
Differential microRNA expression profiles associated with microsatellite status reveal possible epigenetic regulation of microsatellite instability in gastric adenocarcinoma
Introduction
More than one million patients suffered from gastric carcinoma (GCa) with an estimated 783,000 GCa-related deaths in 2018, making it the fifth most common and the third most deadly cancer worldwide (1). For advanced GCa, the palliative and systemic chemotherapies are the mainstay of treatments, and its median overall survival (OS) is only 10–12 months (2). Recently, the immune checkpoint inhibitors have been used to treat advanced GCa with a high-frequency microsatellite instability (MSI-H) or mismatch repair defects (dMMR) (3,4). The Food and Drug Administration (FDA) has granted an accelerated approval to pembrolizumab for pediatric and adult solid tumor patients with MSI-H or dMMR, and MSI-H has emerged as a key predictive biomarker for immunotherapy in GCa. Therefore, it is critical to identify the mechanism underlying the MSI-H formation in GCa.
Microsatellites are short tandem repeats of DNA, which are widely distributed in the eukaryotic genome, and they are mostly located in the non-coding regions of genes or near the telomere regions of chromosomes, likely caused by defects in mismatch repair (MMR) that plays important roles in maintaining genome stability. The gain or loss of tandem repeats resulting in the alteration of microsatellite length is called microsatellite instability (MSI) (5). It is generally considered that MSI arises from the impairment of MMR machinery and is associated with tumorigenesis (6), while dMMR originates from germline mutations in the MMR genes commonly seen in the Lynch syndrome (7), but the majority of sporadic MSI result from somatic mutational inactivation or epigenetic silencing of the MMR genes (8,9). Previous studies demonstrated that more than half of MSI-positive GCa manifested hypermethylation in the promoter of MLH1, a key member of the MMR genes, while another nearly 40% of MSI-positive GCa originated from unknown genetic or epigenetic alterations (10).
As an integral part of epigenetic regulators, non-coding RNAs, including microRNAs, play irreplaceable roles in RNA degradation and post-transcriptional regulation of gene expression. It has been shown that microRNAs are aberrantly expressed in various types of malignancies, functioning either as oncogenes or tumor suppressor genes (11,12). Therefore, whether microRNAs play a role in epigenetic regulation of MSI-H formation needs further exploration.
A previous study has explored the relationship between the expression of certain microRNAs and MSI-H in colorectal cancer with a small set of 39 samples (13), but the relationship between microRNAs and the MSI status in GCa has not been fully investigated yet. Therefore, additional studies on the relationship between microRNAs and microsatellite status in GCa may help elaborate the molecular mechanism underlying MSI formation and the efficacy of immunotherapy. Such a relationship also likely provides new biological markers for immunotherapy in GCa.
Because The Cancer Genome Atlas (TCGA) database provides a large number of microRNA sequencing dataset of GCa tissue samples (14), we evaluated differential expression of microRNAs in GCa with different microsatellite status by analyzing the available high-throughput microRNA data in the TCGA database. Furthermore, we used additional data from Gene Ontology (GO), Kyoto Encyclopedia of Genes and Genomes pathway enrichment (KEGG) databases to identify the pathways that may be regulated by differentially expressed microRNAs, which may provide possible molecular mechanisms underlying the MSI-H formation.
Methods
Data acquisition
The raw microRNA sequencing data and clinical information were downloaded from the FireBrowse database (http://www.firebrowse.org/). The inclusion criteria of GCa tissue samples were as follows: (I) the samples with pathologically confirmed diagnosis of GCa; (II) the samples with both microRNA sequencing data and clinical information; and (III) the samples with microsatellite status information. As a result, a total of 386 GCa samples were included in the analysis. The relationship between the microsatellite status and clinical features of the samples were assessed by the Chi square test, and P<0.05 was considered statistically significant.
