC Immunol 2019;20:4. [Crossref] [PubMed]
- Ritchie ME, Belinda P, Di W, et al. limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res 2015;43:e47. [Crossref] [PubMed]
- Yu G, Wang L, Han Y, et al. clusterProfiler: an R Package for Comparing Biological Themes Among Gene Clusters. OMICS 2012;16:284-7. [Crossref] [PubMed]
- Szklarczyk D, Franceschini A, Wyder S, et al. STRING v10: protein-protein interaction networks, integrated over the tree of life. Nucleic Acids Res 2015;43:D447-52. [Crossref] [PubMed]
- Newman AM, Liu CL, Green MR, et al. Robust enumeration of cell subsets from tissue expression profiles. Nat Methods 2015;12:453-7. [Crossref] [PubMed]
- Lamb J, Crawford ED, Peck D, et al. The Connectivity Map: using gene-expression signatures to connect small molecules, genes, and disease. Science 2006;313:1929-35. [Crossref] [PubMed]
- Cancer Genome Atlas Network. Comprehensive molecular portraits of human breast tumours. Nature 2012;490:61-70. [Crossref] [PubMed]
- Silvestri V, Rizzolo P, Zelli V, et al. A possible role of FANCM mutations in male breast cancer susceptibility: Results from a multicenter study in Italy. Breast 2018;38:92-7. [Crossref] [PubMed]
- Zhang X, Rice M, Tworoger SS, et al. Addition of a polygenic risk score, mammographic density, and endogenous hormones to existing breast cancer risk prediction models: A nested case-control study. PLoS Med 2018;15:e1002644. [Crossref] [PubMed]
- Goodman AM, Kato S, Bazhenova L, et al. Tumor Mutational Burden as an Independent Predictor of Response to Immunotherapy in Diverse Cancers. Mol Cancer Ther 2017;16:2598-608. [Crossref] [PubMed]
- Park SE, Park K, Lee EC, et al. Clinical implication of tumor mutational burden in patients with HER2-positive refractory metastatic breast cancer. OncoImmunology 2018;7:e1466768. [Crossref] [PubMed]
- Innocenti F, Ou FS, Qu X, et al. Mutational Analysis of Patients With Colorectal Cancer in CALGB/SWOG 80405 Identifies New Roles of Microsatellite Instability and Tumor Mutational Burden for Patient Outcome. J Clin Oncol 2019;37:1217-27. [Crossref] [PubMed]
- Zhang SC, Hu ZQ, Long JH, et al. Clinical Implications of Tumor-Infiltrating Immune Cells in Breast Cancer. J Cancer 2019;10:6175-84. [Crossref] [PubMed]
- Kang S, Maeng H, Kim BG, et al. In situ identification and Localization of IGHA2 in the Breast Tumor Microenvironment by Mass Spectrometry. J Proteome Res 2012;11:4567-74. [Crossref] [PubMed]
- Chan HJ, Li H, Liu Z, et al. SERPINA1 is a direct estrogen receptor target gene and a predictor of survival in breast cancer patients. Oncotarget 2015;6:25815-27. [Crossref] [PubMed]
- Kang F, Ma W, Ma X, et al. Propranolol Inhibits Glucose Metabolism and 18F-FDG Uptake of Breast Cancer Through Posttranscriptional Downregulation of Hexokinase-2. J Nucl Med 2014;55:439-45. [Crossref] [PubMed]
- Golipour A, Myers D, Seagroves T, et al. The Spy1/RINGO Family Represents a Novel Mechanism Regulating Mammary Growth and Tumorigenesis. Cancer Res 2008;68:3591-600. [Crossref] [PubMed]
- Udali S, Guarini P, Ruzzenente A, et al. DNA methylation and gene expression profiles show novel regulatory pathways in hepatocellular carcinoma. Clin Epigenetics 2015;7:43. [Crossref] [PubMed]
- He WA, Berardi E, Cardillo VM, et al. NF-κB-mediated Pax7 dysregulation in the muscle microenvironment promotes cancer cachexia. J Clin Invest 2013;123:4821-35. [Crossref] [PubMed]
- Farshchian M, Kivisaari A, Ala-aho R, et al. Serpin Peptidase Inhibitor Clade A Member 1 (SerpinA1) Is a Novel Biomarker for Progression of Cutaneous Squamous Cell Carcinoma. Am J Pathol 2011;179:1110-9. [Crossref] [PubMed]
- Normandin K, Péant B, Page CL, et al. Protease inhibitor SERPINA1 expression in epithelial ovarian cancer. Clin Exp Metastasis 2010;27:55-69. [Crossref] [PubMed]
- Thomas A, Routh ED, Pullikuth A, et al. Tumor mutational burden is a determinant of immune-mediated survival in breast cancer. Oncoimmunology 2018;7:e1490854. [Crossref] [PubMed]


