microRNAs and cancer metabolism reprogramming: the paradigm of metformin
Introduction
The anti-hyperglycemic agent, metformin, is a synthetic dimethylbiguanide compound designed to reduce the side effects of Galega officinalis (the French lilac), rich in guanidine, which was usually prescribed to treat symptoms of diabetes up until the early 1930s in France (1).
Today, metformin is a world wide prescribed agent for treating patients with type II diabetes (T2D) due to its ability to suppress hepatic gluconeogenesis and reduce blood glucose levels (2). Metformin is also prescribed for treating some metabolic disorders such as insulin resistance and for women affected by polycystic ovary syndrome (PCOS), one of the most common female endocrine disorders.
Although the molecular mechanisms underlying metformin action remains a topic of much debate, the activation of the energy sensing enzyme AMP-activated protein kinase (AMPK), a key enzyme of energy homeostasis, has emerged as playing a crucial role in this process. Indeed, metfomin-mediated AMPK activation leads to the modulation of targets that restore energy homeostasis by enhancing glucose uptake into the skeletal muscle (3) and by inhibiting hepatic gluconeogenesis (4).
Interestingly, diabetic patients treated with metformin had a significantly reduced risk of cancer compared to other patients treated with other hypoglycemic therapies (5). Bowker et al. found a positive correlation between metformin assumption and cancer prevention. T2D patients who use metformin have a lower cancer-related mortality incidence compared with insulin users (6).
There is a huge amount of epidemiological evidence relating to new molecular findings on the role of metabolism in cancer development and progression. Cancer cells, in fact, utilize glucose for glycolitic ATP generation and macromolecule synthesis (7), while metformin activity passes through a direct modulation of metabolic homeostasis keepers, such as AMPK. This condition impairs glycolysis and glucose uptake, mimicking starvation, thus promoting apoptosis in tumor cells.
Moreover, in vivo combination therapy using metformin together with different chemotherapeutic agents, such as carboplatin, cisplatin, doxorubicin and paclitaxel, has been shown to increase the citotoxicity (8). Metformin has also shown to enhance radiation response in vivo (9).
Based on molecular and retrospective epidemiological evidence, numerous trials have been designed to test metformin not only as an anti-neoplastic agent in patients with established cancer, but also as a chemopreventive agent in preventing tumor formation (10).
There are several ongoing studies that are attempting to determine the role of metformin as a chemopreventive agent. These studies include phase I, phase II and phase III trials (clinicaltrials.gov.).
Here, we review the anticancer activity of metformin focusing on its anticancer metabolic effects that occur through a direct modulation of metabolic genes and microRNAs (miRNAs). We first describe metformin’s anticancer molecular mechanisms of action and its role in the inhibition of tumor growth and proliferation. We then provide insights into metabolic reprogramming of cancer cells mediated by metformin through the modulation of non-coding RNAs, emphasizing the crucial role of miRNA-33a in the metabolism of breast cancer cells.
Metformin anticancer mechanisms of action
Although the underlying mechanisms of metformin action are still being elucidated, several studies highlight its influence on the cellular energy balance.
Metformin diffuses into the cells through the organic cation transporters (OCTs) (11). OCT1 null mice show a reduction in hepatic metformin uptake, which is the main target of this drug (12). Once in the citosol, metformim induces an increase in the cellular AMP:ATP ratio inhibiting complex I of mitochondrial respiratory chain (13,14). Consequently, ATP depletion is checked by the AMPK γ subunit that binds specifically to AMP. This causes conformational changes that allosterically activate the enzyme and inhibit dephosphorylation on Thr-172 within the activation loop of the catalytic α subunit (15,16). A subsequent phosphorylation of the catalytic α subunit on thr-172 residue is required to activate AMPK (17).
Metformin requires liver kinase B1 (LKB1), a tumor suppressor gene, to phosphorylate the α subunit of AMPK at Thr-172 and to activate the enzyme (17). This contributes to maintain plasma glucose and insulin homeostasis.
Once activated, AMPK regulates not only a pool of substrates which represents the key enzymes in the catabolic pathways and inhibits ATP-consuming anabolic pathways, but also enzymes involved in cell cycle and protein metabolism.
As described by Hardie et al., LKB1 and AMPK pathway functions act as a cellular energy-sensing checkpoint that controls cell growth and proliferation. This depends also from the availability of fuel supplies (18).
However, there is evidence suggesting that metformin acts via an AMPK-independent way of action. Kalender et al. showed that metformin can enhance glucose uptake in rat skeletal muscle cells and inhibits mTOR signaling independent of AMPK (19). Foretz et al. found that metformin inhibited liver glucose synthesis independent of LKB1 or AMPK status (20).
Interestingly, the ataxia telangiectasia mutated (ATM) gene, a tumor suppressor gene involved in DNA repair and cell cycle control, was discovered to activate AMPK through LKB1 dependent and independent pathways (21,22).
Both AMPK-dependent and independent pathways have been proposed to mediate the anticancer effects of metformin treatment. In the following paragraphs we will discuss both metformin’s mechanisms of action.
AMPK-dependent mechanisms of action
As mentioned above, AMPK activation induces the inhibition of anabolic processes, consequently activating also catabolic processes in order to restore energy homeostasis.
Two canonical anabolic processes such as lipogenesis and cholesterol synthesis are inhibited by metformin through AMPK activation (23,24) (Figure 1). Indeed, tumors such as breast, colon, prostate and ovarian cancer are characterized by a high rate of lipid metabolism. High expression of fatty acid synthase (FAS) is a predictor of recurrence in stage I breast carcinoma (25), lung carcinoma (26), endometrial carcinoma patients (27). This is correlated with a worse prognosis in breast carcinomas (28) and ovarian carcinomas (29). Recently, Cantoria et al., have shown that metformin induced growth arrest of pancreatic tumor cells through a direct inhibition of fatty acid synthesis (30).
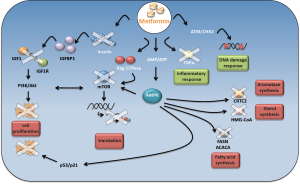
Not only lipid synthesis but also cholesterol synthesis plays a pivotal role in cancer cell metabolism. Based on this set of evidence, metformin induces a shift in lipid and cholesterol metabolism that could deprive pre-malignant and malignant cells of several substrates important for their growth and proliferation.
Indeed, activated AMPK impairs FAS and 3-hydroxy-3-methyl-glutaryl-CoA (HMG-CoA) reductase, thus, reducing the consumption of ATP. As a result, macromolecules are oxidated to produce new ATP molecules, in order to restore the ATP levels reduced by the metformin action.
