
TRIB3 rs6037475 is a potential biomarker for predicting felodipine drug response in Chinese patients with hypertension
Introduction
Hypertension is a major preventable risk-factor that results in cardio-cerebrovascular disease, chronic kidney disease, disability and premature mortality (1). The latest epidemiological data have shown that an added 244.5 million adults in China have suffered from hypertension. Half of them were aware of their conditions but fewer than 40% received treatment. Moreover, less than 15% of the treated patients achieved their blood pressure control goals (2,3). In general, inadequate blood pressure control is one of the leading causes of end-organ damage and the subsequent increase of social and economic cost burden (4). However, whether all patients benefit from treatment according to the existing primary guidelines is still controversial. The pathogenesis of hypertension is a complex process that is influenced by both genetic and environmental factors (5). Substantial inter-individual variation, especially the genetic polymorphisms, can be the leading causes of the low effective control rate in blood pressure response to antihypertensive therapy (6).
Tribbles homolog 3 (TRIB3) interacts with a host of proteins to control many aspects of eukaryotic cell biology. Emerging evidence has revealed that the primary function of TRIB3 is to control the transduction of insulin signal and endothelial vascular function (7). Our previous studies found that single nucleotide polymorphism (SNP) of TRIB3 (rs2295490) could significantly affect the responses of calcium-channel blockers (CCBs) and ACE-inhibitors (8-10). However, these results were limited by the small sample size or the combination of other drug treatments, such as hypoglycemic, lipid-lowering and anticoagulant drugs. Therefore, it is essential to confirm the effect of TRIB3 gene polymorphism on antihypertensive drug sensitivity in a large and long-term follow-up clinical trial cohort.
The European Society of Hypertension (ESH) and European Society of Cardiology (ESC) guidelines emphasized that most patients need the combination of two or more drugs to achieve better blood pressure control (11). Meanwhile, surveys in China showed that CCBs monotherapy were the most commonly used treatment (57.1%), and only those with diuretics monotherapy were able to increase the overall rate of blood pressure control rate by 11% compared to those with CCBs monotherapy (12). These results suggest that the combination of CCBs and diuretics may be more beneficial for Chinese hypertensive patients. FEVER is a double-blind, randomized controlled clinical trial of Chinese hypertensives and was designed to compare the effect of a low-dose felodipine and a low-dose hydrochlorothiazide (HCTZ) combination therapy with that of the matched placebo treatment (13). Using this clinical trial and our previous work as a basis (10), we designed and conducted this study to explore and validate the effect of functional TRIB3 gene polymorphisms on the responses of antihypertensive drugs.
Methods
Patients and treatment
This is a retrospective survey on the FEVER study. Details on the FEVER study design and organization have been published previously (13). In brief, FEVER study is a double-blind, randomized and multi-center clinical trial that was approved by local ethics committees (registered on www.clinicaltrials.gov, No. NCT01136863), and the trial was conducted following the Declaration of Helsinki. All patients were self-reported as Han Chinese and provided written consent. Eligible patients were treated with an open-labeled hydrochlorothiazide 12.5 mg once a day for 6 weeks. After a comprehensive assessment, they were randomly assigned to the felodipine (intensive) and the matched placebo (less intensive) treatment groups. For the intensive treatment group, patients received a combination therapy of a low dose of diuretic (HCTZ: 12.5 mg q.d.) and a low dose of calcium antagonist (felodipine: 5 mg q.d.). For the less intensive treatment group, patients were given a low dose of diuretic (HCTZ: 12.5 mg) combined with the matched placebo therapy. Randomized double-blind treatment was maintained for at least 36 months. Follow-up was conducted at 1-month intervals during the first 6 months, then at 3-month intervals thereafter. In this study, 858 patients’ DNA samples and the matched clinical trial data in the FEVER study cohort were graciously provided by the Beijing Fu Wai Hospital with the collaboration of the Chinese Hypertension League.
DNA isolation
Peripheral venous blood was collected from Chinese patients with hypertension. Genomic DNA was extracted from peripheral venous blood using E.Z.N.A.® SQ blood DNA Kit II (Omega Bio-Tek company, USA) according to the manufacturer’s instructions. Extracted genomic DNA was stored at −80 °C until use.