Analysis of differentially expressed microRNAs in GCa tissues by microsatellite status
We processed microRNA expression data by using R language packages (version 3.5.1) and analyzed the differentially expressed microRNAs in GCa tissues with a microsatellite status, i.e., MSI-H, MSI-low (MSI-L) and microsatellite stable (MSS), by the limma package in R. We calculated the fold changes (FC) of the expression levels of individual microRNAs, and the FCs in differentially expressed microRNAs with |FC| >1 and P<0.05 were considered statistically significant. Because MSI-H and MSS GCa had the most differentially expressed microRNAs (Table S1), we thus focused on MSI-H and MSS in the subsequent analyses. To identify more significantly differentially expressed microRNAs, we calculated the expression levels of microRNAs for both MSI-H and MSS GCa with |FC| >2 and P<0.05.

Full table
To distinguish of MSI-H subtypes from MSS using microRNAs expression profiles
We also used microRNA expression data to distinguish MSI-H from MSS subtype by a stepwise logistic regression analysis, and P<0.05 was considered statistically significant. We then constructed the receiver operating characteristic (ROC) curves to illustrate prediction accuracy of the models containing each of the microRNAs, respectively. We also used ROCs from the models that included all of the microRNAs with P<0.05 using the pROC package of R.
Prediction of genes targeted by MSI-H-related microRNAs and mapping of the target signaling pathway genes
We divided the MSI-H-related microRNAs into two groups of either upregulated or downregulated expression levels and compared their MSS. We selected the top six upregulated and three downregulated microRNAs for the two groups, respectively, by using more stringent criteria (P<0.01 and |FC| >2.175). The genes targeted by upregulated and downregulated microRNAs in GCa with MSI-H were predicted, respectively, according to miRanda (http://www.microrna.org/microrna/home.do) online analytic tools, and the putative genes with a short variable region (SVR) score less than −0.5 were included for further analysis. We further explored the signaling pathways and processes of the predicted genes by using the Annotation, Visualization and Integrated Discovery (DAVID) database (v6.8, https://david.ncifcrf.gov/summary.jsp). Finally, we performed GO and KEGG pathway enrichment analyses for the target genes with P<0.05 and gene counts ≥3 sets as the cut-off criteria for the comparisons.
Statistical analysis
The expression levels of microRNAs in GCa tissues were analyzed and compared by the unpaired t-test. The statistical analyses were performed by using the IBM SPSS statistics software program version 20.0 (IBM Corp., NY, USA) and R language (version 3.5.1). P values were two-sided with a significance level of 0.05.
Results
Different clinicopathological traits of GCa with different MSI status
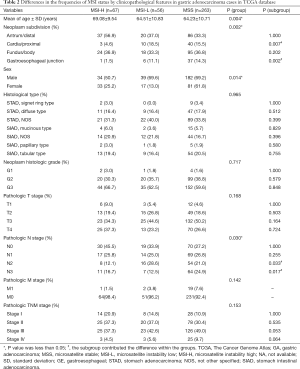
In the present study, we included the data for 386 GCa samples from the TCGA database, and the number of the samples with MSS, MSI-L and MSI-H was 263, 56 and 67, respectively. Their general clinical traits are presented in Table 1. The associations between the MSI status and detailed clinical traits, including age at diagnosis, sex, family history, helicobacter pylori infection, gastric subregion, histologic type, histologic grade and TNM pathological stage are presented in Table 2. We found that the MSI status was significantly associated with age at diagnosis (P=0.049), sex (P=0.014) and gastric subregion (P=0.007). Overall, the proportion of MSI-H positive tumors increased as age increased, while the proportion of MSS tumors decreased as age increased, but no obvious trend was seen for MSI-L tumors. Specifically, 33 of 129 (25.6%) patients with age >70 years had MSI-H positive tumors, 23 of 128 (18.0%) patients with age of 61–70 years had MSI-H positive tumors, and 11 of 129 (8.5%) patients with age ≤61 years had MSI-H positive tumors; female GCa patients (25.2%) were more likely to develop MSI-H tumors than male GCa patients (13.3%); and the MSI-H was more likely found in distal (37.1%) and body (35.5%) GCa than in proximal (13.7%) and junction (11.4%) GCa (Figure 1). No differences were observed for other patients’ traits (Table 2).