Several evidences indicate that metformin impinges on other pathways. One of these is the phosphatidylinositiol 3-kinase (PI3K)/AKT/mammalian target of rapamycin (mTOR) signaling pathway. It is a conserved signaling pathway that tunes cell growth, survival and metabolism. Growth factors and nutrients stimuli impinge this pathway to sustain cell growth and proliferation [85]. Unfortunately, a series of aberrations of this signaling axis promotes a number of malignant and non-malignant diseases. Indeed, the PI3K/Akt/mTOR pathways are often deregulated in a large number of tumors and represent an emerging strategy for tumor treatment. Nowadays, anti mTOR therapies are an alternative strategy for renal cell carcinomas and breast cancer treatment.
Oncogenic activity of mTOR passes, largely, through the constitutive activation of the protein synthesis pathway. In fact, mTOR directly activates two important targets involved in this process, such as S6 kinase (S6K) and translation initiation factor 4E binding protein 1 (4EBP1) (31). Interestingly, mTORC1, one of the two functional complexes on mTOR, is negatively regulated by AMPK (Figure 1). AMPK, in fact, directly phosphorylates and inhibits mTORC1 binding partner Raptor and, indirectly, modulates the activity of tuberous sclerosis complex 2 (TSC2), which together with TSC1 form a tumor suppressor complex that inhibits mTOR (32-34).
Metformin has been reported to inhibit tumor progression of renal cell carcinoma (35), proliferation of bladder cancer cells in vitro and in vivo (36) and skin tumor promotion in overweight and obese mice (37) by inhibiting mTOR activity through AMPK modulation. In addition, osteosarcoma cell lines treated with metformin exhibited a significant cell growth reduction in comparison to untreated cells. This effect appeared to be correlated with an increased expression of AMPK and the consequence inhibition of mTOR downstream targets (38).
The metabolic reprogramming induced by metformin passes, also, through p53 activation. Metformin selectively impairs p53-deficient tumor cell growth unlike those carrying wild type p53 proteins are no longer able to reprogram their metabolism and become apoptotic (39) (Figure 1).
Moreover, activation of AMPK stimulates cell cycle arrest through the p53/p21 axis (40). Indeed, AMPK phosphorylates p53 on serine 15 (40). This phosphorylation is not sufficient for p53 activation (41,42), however, mutation of p53 on serine 18 impairs the ability of AMPK to induce cell cycle arrest (40). He and colleagues demonstrated that AMPK-mediated phosphorylation of human MDMX on Ser342 leads to an enhanced association between MDMX and 14-3-3. This event inhibits p53 ubiquitylation and markedly stabilizes and activates p53 (43).
Recently, metformin was reported to inhibit melanoma cell line proliferation through the activation ofAMPK/p53 signaling (44).
The oncogene c-myc plays an important role during the premalignant and malignant stages of tumor growth, which makes it an ideal prevention and therapeutic target. Akinyeke et al. showed that c-myc protein levels were reduced by metformin treatment in prostate cancer cell lines and in prostate cancer mouse models. They addressed to the AMPK pathway, the down regulation of c-myc (45); however, we reported that the metformin-mediated-c-myc inhibition is a consequence of the up regulation of miR-33a in breast cancer cell lines (46).
Interestingly, metformin-induced activation of AMPK inhibited angiogenesis through the VEGF-dependent activation of ERK1/2. The inhibition of AMPK activity, in fact, abrogated this event (47) (Figure 1).
Metformin impaired aromatase expression in primary human breast adipose stromal cells. This occurred via AMPK and resulted in the inhibition of CREB regulated transcription co-activator 2 (CRTC2) (48) (Figure 1). Altogether these findings might suggest that, metformin could be used in the treatment for hormone-dependent post-menopausal breast carcinomas.
AMPK-independent mechanisms of action
Although, AMPK represents a key target in the metformin anticancer mechanism of action, there is evidence showing that metformin exert its action, also, in an AMPK-independent way. Metformin has been shown to inhibit the PI3K/Akt/mTOR signaling pathway in an AMPK independent way. Metformin modulated the activity of mTOR, in lung tissues, by decreasing activation of insulin like growth factor receptor 1 (IGFR1) and Akt (49). Other evidence highlighted the role of Rag GTPase in modulating mTOR activity as a consequence of metformin treatment (19) (Figure 1).
Metformin has been reported to exert anti-proliferative activity on glioblastoma cells through inhibiting the Akt pathway (50) or by inhibiting mTOR via an increase in PRAS40-RAPTOR binding efficiency, an association known to increase during stressful conditions (51).
Hyperandrogenemia is promoted by an aberrant synergistic action between luteinizing hormone (LH) and estrogen produced by granulose cells under insulin like growth factor 1 (IGF1) stimulation (52). The increase of insulin like growth factor binding protein 1 (IGFBP1), mediated by metformin, has been reported to limit the binding between IGF1 and their receptors (IGF1R) and consequently also the production of androgen (Figure 1). Subsequently, metformin treatment may synergize with common anti-androgen therapies, which are usually prescribed for metastatic prostate cancer treatment (53). Moreover, high levels of IGF-1 impaired chemotherapy-induced apoptosis by activating the PI3K/Akt pathway. Inhibition of IGF1R sensitized small cell lung cancer cell lines to the cytotoxic effects of etoposide and carboplatin combined treatment (54).
Quinn et al. found that metformin decreased lung tumorigenesis in liver IGF-I-deficient (LID) mice. The authors suggested that metformin directly inhibits circulating growth factors and local receptor tyrosine kinase (RTK) signaling in an AMPK/IGF-I independent way (55).
Metformin was, also, found to reduce chronic inflammatory response by inhibiting the tumor necrosis factor alpha (TNFα) production in human monocytes (56). In addition, it was demonstrated that metformin blocks the production of endogenous reactive oxygen species (ROS) by interfering the mitochondrial complex I activity (57) (Figure 1). Metformin modulates the activity of checkpoint homolog kinase 2 (CHK2) which results in an increased sensitivity of cancer cells against DNA damage (58,59) (Figure 1). Recently, Do and colleagues proposed the Raf-ERK-Nrf2 axis and the subsequent down regulation of heme oxygenase-1 to explain a new AMPK-independent metformin anticancer mechanism of action (60).
Ma et al. demonstrated that cancer cell response to metformin was influenced by the K-Ras status (61). In fact, they showed that metformin induced apoptosis in the K-Ras mutant tumors, derived from A549 and PANC 1, but not in the K-Ras wild type tumor, derived from A431 cells (61).