Pharmacogenetics study protocol
The pipeline of this pharmacogenetics study protocol is provided in detail in Figure 1. The candidate SNPs selection and functional prediction in TRIB3 used the Encyclopedia of Deoxyribonucleic Acid (DNA) Elements (ENCODE) database. The ENCODE database is an ongoing international cooperation project that has systematically listed functional elements, chromatin annotations and variation annotations in human genome, intuitively showing whether a SNP is located in any potential functional region, such as transcription factor binding sites, open chromatin regions, micro-ribonucleic acid (miRNA) and long non-coding RNA (lncRNA) transcription regions, miRNA target sites, and DNA methylation sites. For the candidate SNPs, a minimal allele frequency (MAF) of more than 10% of the Chinese population in 1000 Genomes Project was required. To explore the relationship between SNPs and antihypertensive drug response, we implemented a strategy of screening small samples and validating in the remaining large samples. By using computer-generated random numbers, patients were assigned (1:1) to each treatment group according to the study protocol. Candidate SNPs were genotyped for screening by Bioyong Technologies Inc. using a Sequenom MassARRAY® SNP system. The significant SNPs were then genotyped for validation by using the TaqMan fluorescent probe typing method on the ABI-based AppliedBiosystems7500 Instrument platform. Primers information are shown in Table S1. The reaction mixture (20 µL) contained the following: TaqMan genotyping master mix (2×) 10.0 µL, TaqMan genotyping assay mix (2×) 9.0 µL, and g-DNA 1.0 µL. Temperature cycling proceeded as follows: (I) initial denaturation for 10 min at 95 °C; (II) 40 cycles of 5 s at 95 °C; (III) elongation at 60 °C for 1 min. Finally, 5% of the participants were randomly selected for validation via Sanger sequencing.
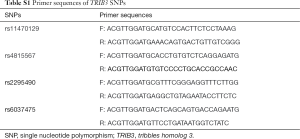
Full table
Statistical analysis
All the statistical analyses were performed using SAS software (version 9.4, SAS Institute) for windows. Allele frequencies were determined by the genotypes of all the participants. Hardy-Weinberg equilibrium analysis was carried out by using the chi-squared or Fisher’s exact test. Baseline characteristics among different phenotypes were assessed by independent-samples T-test or Wilcoxon rank-sum test, as appropriate. A mixed linear model, with adjustment for age, body mass index (BMI) and gender, was used to analyze the effect of SNPs on the antihypertensive drug response during the whole follow-up period. The quantitative data described in the text and figures are presented as means ± standard deviation (SD) and percentages for categorical data. P<0.05 (2-tailed) was considered a statistically significant value.
Results
Baseline characteristics and genotyping results
A total of 858 Chinese patients with hypertension provided clinical data and matched DNA samples. However, 16 (1.9%) DNA samples that did not qualify for genotyping, and 12 (1.4%) patients with incomplete clinical information were excluded. Finally, 830 patients were included in our study. Among them, 397 (47.8%) were assigned to the intensive treatment group, and 433 (52.2%) were assigned to the less intensive treatment group (Figure 1). Except for heart rate, the clinical baseline characteristics between the two groups (age, sex, and BMI, etc.) did not observe a significant difference (Table 1). According to our study protocol, 4 candidate SNPs (rs2295490, rs11470129, rs4815567, and rs6037475) in TRIB3 gene were included and successfully identified. The detailed results are shown in Table S2.