Full table
Full table

MicroRNA expression profiles by MSI status
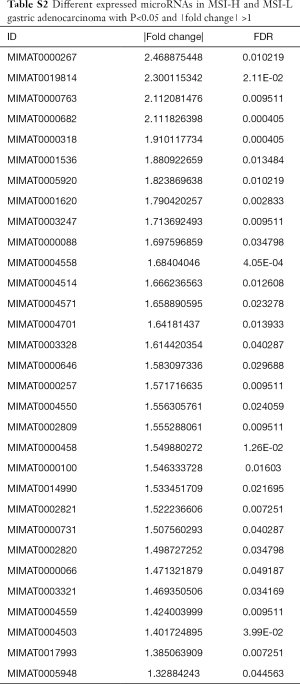
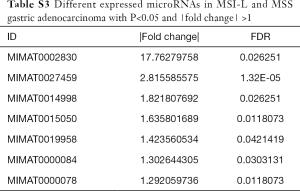
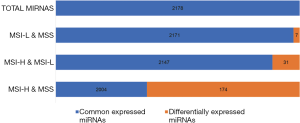
To explore the differences in the frequencies of MSI-H, MSI-L and MSS in the microRNA expression profiles, all the differentially expressed microRNAs (defined as P<0.05 with |FC| >1) among these three groups were assessed and compared with each other (Tables S1-S3). We found that MSI-L and MSS tumors had similar microRNA expression profiles, but MSI-H tumors had the most different expression profiles in comparison with MSS (Figure 2).
Full table
Full table
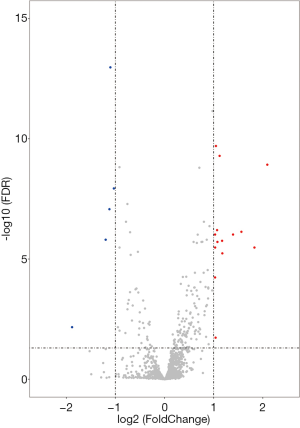
To analyze the association between microRNA expression and MSI, we further analyzed the difference in microRNA expression between the MSI-H and MSS groups. By using a more stringent criterion (P<0.05 and |FC| >2), we found that a total of 19 differentially expressed microRNAs were identified between MSI-H and MSS samples, of which 14 were upregulated and five were downregulated in MSI-H samples, compared with those in MSS samples (Table 3). The Volcano plot is presented to show microRNA expression levels with P<0.05 and |FC| >2 (Figure 3).
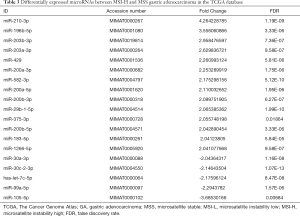
Full table
MicroRNAs that predicted the MSI-H status
By the microRNA expression profiles from the TCGA database, we found that four microRNAs (miR-210-3p, miR-582-3p, miR-30a-3p and miR-105-5p) could accurately distinguish the MSI-H tumors from the MSS tumors (P=4.93×10−10, 5.63×10−10, 3.23×10−9 and 7.64×10−4, respectively). To further validate the accuracy of the prediction models, ROCs of the miR-210-3p, miR-582-3p and miR-30a-3p were constructed, and the area under the curve (AUC) was 0.784, 0.757 and 0.738 for these three microRNAs, respectively, and the increase in these AUCs was statistically significant (P<0.01 for all), while the ROC of miR-105-5p could not be performed due to the missing expression data of some samples. When the three microRNAs were combined, the AUC of the combined prediction model increased to 0.886 (P=0.0004), indicating that the MSI-H subtype could be accurately distinguished from the MSS subtype by this combined prediction model (Figure 4).