MicroRNA, metabolism and cancer
Over the last few years several evidences have shown that metformin can exert its anticancer effects through miRNAs modulation. miRNAs are a family of small non-coding RNAs of about 20-25 nucleotides in length, evolutionarily conserved across species. They regulate gene expression at the post-transcriptional level. Many miRNAs (~50%) are in close proximity to other miRNAs, suggesting that they are transcribed as clusters from a single polycistronic unit (TU) (62-65). MiRNAs can be generated from both non-coding TUs and protein-coding TUs. In particular, ~40% of miRNA loci are present in the intronic region of non-coding transcripts, whereas ~10% are placed in the exonic region of non-coding TUs. miRNAs in protein-coding TUs are usually found in intronic regions (~40%) but a minor fraction of them (~10%) can also be present in exonic/intronic regions, depending on the alternative splicing patterns (66).
RNA Polymerase II is the enzyme that transcribes miRNA genes in a 170 bp primary transcript (pri-miRNA), containing both a 5’cap and a poly (A) tail, folding into a hairpin-shaped structure (67). The first step of miRNA maturation is a cleavage at the stem of the hairpin performed by the “Microprocessor complex”. This large nuclear complex contains two proteins, Drosha and DGCR8, conserved only in animals. These two proteins recognize and crop the stem structure releasing a small hairpin (~70 nt) termed “pre-miRNA” (68). After nuclear cropping, pre-miRNAs are exported to the cytosol by exportin 5, a member of the nuclear transport receptor family (69,70). In the cytoplasm, the hairpin precursors are cleaved by Dicer, a highly conserved RNAse III endonuclease, thus releasing ~22 nt miRNA duplexes (71,72). One of the two strands, the mature miRNA, is loaded onto an Ago protein to generate the effector complex of the miRNA pathway, the RISC (or miRISC). Once in the miRISC, partial pairing between a miRNA and the 3'UTR of target mRNA occurs, resulting in repression of protein translation or in case of complete pairing, in mRNA degradation (73).
MiRNAs are key regulators of many biological processes, such as cell proliferation, differentiation, apoptosis, stress response and angiogenesis due to their ability to bind 3'UTR of multiple target mRNAs. This is why any deregulation of miRNAs expression can contribute to diverse human pathologies, including cancer (74). Indeed, miRNAs behave either as oncogenes or tumor suppressor genes thereby promoting or inhibiting cancer progression.
Growing evidences have highlighted the role of miRNAs as master regulators of metabolic processes, such as lipid and cholesterol-synthesis. As mentioned before, perturbation of these processes represents an important step in tumor development, but also a strategic opportunity to block the activity of specific miRNAs by using synthetic antagomirs. MiRNA-122 family are the most abundant miRNAs in the liver and modulate cholesterol and lipid metabolism. Interestingly, the antisense targeting of miR-122 resulted in a decrease of plasma cholesterol levels in mouse models (75,76). This occurs through the indirect modulation of genes involved in cholesterol biosynthesis, such as 3-hydroxy-3-methylglutaryl-CoA synthase 1 (Hmgcs1), 3-hydroxy-3-methylglutaryl-CoA reductase (Hmgcr) and 7-dehydrocholesterol reductase (Dhcr7) (76).
Similarly, the miR-33 family represents a potential target for the treatment of metabolic disorders. MiR-33a and miR-33b are intronic miRNAs that are encoded together with their host genes, the sterol-regulatory element-binding protein 1 (Srebp1) and 2 (Srebp2). The Srebp network which includes two genes and two intragenic miRNAs act synergistically to regulate cholesterol, triglyceride and lipid homeostasis.
While Srebp1 and miR-33b regulate lipid homeostasis and insulin signaling by modulating the activity of key genes involved in these processes, Srebp2 and its intronic miRNA, miR-33a, regulate cholesterol homeostasis by modulating transcriptionally and post-transcriptionally the activity of genes involved in the cellular cholesterol export, such as the ATP-binding cassette (ABC) transporter ABCA1 and ABCG1 (77-79). However, there is no specific separation of function made between miR-33a and miR-33b. The two miRNAs share overlapping gene targets through their similar mature sequence that differs only for two nucleotides.
Inhibition of ABCA1 by miR-33 induced an efflux of cholesterol from peripheral tissues to the liver and a consequent reduction of circulating high-density lipoprotein-cholesterol (HDL-C) (78). In addition, this condition induced fatty acid degradation in human liver cells, through the lack of modulation in the expression of genes involved in the oxidation of fatty acid (77,80). MiR-33 is also involved in the post-transcriptional modulation of other mRNAs that modulate lipid and glucose metabolism, such as the α1 subunit of AMP-activated protein kinase (AMPKα1) (77,81,82).
Interestingly, miR33a has been found to interact with the 3'-utr region of c-Myc mRNA in breast cancer cell lines (46). c-Myc is an important oncogene deregulated in several tumors. In particular, it is able to directly enhance the glycolytic pathway modulating the expression of lactate dehydrogenase A (LDHA), the enzyme that converts pyruvate to lactate (83), and promote mitochondrial biogenesis, hence increasing mitochondrial function (83,84). These aspects are important in generating substrates for macromolecular biosynthesis that guarantee rapid cell proliferation, which is a typical hallmark of cancer cells.
The anticancer metabolic effects induced by metformin in breast cancer cell lines were, also, a direct consequence of the c-Myc inhibition mediated by the up regulation of miR-33a. Metformin was no longer able to induce metabolic changes in breast cancer cell lines over expressing c-Myc protein (46).
All these evidences have drawn researchers’ attention to studying the role that miRNAs play in the development of pathologies. This also highlights novel target therapies that can reverse either aberrant or loss of miRNA function.
MicroRNAs as mediators of the metformin anticancer activity
Growing evidence show that metformin can exert anticancer effects through miRNA modulation. It has recently been shown that metformin inhibited the growth of hepatocellular carcinoma by inducing G1 cell cycle arrest. This occurred through the aberrant modulation of miRNA expression (85). In addition, metformin treatment modulates differently the expression of miRNAs in human pancreatic cancer (86), esophageal squamous carcinoma (87), gastric cancer (88) and prostate cancer (89).
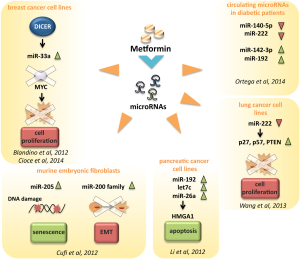
In pancreatic cancer cell lines, metformin up-regulated the expression of miR-26a, miR-192 and let-7c (86). Over expression of miR-26a inhibited cell proliferation, invasion, migration and increased cell apoptosis through a direct modulation of HMGA1 (86) (Figure 2 and Table 1).