Full table

Full table
Association analysis of the candidate SNPs
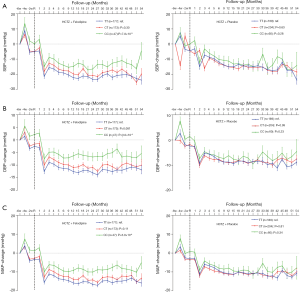
In the screening stage, 259 patients were included in our study. Among them 136 (52.5%) were assigned to the intensive treatment group, and 123 (47.5%) were assigned to the less intensive treatment group. A mixed linear regression model was used for analyzing the influence of rs2295490, rs11470129, rs4815567 and rs6037475 on antihypertensive efficacy of the two afore-mentioned regimens. This revealed that compared with the placebo treatment group, rs6037475 genetic variation could significantly influence the effect of felodipine on diastolic blood pressure (DBP) reduction (P=5.5×10−3) (Table 2). As for the reductions influenced by the rs2295490, rs11470129 and rs4815567 genetic variations, however, the adjusted P-values did not reach statistical significance. Further analysis, as shown in Figure 2, confirmed that TRIB3 rs6037475 CC genotype was associated with the lower DBP (P=6.3×10−3) compared with TT genotype in the felodipine treatment group during the follow-up period. Collectively, these results suggest that TRIB3 rs6037475 may be associated with the effects of felodipine. Therefore, we carried out further research focused on this genetic variation locus.
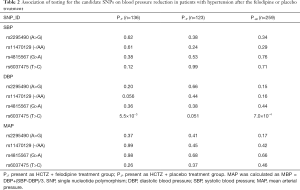
Full table
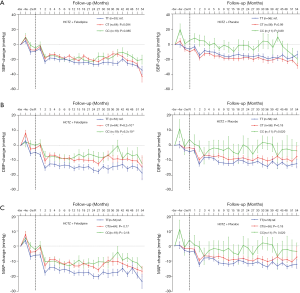
Validation and combined analysis of TRIB3 rs6037475
To validate whether rs6037475 is a biomarker for predicting felodipine drug response, a validation study that included 261 cases of HCTZ combined with the felodipine therapy and 310 HCTZ combined with the matched placebo controls was performed. As it is shown in Table S3, rs6037475 genetic variation was still significantly associated with DBP reduction (P=0.021) in the intensive treatment group and exhibited a marginal association with systolic blood pressure (SBP) (P=0.062) and mean arterial pressure (MAP) (P=0.061) reduction. In the less intensive treatment group, however, no correlation was observed between rs6037475 genetic variation with SBP, DBP and MAP reduction. We also found that TRIB3 rs6037475 CC genotype was significantly associated with the reduction of SBP (P=0.021), DBP (P=6.0×10−3) and MAP (P=0.021) in the felodipine treatment group (Figure 3).
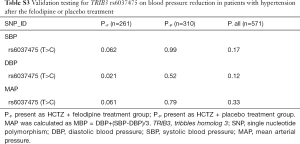
Full table
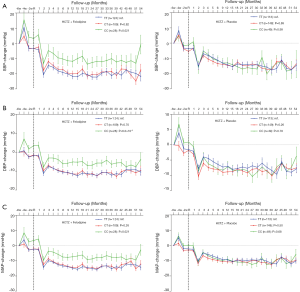
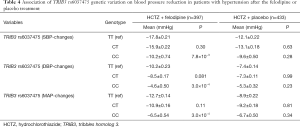
Combined screening and validation set analysis found that TRIB3 rs6037475 genetic variation significantly influenced the effect of felodipine on SBP (P=0.028), DBP (P=2.1×10−3) and MAP (P=1.3×10−3), but a significant difference was not observed in the matched placebo treatment group (Table 3). Further analysis found that patients with TRIB3 rs6037475 CC genotype had a significant lower reduction of mean SBP (CC vs. TT −10.2±0.74 vs. −17.8±0.21), DBP (CC vs. TT −4.6±0.50 vs. −10.2±0.23) and MAP (CC vs. TT −6.5±0.54 vs. −12.7±0.14) than those with TT genotype in the felodipine treatment group, but not in the matched placebo group (Table 4). Meanwhile, compared with the TT genotype carriers, patients with TRIB3 rs6037475 CC genotype had a significant higher mean SBP (P=7.8×10−3), DBP (P=3.0×10−4) and MAP (P=3.0×10-4) than those with TT genotype in the felodipine treatment group (Figure 4).