Biological signaling pathway enrichment for MSI-H related microRNAs
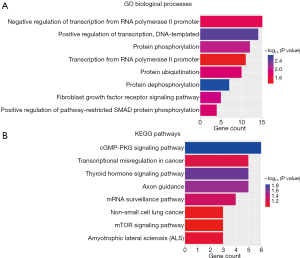
According to the cut-off criteria (P<0.01 and |FC| >2.175), we considered the top six microRNAs of the 14 upregulated microRNAs and the top three microRNAs of the five downregulated microRNAs as the MSI-H-related microRNAs. By using the miRanda online analysis tools, we identified a total of 171 genes of upregulated microRNAs and 119 genes of downregulated microRNAs. Then, we performed an enrichment analysis to elucidate biological functions of these target genes. We found that the GO biological process (BP) terms were mainly enriched in the regulation of transcription (DNA templated); positive regulation of transcription (DNA templated); positive regulation of transcription from RNA polymerase II promoter, and negative regulation of transcription from polymerase II promoter (Figure 5A). In addition, the KEGG pathways were significantly enriched in those for transcription mis-regulation in cancer (Figure 5B).
Discussion
Current choice of therapies for the advanced GCa are limited, and the prognosis is still relatively poor. For GCa patients with MSI-H or dMMR, however, recent therapeutic regimes of using PD-1/PD-L1 inhibitors alone or a combination with chemotherapy have achieved a remarkable progress (15-17). Based on the findings from the present study, 17.1% of the GCa patients had MSI-H tumors (18), which means nearly 1/6 of the GCa patients may benefit from the PD-1/PD-L1 mono-antibody therapy.
Only a small proportion of MSI-H GCa arises from germline mutations of the MMR genes (19). It is known that microRNAs play important roles in epigenetic regulation and that among the sporadic GCa, MSI-H is associated with epigenetic regulation, but the mechanism of MSI-H formation remains ambiguous (10,20,21). Previous studies have revealed that some microRNAs had a consistent expression pattern in both tumor tissues and circulatory plasma, serving as important predictive biomarkers for various types of malignant tumors (22,23). Therefore, the present study focused on the relationship between microRNA expression profiles and the MSI status in GCa, aiming at revealing the mechanism underlying the MSI-H formation.
Firstly, we found that the MSI-H status in 386 GCa patients was correlated with some clinicopathological features, e.g., the MSI-H status increased as age increased, with a higher frequency in female patients and patients with distal GCa located in the pylorus or body of stomach. These findings are consistent with those described in a review of other previously published results from fewer tumor samples (24).
Secondly, the present study also suggests that the microRNA expression profiles of MSS, MSI-L and MSI-H showed a trend change in GCa tumor samples. Although the difference between MSS and MSI-L was rather small, the difference between MSI-H and MSI-L was relatively remarkable and associated with aging. These trends indicate that it is a continuous change from MSS to MSI-H, consistent with the dividing method of MSI in colon cancer (25). Furthermore, we found that both MSI-H and MSS were significantly associated with microRNA expression levels.
MiR-210, which ranks the top of the most significantly differentially expressed microRNAs, has been reported to impair the functions of DNA damage-repair genes, possibly causing DNA replication errors (26,27), which may lead to the MSI formation (28). As for miR-196b, there are a few reports on the role of miR-196b in GCa. For example, a couple of studies have suggested that miR-196b promotes the metastasis and invasion of GCa cells (29,30). Other studies had shown that the high expression of miR-196b significantly impaired DNA damage-repair functions (31). Hence, we speculate that the high expression levels of miR-196b in the MSI-H-related GCa may affect the stability of the genome through the impairment of DNA damage-repair functions.