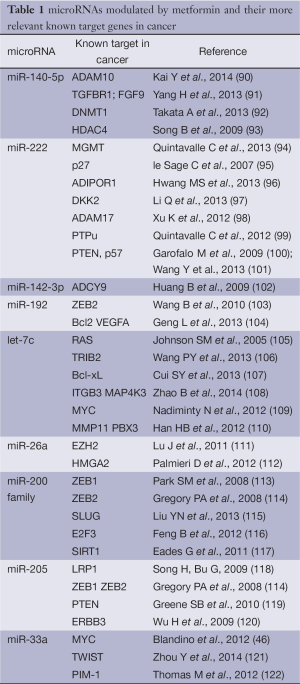
Full table
In A549 and NCI-H358 human lung cancer cell lines metformin reduced the expression of miR-222, thus inhibiting cell growth and cell cycle progression via direct targeting of p27, p57 and PTEN, whose expression was up-regulated in cells exposed to metformin (101) (Figure 2 and Table 1).
Metformin was also shown to significantly increase the number of senescence-prone murine embryonic fibroblasts (MEFs) entering a senescent stage in response to doxorubicin treatment. This might in part occur through metformin-induced modulation of the miRNAs 200 family and miR-205 (123) (Figure 2 and Table 1).
Recent evidences suggest that metformin impacts of the miRNA profile of type 2 diabetes patients.
The analysis of circulating miRNAs in plasma samples collected from type 2 diabetes and healthy individuals revealed a specific miRNAs profile linked to T2D. Interestingly plasma concentrations of four circulating miRNAs, miR-140-5p, miR-222, miR-142-3p and miR-192 were significantly modulated by metformin, but not by placebo (124) (Figure 2 and Table 1). Coleman et al., performed a miRNA profiling of internal mammary artery segments collected from non-diabetic subjects, diabetic subjects treated with metformin (DMMet+), and diabetic subjects not treated with metformin (DMMet–). They found that the DMMet– group exhibited a significant increase in miR-221/222 which led to a decrease in p27 mRNA compared to both the ND and DMMet+ groups. Vascular smooth muscle cells isolated from the internal mammary artery of the DMMet- group showed a high proliferation rate when compared to that of the ND and DMMet+ groups (125).
We reported that metformin modulated miRNA expression through the transcriptional modulation of Dicer in human breast cancer cell lines (Figure 2). Metformin was no longer able to affect tumor engraftment of breast cancer cells whose endogenous expression of DICER was selectively knocked-down (46). Low expression of DICER is correlated with a worst prognosis of breast, lung and ovarian cancer patients (126,127). Altogether these evidences indicate that the transcriptional axis including DICER and miRNAs could be one of molecular mechanisms underlying the anticancer effects exerted by metformin.
Metformin was also shown to affect cancer stem cells (CSCs). It increased the cytotoxicity of many chemotherapeutic agents and prolonged tumor remission through its ability to selectively kill CSCs (8,128). Indeed, metformin sensitized breast cancer xenografts to doxorubicin treatment by targeting CD44+/CD24− CSCs–. This could potentially lead to a reduction in the dose of chemotherapy used and thereby reducing complications due to toxicity (8).
Interestingly, Bao and colleagues reported that pancreatic cell lines treated with metformin re-expressed several miRNAs usually switched off in pancreatic cancer, such as the miR-200 family of miRNA which plays a major role in epithelial to mesenchymal transition and in maintaining the stem cell state (129). They also showed that metformin inhibited tumor sphere formation by deregulating several CSC markers (CD44, EpCAM, EZH2, Notch-1, Nanog, and Oct4) partially through the up regulation of miRNAs (130).
We recently showed that metformin reprograms of ALDH bright cells. These represent a chemoresistant breast cancer cell subpopulation endowing tumor initiating cell properties. We demonstrated that metformin down-regulates the c-MYC levels, possibly by up-regulating the mir-33a levels (131). Altogether these findings depict the close interplay between non-coding factors that exert a profound impact on the coding-derived proteins and reprogramming of cell metabolism.
Conclusions
In the last decades an extraordinary amount of experimental work has been performed to uncover most of the messages contained in the coding genome. The recent advent of non-coding RNAs, of which miRNAs, among the diverse non-coding RNA populations represent that mostly investigated, adds a new layer of complexity toward the fully comprehension of the aberrant molecular mechanism underlying the diverse cancer hallmarks. In line with this, the fine deciphering of the anticancer metabolic effects exerted by metformin, at least partially, through the interplay with miRNAs might provide powerful insights to decode cancer metabolism. The principal aim is still to reprogram cancer metabolism toward that of untransformed cell through the re-wiring of the altered functional nodes. Much more experimental evidence is required to fulfill it, but the ongoing studies hold great promise.
Acknowledgements
We thank MIUR-FIRB (RBAP11LP2W, H81J10000030001) and Italian Health Office (TEVERE project) for generous support. We are grateful to Mrs Tania Merlino for proofreading the manuscript.
Disclosure: The authors declare no conflict of interest.
References
- Parturier G, Hugnot G. Le galega dans le traitement du diabete. Paris: Massons, 1935.