Full table
Full table
Discussion
The results from our previous studies, which were based on a well-controlled drug clinical trial, suggested that TRIB3 (rs2295490, A>G) genetic variation was closely related to blood pressure regulation and antihypertensive drugs responses (8-10). Our data indicated that a novel variant of TRIB3 rs6037475, which was located 2 kb upstream of the gene, was significantly associated with the felodipine effect on DBP-lowering. However, in this study, TRIB3 rs2295490 genetic variation did not affect the antihypertensive efficacy of felodipine or hydrochlorothiazide. A systematic review elaborated that the abnormal expression of TRIB3 is related to insulin resistance, impaired insulin secretion, endothelial dysfunction, and eventually led to type 2 diabetes mellitus (T2DM) and cardiovascular disease (7). Andreozzi et al. reported that TRIB3 rs2295490 mutation altered the structure rather than the expression of TRIB3, and enhanced the regulatory effect of TRIB3 and Akt (14). Another study found that TRIB3 rs2295490 mutation resulted in a significant reduction in insulin-induced nitric oxide (NO) release compared with the wild type in vitro (15). In view of this, TRIB3 rs6037475 may regulate felodipine response by affecting the expression of TRIB3 or mediating NO release. Further investigations are warranted to confirm these possible mechanisms. Our previous studies also found that patients with TRIB3 rs2295490 G allele had a weaker DBP response to calcium channel blockers (azelnidipine and compound nitrendipine), which was inversed by angiotensin receptor blocker (candesartan and irbesartan) or ACE-inhibitors (imidapril) prescription (8-10). In short, these findings suggested that TRIB3 genetic variations were involved in the regulation of blood pressure. Furthermore, TRIB3 genetic variations could affect hypotensive process, due to TRIB3s’ unique pseudokinase plastic domains (16).
At present, no report on the regulatory effect of felodipine and hydrochlorothiazide on TRIB3 exists. However, Ding et al. revealed that felodipine could significantly increase NO production, Ca2+-dependent nitric oxide synthase (NOS) activity and endothelial nitric oxide synthase (eNOS) protein expression by a NO-cGMP mediated mechanism in human umbilical vein endothelial cells (HUVEC) cells (17). Emerging data also demonstrated that a CCB potentiates the vascular protective effects of angiotensin receptor blockers (ARBs) in salt-sensitive hypertension as compared with a diuretic. For instance, olmesartan combined with azelnidipine resulted in a more significant reversal of the decrease in p-eNOS, total eNOS and p-Akt than olmesartan combined with hydrochlorothiazide (18). However, monotherapy of diuretics, such as hydrochlorothiazide and indapamide, did not affect NOS activity, eNOS and inducible nitric oxide synthase (iNOS) protein expressions (18,19). Jiang et al. reported tag-SNPs in four insulin resistance genes (ADIPOQ, LEPR, RETN and TRIB3). Among them, gene polymorphisms in LEPR and ADIPOQ were significantly associated with hypertension, while the other two (RETN and TRIB3) were not (20). However, the tag-SNPs study was limited, because it did not contain the most classical mutation of TRIB3, such as rs2295490. Therefore, these results may indirectly favor our findings that TRIB3 rs6037475 genetic variation was significantly associated with DBP-lowering in the HCTZ therapy combined with felodipine, but not in the place-combined treatment.
Studies have found that when a 10 mmHg is decreased in SBP, major cardiovascular disease events can be markedly reduced by about 10–35% (21-23). In the A Coronary disease Trial Investigating Outcome with Nifedipine GITS (ACTION), a blood pressure reduction of 14.6/7.6 mmHg in the nifedipine administration group was associated with a 38% reduction in the incidence of heart failure as compared with the matched placebo treatment group (24,25). In the FEVER trial, for the felodipine treated group, in which blood pressure achieved slightly lower values than in the placebo group, a blood pressure reduction of 3.5/1.5 mmHg was found to significantly reduce the incidence of all cardiovascular events by about 28% (13). In our study, there was no significant difference in SBP, DBP and MAP reduction between the two groups in patients who carried TRIB3 rs6037475 TC and CC genotypes (shown in Table S4). A significant antihypertensive benefit after felodipine treatment was observed in those patients with TT genotype. Therefore, we believe that TRIB3 rs6037475 genetic variation may be a potential biomarker for predicting the efficacy of felodipine in patients with hypertension.