Studies have revealed that miR-203 also inhibits invasion and metastasis of GCa cells. For example, one study found that the expression of miR-203 was negatively correlated with expression of ataxia-telangiectasia mutated (ATM) protein (32), while another study demonstrated that the ATM gene was highly mutated and that the expression of the ATM protein was downregulated in MSI-H-related GCa tissues (33). Since ATM plays a critical role in DNA damage-induced signaling and initiation of cell cycle checkpoint signaling, it is reasonable to assume that miR-203 may contribute to MSI-H by targeting the ATM gene.
miR-429 and miR-200a, as the members of the miR-200 family, were significantly upregulated in MSI-H GCa tissues than in the MSS subtype. One study demonstrated that expression levels of the miR-200 family increased substantially in GCa tumor tissues, compared with that of normal tissues, indicating that the miR-200 family may play an important role in promoting GCa cell growth (34).
The miR-105, miR-99a and hsa-let-7c were the three microRNAs downregulated the most in MSI-H GCa, compared with the MSS subtype. Few studies reported the roles of miR-105 and has-let-7c in GCa. One study reported, however, that the miR-99 family of microRNAs could regulate DNA damage response by targeting SNF2H (35), while other studies showed that overexpression of the miR-99 family in prostatic cancer cells could inhibit the expression of SNF2H and reduce DNA damage-repair rate and overall repair efficiency (36,37), although the role of miR-99 in GCa has not been reported yet.
To further explore the functions of the above-mentioned nine microRNAs, we searched for the predicted target genes of these microRNAs and analyzed their related pathways and GO annotations by using bioinformatics online tools. We found that these nine microRNAs could regulate a variety of genes in several key signaling pathways, including regulation of transcription (DNA templated), positive regulation of transcription from RNA polymerase II promoter, positive regulation of transcription (DNA templated) and negative regulation of transcription from polymerase II promoter. It has been suggested that abnormal signaling pathways, such as the KRAS signaling pathway and the base-excision repair pathway, may contribute to the formation of MSI-H in gastrointestinal and endometrial cancers (38-40). Therefore, we assume that other DNA damage repair pathways may also play important roles in the formation of MSI-H, in addition to the impairment of the MMR pathway; however, further investigations are needed to test this hypothesis and unravel the underlying molecular mechanisms.
Conclusions
In the present study, we identified nine significantly differentially expressed microRNAs in GCa tumor tissues, and the results suggested that the pathways related to DNA damage-repair functions, other than MMR, were associated with MSI formation in GCa. Because of limited sample size and the limitations in bioinformatics analysis, further rigorous laboratory experiments in molecular and functional investigations are needed to substantiate these results.
Acknowledgments
Funding: This work was supported by the Natural Science Foundation of China (No. 81871948).
Footnote
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/atm.2020.03.54). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. Ethics Approval was exempt, because all the raw data were from the TCGA database that is publicly available for all interested researchers, and the patients’ privacy was strictly protected due to deidentification in the TCGA database.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Bray F, Ferlay J, Soerjomataram I, et al. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin 2018;68:394-424. [Crossref] [PubMed]