- Hundal RS, Krssak M, Dufour S, et al. Mechanism by which metformin reduces glucose production in type 2 diabetes. Diabetes 2000;49:2063-9. [PubMed]
- Lee JO, Lee SK, Jung JH, et al. Metformin Induces Rab4 Through AMPK and Modulates GLUT4 Translocation in Skeletal Muscle Cells. Journal of Cellular Physiology 2011;226:974-81. [PubMed]
- Stephenne X, Foretz M, Taleux N, et al. Metformin activates AMP-activated protein kinase in primary human hepatocytes by decreasing cellular energy status. Diabetologia 2011;54:3101-10. [PubMed]
- Evans JMM, Donnelly LA, Emslie-Smith AM, et al. Metformin and reduced risk of cancer in diabetic patients. British Medical Journal 2005;330:1304-5. [PubMed]
- Bowker SL, Majumdar SR, Veugelers P, et al. Increased cancer-related mortality for patients with type 2 diabetes who use sulfonylureas or insulin - Response to Farooki and Schneider. Diabetes Care 2006;29:1990-1. [PubMed]
- Menendez JA, Lupu R. Fatty acid synthase and the lipogenic phenotype in cancer pathogenesis. Nature Reviews Cancer 2007;7:763-77. [PubMed]
- Hirsch HA, Iliopoulos D, Tsichlis PN, et al. Metformin selectively targets cancer stem cells, and acts together with chemotherapy to block tumor growth and prolong remission. Cancer Res 2009;69:7507-11. [PubMed]
- Storozhuk Y, Hopmans SN, Sanli T, et al. Metformin inhibits growth and enhances radiation response of non-small cell lung cancer (NSCLC) through ATM and AMPK. British Journal of Cancer 2013;108:2021-32. [PubMed]
- Pulito C, Sanli T, Rana P, et al. Metformin: On Ongoing Journey across Diabetes, Cancer Therapy and Prevention. Metabolites 2013;3:1051-75. [PubMed]
- Viollet B, Guigas B, Sanz Garcia N, et al. Cellular and molecular mechanisms of metformin: an overview. Clin Sci (Lond) 2012;122:253-70. [PubMed]
- Shu Y, Sheardown SA, Brown C, et al. Effect of genetic variation in the organic cation transporter 1 (OCT1) on metformin action. J Clin Invest 2007;117:1422-31. [PubMed]
- Owen MR, Doran E, Halestrap AP. Evidence that metformin exerts its anti-diabetic effects through inhibition of complex 1 of the mitochondrial respiratory chain. Biochem J 2000;348:607-14. [PubMed]
- El-Mir MY, Nogueira V, Fontaine E, et al. Dimethylbiguanide inhibits cell respiration via an indirect effect targeted on the respiratory chain complex I. J Biol Chem 2000;275:223-8. [PubMed]
- Sanders MJ, Grondin PO, Hegarty BD, et al. Investigating the mechanism for AMP activation of the AMP-activated protein kinase cascade. Biochem J 2007;403:139-48. [PubMed]
- Xiao B, Heath R, Saiu P, et al. Structural basis for AMP binding to mammalian AMP-activated protein kinase. Nature 2007;449:496-500. [PubMed]
- Shaw RJ, Lamia KA, Vasquez D, et al. The kinase LKB1 mediates glucose homeostasis in liver and therapeutic effects of metformin. Science 2005;310:1642-6. [PubMed]
- Hardie DG. The LKB1-AMPK Pathway-Friend or Foe in Cancer? Cancer Cell 2013;23:131-2. [PubMed]
- Kalender A, Selvaraj A, Kim SY, et al. Metformin, independent of AMPK, inhibits mTORC1 in a rag GTPase-dependent manner. Cell Metab 2010;11:390-401. [PubMed]
- Foretz M, Hebrard S, Leclerc J, et al. Metformin inhibits hepatic gluconeogenesis in mice independently of the LKB1/AMPK pathway via a decrease in hepatic energy state. J Clin Invest 2010;120:2355-69. [PubMed]
- Sun Y, Connors KE, Yang DQ. AICAR induces phosphorylation of AMPK in an ATM-dependent, LKB1-independent manner. Mol Cell Biochem 2007;306:239-45. [PubMed]
- Suzuki A, Kusakai G, Kishimoto A, et al. IGF-1 phosphorylates AMPK-alpha subunit in ATM-dependent and LKB1-independent manner. Biochem Biophys Res Commun 2004;324:986-92. [PubMed]
- Algire C, Amrein L, Bazile M, et al. Diet and tumor LKB1 expression interact to determine sensitivity to anti-neoplastic effects of metformin in vivo. Oncogene 2011;30:1174-82. [PubMed]
- Brunet J, Vazquez-Martin A, Colomer R, et al. BRCA1 and acetyl-CoA carboxylase: the metabolic syndrome of breast cancer. Mol Carcinog 2008;47:157-63. [PubMed]
- Alo’ PL, Visca P, Marci A, et al. Expression of fatty acid synthase (FAS) as a predictor of recurrence in stage I breast carcinoma patients. Cancer 1996;77:474-82. [PubMed]
- Visca P, Sebastiani V, Botti C, et al. Fatty acid synthase (FAS) is a marker of increased risk of recurrence in lung carcinoma. Anticancer Res 2004;24:4169-73. [PubMed]
- Sebastiani V, Visca P, Botti C, et al. Fatty acid synthase is a marker of increased risk of recurrence in endometrial carcinoma. Gynecol Oncol 2004;92:101-5. [PubMed]
- Alò PL, Visca P, Trombetta G, et al. Fatty acid synthase (FAS) predictive strength in poorly differentiated early breast carcinomas. Tumori 1999;85:35-40. [PubMed]
- Gansler TS, Hardman W 3rd, Hunt DA, et al. Increased expression of fatty acid synthase (OA-519) in ovarian neoplasms predicts shorter survival. Hum Pathol 1997;28:686-92. [PubMed]
- Cantoria MJ, Boros LG, Meuillet EJ. Contextual inhibition of fatty acid synthesis by metformin involves glucose-derived acetyl-CoA and cholesterol in pancreatic tumor cells. Metabolomics 2014;10:91-104. [PubMed]
- Dowling RJ, Zakikhani M, Fantus IG, et al. Metformin inhibits mammalian target of rapamycin-dependent translation initiation in breast cancer cells. Cancer Res 2007;67:10804-12. [PubMed]
- Gwinn DM, Shackelford DB, Egan DF, et al. AMPK phosphorylation of raptor mediates a metabolic checkpoint. Mol Cell 2008;30:214-26. [PubMed]
- Inoki K, Corradetti MN, Guan KL. Dysregulation of the TSC-mTOR pathway in human disease. Nat Genet 2005;37:19-24. [PubMed]
- Inoki K, Zhu T, Guan KL. TSC2 mediates cellular energy response to control cell growth and survival. Cell 2003;115:577-90. [PubMed]