Full table
The strength of our study is that the FEVER is a well-controlled clinical drug trial with long-term follow-up. Nevertheless, certain limitations should be considered when interpreting our findings. First, due to the quality of peripheral blood samples, we did not obtain enough qualified DNA samples. Therefore, sampling errors might have occurred that were factored into the statistical analysis. Second, other SNPs (e.g., rs4813620 variant), which may also be involved in the interaction between TRIB3 expression and drug efficacy or affect vascular function (26), were not examined in our study. Besides, the TRIB3 rs6037475 genetic variation in the promoter region of TRIB3 may have a functional effect on transcriptional activity, but this speculation was only derived from database prediction. Overall, our results strongly suggest that although felodipine is more effective in the treatment of patients with TRIB3 rs6037475 TT genotype than those with CC genotype, its clinical application merits further investigation.
Conclusions
The results we present here reveal that TRIB3 rs6037475 genetic variation is significantly associated with blood pressure reduction in patients with felodipine treatment and can be useful as a potential biomarker for predicting felodipine drug response in Chinese hypertensive patients.
Acknowledgments
We also thank the many patients and the FEVER collaborative group that participated in this study.
Funding: This research was supported by National Scientific Foundation of China (No. 81573511, No. 81903715 and No. 81874329), the National Key R&D Plan (No. 2016YFC0905000), the Science and Technology Program of Guangdong Province (2016A020215130).
Footnote
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/atm.2020.03.176). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. The study protocol was approved by local ethics committees (registered on www.clinicaltrials.gov, No. NCT01136863), and all patients provided written consent.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Mills KT, Bundy JD, Kelly TN, et al. Global Disparities of Hypertension Prevalence and Control: A Systematic Analysis of Population-Based Studies From 90 Countries. Circulation 2016;134:441-50. [Crossref] [PubMed]
- Wang Z, Chen Z, Zhang L, et al. Status of Hypertension in China: Results From the China Hypertension Survey, 2012-2015. Circulation 2018;137:2344-56. [Crossref] [PubMed]
- Lu J, Lu Y, Wang X, et al. Prevalence, awareness, treatment, and control of hypertension in China: data from 1.7 million adults in a population-based screening study (China PEACE Million Persons Project). Lancet 2017;390:2549-58. [Crossref] [PubMed]
- Lawes CM, Vander Hoorn S, Rodgers A. Global burden of blood-pressure-related disease, 2001. Lancet 2008;371:1513-8. [Crossref] [PubMed]
- Ng FL, Warren HR, Caulfield MJ. Hypertension genomics and cardiovascular prevention. Ann Transl Med 2018;6:291. [Crossref] [PubMed]
- Johnson JA. Advancing management of hypertension through pharmacogenomics. Ann Med 2012;44 Suppl 1:S17-22. [Crossref] [PubMed]
- Prudente S, Sesti G, Pandolfi A, et al. The mammalian tribbles homolog TRIB3, glucose homeostasis, and cardiovascular diseases. Endocr Rev 2012;33:526-46. [Crossref] [PubMed]
- Zhou J, He F, Sun B, et al. Polytropic Influence of TRIB3 rs2295490 Genetic Polymorphism on Response to Antihypertensive Agents in Patients With Essential Hypertension. Front Pharmacol 2019;10:236. [Crossref] [PubMed]
- He F, Shu Y, Wang X, et al. Intensive Glucose Control Reduces the Risk Effect of TRIB3, SMARCD3, and ATF6 Genetic Variation on Diabetic Vascular Complications. Front Pharmacol 2018;9:1422. [Crossref] [PubMed]