- Digklia A, Wagner AD. Advanced gastric cancer: Current treatment landscape and future perspectives. World J Gastroenterol 2016;22:2403-14. [Crossref] [PubMed]
- Le DT, Durham JN, Smith KN, et al. Mismatch repair deficiency predicts response of solid tumors to PD-1 blockade. Science 2017;357:409-13. [Crossref] [PubMed]
- Muro K, Bang YJ, Shankaran V, et al. Relationship between PD-L1 expression and clinical outcomes in patients (Pts) with advanced gastric cancer treated with the anti-PD-1 monoclonal antibody pembrolizumab (Pembro; MK-3475) in KEYNOTE-012. J Clin Oncol 2015;33:3. [Crossref]
- Yamamoto H, Imai K. Microsatellite instability: an update. Arch Toxicol 2015;89:899-921. [Crossref] [PubMed]
- Tamura K, Kaneda M, Futagawa M, et al. Genetic and genomic basis of the mismatch repair system involved in Lynch syndrome. Int J Clin Oncol 2019;24:999-1011. [Crossref] [PubMed]
- Sinicrope FA. Lynch Syndrome-Associated Colorectal Cancer. N Engl J Med 2018;379:764-73. [Crossref] [PubMed]
- Vilar E, Gruber SB. Microsatellite instability in colorectal cancer-the stable evidence. Nat Rev Clin Oncol 2010;7:153-62. [Crossref] [PubMed]
- Zighelboim I, Goodfellow PJ, Gao F, et al. Microsatellite instability and epigenetic inactivation of MLH1 and outcome of patients with endometrial carcinomas of the endometrioid type. J Clin Oncol 2007;25:2042-8. [Crossref] [PubMed]
- Ottini L, Falchetti M, Lupi R, et al. Patterns of genomic instability in gastric cancer: clinical implications and perspectives. Ann Oncol 2006;17 Suppl 7:vii97-102. [Crossref] [PubMed]
- Ishiguro H, Kimura M, Takeyama H. Role of microRNAs in gastric cancer. World J Gastroenterol 2014;20:5694-9. [Crossref] [PubMed]
- Ma J, Hong L, Chen Z, et al. Epigenetic regulation of microRNAs in gastric cancer. Dig Dis Sci 2014;59:716-23. [Crossref] [PubMed]
- Lanza G, Ferracin M, Gafa R, et al. mRNA/microRNA gene expression profile in microsatellite unstable colorectal cancer. Mol Cancer 2007;6:54. [Crossref] [PubMed]
- Chu A, Liu J, Yuan Y, et al. Comprehensive Analysis of Aberrantly Expressed ceRNA network in gastric cancer with and without H.pylori infection. J Cancer 2019;10:853-63. [Crossref] [PubMed]
- Le DT, Uram JN, Wang H, et al. PD-1 Blockade in Tumors with Mismatch-Repair Deficiency. N Engl J Med 2015;372:2509-20. [Crossref] [PubMed]
- Boku N, Ryu MH, Kato K, et al. Safety and efficacy of nivolumab in combination with S-1/capecitabine plus oxaliplatin in patients with previously untreated, unresectable, advanced, or recurrent gastric/gastroesophageal junction cancer: interim results of a randomized, phase II trial (ATTRACTION-4). Ann Oncol 2019;30:250-8. [Crossref] [PubMed]
- Bang YJ, Kang YK, Catenacci DV, et al. Pembrolizumab alone or in combination with chemotherapy as first-line therapy for patients with advanced gastric or gastroesophageal junction adenocarcinoma: results from the phase II nonrandomized KEYNOTE-059 study. Gastric Cancer 2019;22:828-37. [Crossref] [PubMed]
- Polkowski W, van Sandick JW, Offerhaus GJ, et al. Prognostic value of Lauren classification and c-erbB-2 oncogene overexpression in adenocarcinoma of the esophagus and gastroesophageal junction. Ann Surg Oncol 1999;6:290-7. [Crossref] [PubMed]
- Boland PM, Yurgelun MB, Boland CR. Recent progress in Lynch syndrome and other familial colorectal cancer syndromes. CA Cancer J Clin 2018;68:217-31. [Crossref] [PubMed]
- Keller G, Grimm V, Vogelsang H, et al. Analysis for microsatellite instability and mutations of the DNA mismatch repair gene hMLH1 in familial gastric cancer. Int J Cancer 1996;68:571-6. [Crossref] [PubMed]
- Toyota M, Ahuja N, Suzuki H, et al. Aberrant methylation in gastric cancer associated with the CpG island methylator phenotype. Cancer Res 1999;59:5438-42. [PubMed]