- Woodard J, Joshi S, Viollet B, et al. AMPK as a therapeutic target in renal cell carcinoma. Cancer Biol Ther 2010;10:1168-77. [PubMed]
- Zhang T, Guo P, Zhang Y, et al. The antidiabetic drug metformin inhibits the proliferation of bladder cancer cells in vitro and in vivo. Int J Mol Sci 2013;14:24603-18. [PubMed]
- Checkley LA, Rho O, Angel JM, et al. Metformin inhibits skin tumor promotion in overweight and obese mice. Cancer Prev Res (Phila) 2014;7:54-64. [PubMed]
- Quattrini I, Conti A, Pazzaglia L, et al. Metformin inhibits growth and sensitizes osteosarcoma cell lines to cisplatin through cell cycle modulation. Oncol Rep 2014;31:370-5. [PubMed]
- Buzzai M, Jones RG, Amaravadi RK, et al. Systemic treatment with the antidiabetic drug metformin selectively impairs p53-deficient tumor cell growth. Cancer Res 2007;67:6745-52. [PubMed]
- Jones RG, Plas DR, Kubek S, et al. AMP-activated protein kinase induces a p53-dependent metabolic checkpoint. Mol Cell 2005;18:283-93. [PubMed]
- Ashcroft M, Kubbutat MH, Vousden KH. Regulation of p53 function and stability by phosphorylation. Mol Cell Biol 1999;19:1751-8. [PubMed]
- Chao C, Saito S, Anderson CW, et al. Phosphorylation of murine p53 at ser-18 regulates the p53 responses to DNA damage. Proc Natl Acad Sci U S A 2000;97:11936-41. [PubMed]
- He G, Zhang YW, Lee JH, et al. AMP-activated protein kinase induces p53 by phosphorylating MDMX and inhibiting its activity. Mol Cell Biol 2014;34:148-57. [PubMed]
- Cerezo M, Tichet M, Abbe P, et al. Metformin Blocks Melanoma Invasion and Metastasis Development in AMPK/p53-Dependent Manner. Mol Cancer Ther 2013;12:1605-15. [PubMed]
- Akinyeke T, Matsumura S, Wang X, et al. Metformin targets c-MYC oncogene to prevent prostate cancer. Carcinogenesis 2013;34:2823-32. [PubMed]
- Blandino G, Valerio M, Cioce M, et al. Metformin elicits anticancer effects through the sequential modulation of DICER and c-MYC. Nat Commun 2012;3:865. [PubMed]
- Dallaglio K, Bruno A, Cantelmo AR, et al. Paradoxic effects of metformin on endothelial cells and angiogenesis. Carcinogenesis 2014;35:1055-66. [PubMed]
- Samarajeewa NU, Ham S, Yang F, et al. Promoter-specific effects of metformin on aromatase transcript expression. Steroids 2011;76:768-71. [PubMed]
- Memmott RM, Mercado JR, Maier CR, et al. Metformin prevents tobacco carcinogen--induced lung tumorigenesis. Cancer Prev Res (Phila) 2010;3:1066-76. [PubMed]
- Würth R, Pattarozzi A, Gatti M, et al. Metformin selectively affects human glioblastoma tumor-initiating cell viability: A role for metformin-induced inhibition of Akt. Cell Cycle 2013;12:145-56. [PubMed]
- Liu X, Chhipa RR, Pooya S, et al. Discrete mechanisms of mTOR and cell cycle regulation by AMPK agonists independent of AMPK. Proc Natl Acad Sci U S A 2014;111:E435-44. [PubMed]
- Adashi EY. The IGF family and folliculogenesis. J Reprod Immunol 1998;39:13-9. [PubMed]
- Smith MR, Lee H, Nathan DM. Insulin sensitivity during combined androgen blockade for prostate cancer. J Clin Endocrinol Metab 2006;91:1305-8. [PubMed]
- Warshamana-Greene GS, Litz J, Buchdunger E, et al. The insulin-like growth factor-I receptor kinase inhibitor, NVP-ADW742, sensitizes small cell lung cancer cell lines to the effects of chemotherapy. Clin Cancer Res 2005;11:1563-71. [PubMed]
- Quinn BJ, Dallos M, Kitagawa H, et al. Inhibition of lung tumorigenesis by metformin is associated with decreased plasma igf-I and diminished receptor tyrosine kinase signaling. Cancer Prev Res (Phila) 2013;6:801-10. [PubMed]
- Arai M, Uchiba M, Komura H, et al. Metformin, an antidiabetic agent, suppresses the production of tumor necrosis factor and tissue factor by inhibiting early growth response factor-1 expression in human monocytes in vitro. J Pharmacol Exp Ther 2010;334:206-13. [PubMed]
- Algire C, Moiseeva O, Deschenes-Simard X, et al. Metformin reduces endogenous reactive oxygen species and associated DNA damage. Cancer Prev Res (Phila) 2012;5:536-43. [PubMed]
- Menendez JA, Cufi S, Oliveras-Ferraros C, et al. Metformin and the ATM DNA damage response (DDR): accelerating the onset of stress-induced senescence to boost protection against cancer. Aging (Albany NY) 2011;3:1063-77. [PubMed]
- Vazquez-Martin A, Oliveras-Ferraros C, Cufi S, et al. Metformin activates an ataxia telangiectasia mutated (ATM)/Chk2-regulated DNA damage-like response. Cell Cycle 2011;10:1499-501. [PubMed]
- Do MT, Kim HG, Khanal T, et al. Metformin inhibits heme oxygenase-1 expression in cancer cells through inactivation of Raf-ERK-Nrf2 signaling and AMPK-independent pathways. Toxicol Appl Pharmacol 2013;271:229-38. [PubMed]
- Ma Y, Guo FC, Wang W, et al. K-ras gene mutation as a predictor of cancer cell responsiveness to metformin. Mol Med Rep 2013;8:763-8. [PubMed]
- Bartel DP. MicroRNAs: Genomics, biogenesis, mechanism, and function. Cell 2004;116:281-97. [PubMed]
- Lagos-Quintana M, Rauhut R, Lendeckel W, et al. Identification of novel genes coding for small expressed RNAs. Science 2001;294:853-8. [PubMed]
- Lagos-Quintana M, Rauhut R, Meyer J, et al. New microRNAs from mouse and human. RNA 2003;9:175-9. [PubMed]
- Lau NC, Lim LP, Weinstein EG, et al. An abundant class of tiny RNAs with probable regulatory roles in Caenorhabditis elegans. Science 2001;294:858-62. [PubMed]
- Kim VN, Han J, Siomi MC. Biogenesis of small RNAs in animals. Nature Reviews Molecular Cell Biology 2009;10:126-39. [PubMed]
- Lee Y, Kim M, Han JJ, et al. MicroRNA genes are transcribed by RNA polymerase II. Embo Journal 2004;23:4051-60. [PubMed]
- Han J, Lee Y, Yeom KH, et al. Molecular basis for the recognition of primary microRNAs by the Drosha-DGCR8 complex. Cell 2006;125:887-901. [PubMed]