- He F, Liu M, Chen Z, et al. Assessment of Human Tribbles Homolog 3 Genetic Variation (rs2295490) Effects on Type 2 Diabetes Patients with Glucose Control and Blood Pressure Lowering Treatment. EBioMedicine 2016;13:181-9. [Crossref] [PubMed]
- Mancia G, Fagard R, Narkiewicz K, et al. 2013 ESH/ESC guidelines for the management of arterial hypertension: the Task Force for the Management of Arterial Hypertension of the European Society of Hypertension (ESH) and of the European Society of Cardiology (ESC). Eur Heart J 2013;34:2159-219. [Crossref] [PubMed]
- Hou L, Chen X, Chen B, et al. Pharmacological therapy and blood pressure control in primary health care sites in China: data from 254,848 hypertensive patients. Clin Epidemiol 2018;10:1467-78. [Crossref] [PubMed]
- Liu L, Zhang Y, Liu G, et al. The Felodipine Event Reduction (FEVER) Study: a randomized long-term placebo-controlled trial in Chinese hypertensive patients. J Hypertens 2005;23:2157-72. [Crossref] [PubMed]
- Andreozzi F, Formoso G, Prudente S, et al. TRIB3 R84 variant is associated with impaired insulin-mediated nitric oxide production in human endothelial cells. Arterioscler Thromb Vasc Biol 2008;28:1355-60. [Crossref] [PubMed]
- Prudente S, Trischitta V.. The TRIB3 Q84R polymorphism, insulin resistance and related metabolic alterations. Biochem Soc Trans 2015;43:1108-11. [Crossref] [PubMed]
- Eyers PA, Keeshan K, Kannan N. Tribbles in the 21st Century: The Evolving Roles of Tribbles Pseudokinases in Biology and Disease. Trends Cell Biol 2017;27:284-98. [Crossref] [PubMed]
- Ding Y, Vaziri ND. Calcium channel blockade enhances nitric oxide synthase expression by cultured endothelial cells. Hypertension 1998;32:718-23. [Crossref] [PubMed]
- Braeuning A.. The connection of beta-catenin and phenobarbital in murine hepatocarcinogenesis: a critical discussion of Awuah et al., PLoS ONE 7(6):e39771, 2012. Arch Toxicol 2013;87:401-2. [Crossref] [PubMed]
- Janega P, Kojsova S, Jendekova L, et al. Indapamide-induced prevention of myocardial fibrosis in spontaneous hypertension rats is not nitric oxide-related. Physiol Res 2007;56:825-8. [PubMed]
- Jiang B, Liu Y, Liu Y, et al. Association of four insulin resistance genes with type 2 diabetes mellitus and hypertension in the Chinese Han population. Mol Biol Rep 2014;41:925-33. [Crossref] [PubMed]
- 2013 Practice guidelines for the management of arterial hypertension of the European Society of Hypertension (ESH) and the European Society of Cardiology (ESC): ESH/ESC Task Force for the Management of Arterial Hypertension. J Hypertens 2013;31:1925-38. [Crossref] [PubMed]
- Turnbull F, Neal B, Pfeffer M, et al. Blood pressure-dependent and independent effects of agents that inhibit the renin-angiotensin system. J Hypertens 2007;25:951-8. [Crossref] [PubMed]
- Ettehad D, Emdin CA, Kiran A, et al. Blood pressure lowering for prevention of cardiovascular disease and death: a systematic review and meta-analysis. Lancet 2016;387:957-67. [Crossref] [PubMed]
- Poole-Wilson PA, Lubsen J, Kirwan BA, et al. Effect of long-acting nifedipine on mortality and cardiovascular morbidity in patients with stable angina requiring treatment (ACTION trial): randomised controlled trial. Lancet 2004;364:849-57. [Crossref] [PubMed]
- Lubsen J, Wagener G, Kirwan BA, et al. Effect of long-acting nifedipine on mortality and cardiovascular morbidity in patients with symptomatic stable angina and hypertension: the ACTION trial. J Hypertens 2005;23:641-8. [Crossref] [PubMed]
- Liu G, Jin S, Hu Y, et al. Disease status affects the association between rs4813620 and the expression of Alzheimer’s disease susceptibility gene TRIB3. Proc Natl Acad Sci U S A 2018;115:E10519-20. [Crossref] [PubMed]