- Yu X, Liang J, Xu J, et al. Identification and Validation of Circulating MicroRNA Signatures for Breast Cancer Early Detection Based on Large Scale Tissue-Derived Data. J Breast Cancer 2018;21:363-70. [Crossref] [PubMed]
- Zhang R, Wang W, Li F, et al. MicroRNA-106b~25 expressions in tumor tissues and plasma of patients with gastric cancers. Med Oncol 2014;31:243. [Crossref] [PubMed]
- Ratti M, Lampis A, Hahne JC, et al. Microsatellite instability in gastric cancer: molecular bases, clinical perspectives, and new treatment approaches. Cell Mol Life Sci 2018;75:4151-62. [Crossref] [PubMed]
- Benson AB 3rd, Venook AP, Cederquist L, et al. Colon Cancer, Version 1.2017, NCCN Clinical Practice Guidelines in Oncology. J Natl Compr Canc Netw 2017;15:370-98. [Crossref] [PubMed]
- Bavelloni A, Ramazzotti G, Poli A, et al. MiRNA-210: A Current Overview. Anticancer Res 2017;37:6511-21. [PubMed]
- Crosby ME, Kulshreshtha R, Ivan M, et al. MicroRNA regulation of DNA repair gene expression in hypoxic stress. Cancer Res 2009;69:1221-9. [Crossref] [PubMed]
- Miquel C, Jacob S, Grandjouan S, et al. Frequent alteration of DNA damage signalling and repair pathways in human colorectal cancers with microsatellite instability. Oncogene 2007;26:5919-26. [Crossref] [PubMed]
- Shao L, Chen Z, Peng D, et al. Methylation of the HOXA10 Promoter Directs miR-196b-5p-Dependent Cell Proliferation and Invasion of Gastric Cancer Cells. Mol Cancer Res 2018;16:696-706. [Crossref] [PubMed]
- Lim JY, Yoon SO, Seol SY, et al. Overexpression of miR-196b and HOXA10 characterize a poor-prognosis gastric cancer subtype. World J Gastroenterol 2013;19:7078-88. [Crossref] [PubMed]
- Shen YN, Bae IS, Park GH, et al. MicroRNA-196b enhances the radiosensitivity of SNU-638 gastric cancer cells by targeting RAD23B. Biomed Pharmacother 2018;105:362-9. [Crossref] [PubMed]
- Zhou P, Jiang N, Zhang GX, et al. MiR-203 inhibits tumor invasion and metastasis in gastric cancer by ATM. Acta Biochim Biophys Sin (Shanghai) 2016;48:696-703. [Crossref] [PubMed]
- Kim HS, Choi SI, Min HL, et al. Mutation at intronic repeats of the ataxia-telangiectasia mutated (ATM) gene and ATM protein loss in primary gastric cancer with microsatellite instability. PLoS One 2013;8:e82769. [Crossref] [PubMed]
- Chang L, Guo F, Huo B, et al. Expression and clinical significance of the microRNA-200 family in gastric cancer. Oncol Lett 2015;9:2317-24. [Crossref] [PubMed]
- Mueller AC, Sun D, Dutta A. The miR-99 family regulates the DNA damage response through its target SNF2H. Oncogene 2013;32:1164-72. [Crossref] [PubMed]
- Sun D, Lee YS, Malhotra A, et al. miR-99 family of MicroRNAs suppresses the expression of prostate-specific antigen and prostate cancer cell proliferation. Cancer Res 2011;71:1313-24. [Crossref] [PubMed]
- Rane JK, Erb HH, Nappo G, et al. Inhibition of the glucocorticoid receptor results in an enhanced miR-99a/100-mediated radiation response in stem-like cells from human prostate cancers. Oncotarget 2016;7:51965-80. [Crossref] [PubMed]
- Bosse T, ter Haar NT, Seeber LM, et al. Loss of ARID1A expression and its relationship with PI3K-Akt pathway alterations, TP53 and microsatellite instability in endometrial cancer. Mod Pathol 2013;26:1525-35. [Crossref] [PubMed]
- Garre P, Briceno V, Xicola RM, et al. Analysis of the oxidative damage repair genes NUDT1, OGG1, and MUTYH in patients from mismatch repair proficient HNPCC families (MSS-HNPCC). Clin Cancer Res 2011;17:1701-12. [Crossref] [PubMed]
- Velho S, Corso G, Oliveira C, et al. KRAS signaling pathway alterations in microsatellite unstable gastrointestinal cancers. Adv Cancer Res 2010;109:123-43. [Crossref] [PubMed]