- Yi R, Qin Y, Macara IG, et al. Exportin-5 mediates the nuclear export of pre-microRNAs and short hairpin RNAs. Genes & Development 2003;17:3011-6. [PubMed]
- Bohnsack MT, Czaplinski K, Gorlich D. Exportin 5 is a RanGTP-dependent dsRNA-binding protein that mediates nuclear export of pre-miRNAs. RNA 2004;10:185-91. [PubMed]
- Chendrimada TP, Gregory RI, Kumaraswamy E, et al. TRBP recruits the Dicer complex to Ago2 for microRNA processing and gene silencing. Nature 2005;436:740-4. [PubMed]
- Gregory RI, Chendrimada TP, Cooch N, et al. Human RISC couples microRNA biogenesis and posttranscriptional gene silencing. Cell 2005;123:631-40. [PubMed]
- Pasquinelli AE. NON-CODING RNA MicroRNAs and their targets: recognition, regulation and an emerging reciprocal relationship. Nature Reviews Genetics 2012;13:271-82. [PubMed]
- Iorio MV, Croce CM. MicroRNA dysregulation in cancer: diagnostics, monitoring and therapeutics. A comprehensive review. Embo Molecular Medicine 2012;4:143-59. [PubMed]
- Esau C, Davis S, Murray SF, et al. miR-122 regulation of lipid metabolism revealed by in vivo antisense targeting. Cell Metab 2006;3:87-98. [PubMed]
- Krützfeldt J, Rajewsky N, Braich R, et al. Silencing of microRNAs in vivo with ‘antagomirs’. Nature 2005;438:685-9. [PubMed]
- Dávalos A, Goedeke L, Smibert P, et al. miR-33a/b contribute to the regulation of fatty acid metabolism and insulin signaling. Proc Natl Acad Sci U S A 2011;108:9232-7. [PubMed]
- Marquart TJ, Allen RM, Ory DS, et al. miR-33 links SREBP-2 induction to repression of sterol transporters. Proc Natl Acad Sci U S A 2010;107:12228-32. [PubMed]
- Najafi-Shoushtari SH, Kristo F, Li Y, et al. MicroRNA-33 and the SREBP host genes cooperate to control cholesterol homeostasis. Science 2010;328:1566-9. [PubMed]
- Gerin I, Clerbaux LA, Haumont O, et al. Expression of miR-33 from an SREBP2 intron inhibits cholesterol export and fatty acid oxidation. J Biol Chem 2010;285:33652-61. [PubMed]
- Rayner KJ, Esau CC, Hussain FN, et al. Inhibition of miR-33a/b in non-human primates raises plasma HDL and lowers VLDL triglycerides. Nature 2011;478:404-7. [PubMed]
- Rottiers V, Najafi-Shoushtari SH, Kristo F, et al. MicroRNAs in metabolism and metabolic diseases. Cold Spring Harb Symp Quant Biol 2011;76:225-33. [PubMed]
- Lewis BC, Shim H, Li Q, et al. Identification of putative c-Myc-responsive genes: characterization of rcl, a novel growth-related gene. Mol Cell Biol 1997;17:4967-78. [PubMed]
- Menssen A, Hermeking H.. Characterization of the c-MYC-regulated transcriptome by SAGE: identification and analysis of c-MYC target genes. Proc Natl Acad Sci U S A 2002;99:6274-9. [PubMed]
- Miyoshi H, Kato K, Iwama H, et al. Effect of the anti-diabetic drug metformin in hepatocellular carcinoma in vitro and in vivo. Int J Oncol 2013. [Epub ahead of print]. [PubMed]
- Li W, Yuan Y, Huang L, et al. Metformin alters the expression profiles of microRNAs in human pancreatic cancer cells. Diabetes Res Clin Pract 2012;96:187-95. [PubMed]
- Kobayashi M, Kato K, Iwama H, et al. Antitumor effect of metformin in esophageal cancer: in vitro study. Int J Oncol 2013;42:517-24. [PubMed]
- Kato K, Gong J, Iwama H, et al. The antidiabetic drug metformin inhibits gastric cancer cell proliferation in vitro and in vivo. Mol Cancer Ther 2012;11:549-60. [PubMed]
- Avci CB, Harman E, Dodurga Y, et al. Therapeutic potential of an anti-diabetic drug, metformin: alteration of miRNA expression in prostate cancer cells. Asian Pac J Cancer Prev 2013;14:765-8. [PubMed]
- Kai Y, Peng W, Ling W, et al. Reciprocal effects between microRNA-140-5p and ADAM10 suppress migration and invasion of human tongue cancer cells. Biochem Biophys Res Commun 2014;448:308-14. [PubMed]
- Yang H, Fang F, Chang R, et al. MicroRNA-140-5p suppresses tumor growth and metastasis by targeting transforming growth factor beta receptor 1 and fibroblast growth factor 9 in hepatocellular carcinoma. Hepatology 2013;58:205-17. [PubMed]
- Takata A, Otsuka M, Yoshikawa T, et al. MicroRNA-140 acts as a liver tumor suppressor by controlling NF-kappaB activity by directly targeting DNA methyltransferase 1 (Dnmt1) expression. Hepatology 2013;57:162-70. [PubMed]
- Song B, Wang Y, Xi Y, et al. Mechanism of chemoresistance mediated by miR-140 in human osteosarcoma and colon cancer cells. Oncogene 2009;28:4065-74. [PubMed]
- Quintavalle C, Mangani D, Roscigno G, et al. MiR-221/222 target the DNA methyltransferase MGMT in glioma cells. PLoS One 2013;8:e74466. [PubMed]
- le Sage C, Nagel R, Egan DA, et al. Regulation of the p27(Kip1) tumor suppressor by miR-221 and miR-222 promotes cancer cell proliferation. EMBO J 2007;26:3699-708. [PubMed]
- Hwang MS, Yu N, Stinson SY, et al. miR-221/222 targets adiponectin receptor 1 to promote the epithelial-to-mesenchymal transition in breast cancer. PLoS One 2013;8:e66502. [PubMed]
- Li Q, Shen K, Zhao Y, et al. MicroRNA-222 promotes tumorigenesis via targeting DKK2 and activating the Wnt/beta-catenin signaling pathway. FEBS Lett 2013;587:1742-8. [PubMed]
- Xu K, Liang X, Shen K, et al. MiR-222 modulates multidrug resistance in human colorectal carcinoma by down-regulating ADAM-17. Exp Cell Res 2012;318:2168-77. [PubMed]
- Quintavalle C, Garofalo M, Zanca C, et al. miR-221/222 overexpession in human glioblastoma increases invasiveness by targeting the protein phosphate PTPmu. Oncogene 2012;31:858-68. [PubMed]
- Garofalo M, Di Leva G, Romano G, et al. miR-221&222 Regulate TRAIL Resistance and Enhance Tumorigenicity through PTEN and TIMP3 Downregulation. Cancer Cell 2009;16:498-509. [PubMed]
- Wang Y, Dai W, Chu X, et al. Metformin inhibits lung cancer cells proliferation through repressing microRNA-222. Biotechnol Lett 2013;35:2013-9. [PubMed]
- Huang B, Zhao J, Lei Z, et al. miR-142-3p restricts cAMP production in CD4+CD25- T cells and CD4+CD25+ TREG cells by targeting AC9 mRNA. EMBO Rep 2009;10:180-5. [PubMed]
- Wang B, Herman-Edelstein M, Koh P, et al. E-cadherin expression is regulated by miR-192/215 by a mechanism that is independent of the profibrotic effects of transforming growth factor-beta. Diabetes 2010;59:1794-802. [PubMed]
- Geng L, Chaudhuri A, Talmon G, et al. MicroRNA-192 suppresses liver metastasis of colon cancer. Oncogene 2013. [Epub ahead of print]. [PubMed]
- Johnson SM, Grosshans H, Shingara J, et al. RAS is regulated by the let-7 microRNA family. Cell 2005;120:635-47. [PubMed]
- Wang PY, Sun YX, Zhang S, et al. Let-7c inhibits A549 cell proliferation through oncogenic TRIB2 related factors. FEBS Lett 2013;587:2675-81. [PubMed]
- Cui SY, Huang JY, Chen YT, et al. Let-7c governs the acquisition of chemo- or radioresistance and epithelial-to-mesenchymal transition phenotypes in docetaxel-resistant lung adenocarcinoma. Mol Cancer Res 2013;11:699-713. [PubMed]
- Zhao B, Han H, Chen J, et al. MicroRNA let-7c inhibits migration and invasion of human non-small cell lung cancer by targeting ITGB3 and MAP4K3. Cancer Lett 2014;342:43-51. [PubMed]
- Nadiminty N, Tummala R, Lou W, et al. MicroRNA let-7c suppresses androgen receptor expression and activity via regulation of Myc expression in prostate cancer cells. J Biol Chem 2012;287:1527-37. [PubMed]
- Han HB, Gu J, Zuo HJ, et al. Let-7c functions as a metastasis suppressor by targeting MMP11 and PBX3 in colorectal cancer. J Pathol 2012;226:544-55. [PubMed]
- Lu J, He ML, Wang L, et al. MiR-26a inhibits cell growth and tumorigenesis of nasopharyngeal carcinoma through repression of EZH2. Cancer Res 2011;71:225-33. [PubMed]
- Palmieri D, D’Angelo D, Valentino T, et al. Downregulation of HMGA-targeting microRNAs has a critical role in human pituitary tumorigenesis. Oncogene 2012;31:3857-65. [PubMed]
- Park SM, Gaur AB, Lengyel E, et al. The miR-200 family determines the epithelial phenotype of cancer cells by targeting the E-cadherin repressors ZEB1 and ZEB2. Genes Dev 2008;22:894-907. [PubMed]
- Gregory PA, Bert AG, Paterson EL, et al. The miR-200 family and miR-205 regulate epithelial to mesenchymal transition by targeting ZEB1 and SIP1. Nat Cell Biol 2008;10:593-601. [PubMed]
- Liu YN, Yin JJ, Abou-Kheir W, et al. MiR-1 and miR-200 inhibit EMT via Slug-dependent and tumorigenesis via Slug-independent mechanisms. Oncogene 2013;32:296-306. [PubMed]
- Feng B, Wang R, Song HZ, et al. MicroRNA-200b reverses chemoresistance of docetaxel-resistant human lung adenocarcinoma cells by targeting E2F3. Cancer 2012;118:3365-76. [PubMed]
- Eades G, Yao Y, Yang M, et al. miR-200a regulates SIRT1 expression and epithelial to mesenchymal transition (EMT)-like transformation in mammary epithelial cells. J Biol Chem 2011;286:25992-6002. [PubMed]
- Song H, Bu G.. MicroRNA-205 inhibits tumor cell migration through down-regulating the expression of the LDL receptor-related protein 1. Biochem Biophys Res Commun 2009;388:400-5. [PubMed]
- Greene SB, Herschkowitz JI, Rosen JM. The ups and downs of miR-205: identifying the roles of miR-205 in mammary gland development and breast cancer. RNA Biol 2010;7:300-4. [PubMed]
- Wu H, Zhu S, Mo YY. Suppression of cell growth and invasion by miR-205 in breast cancer. Cell Res 2009;19:439-48. [PubMed]
- Zhou Y, Huang Z, Wu S, et al. miR-33a is up-regulated in chemoresistant osteosarcoma and promotes osteosarcoma cell resistance to cisplatin by down-regulating TWIST. J Exp Clin Cancer Res 2014;33:12. [PubMed]
- Thomas M, Lange-Grunweller K, Weirauch U, et al. The proto-oncogene Pim-1 is a target of miR-33a. Oncogene 2012;31:918-28. [PubMed]
- Cufí S, Vazquez-Martin A, Oliveras-Ferraros C, et al. Metformin lowers the threshold for stress-induced senescence: a role for the microRNA-200 family and miR-205. Cell Cycle 2012;11:1235-46. [PubMed]
- Ortega FJ, Mercader JM, Moreno-Navarrete JM, et al. Profiling of Circulating MicroRNAs Reveals Common MicroRNAs Linked to Type 2 Diabetes That Change With Insulin Sensitization. Diabetes Care 2014.
- Coleman CB, Lightell DJ Jr, Moss SC, et al. Elevation of miR-221 and -222 in the internal mammary arteries of diabetic subjects and normalization with metformin. Mol Cell Endocrinol 2013;374:125-9. [PubMed]
- Karube Y, Tanaka H, Osada H, et al. Reduced expression of Dicer associated with poor prognosis in lung cancer patients. Cancer Sci 2005;96:111-5. [PubMed]
- Merritt WM, Lin YG, Han LY, et al. Dicer, Drosha, and outcomes in patients with ovarian cancer. N Engl J Med 2008;359:2641-50. [PubMed]
- Ning X, Shu J, Du Y, et al. Therapeutic strategies targeting cancer stem cells. Cancer Biol Ther 2013;14:295-303. [PubMed]
- Bao B, Wang Z, Ali S, et al. Metformin inhibits cell proliferation, migration and invasion by attenuating CSC function mediated by deregulating miRNAs in pancreatic cancer cells. Cancer Prev Res (Phila) 2012;5:355-64. [PubMed]
- Wellner U, Schubert J, Burk UC, et al. The EMT-activator ZEB1 promotes tumorigenicity by repressing stemness-inhibiting microRNAs. Nat Cell Biol 2009;11:1487-95. [PubMed]
- Cioce M, Valerio M, Casadei L, et al. Metformin-induced metabolic reprogramming of chemoresistant ALDH bright breast cancer cells. Oncotarget, 2014. Available online: http://www.impactjournals.com/oncotarget/index.php?journal=oncotarget&page=article&op=view&path%5B%5D=1864


