RNA-sequencing and microarray data mining revealing: the aberrantly expressed mRNAs were related with a poor outcome in the triple negative breast cancer patients
Introduction
Breast cancer (BC) was the major cause of cancer death in women worldwide, with an estimated 268,600 new cases (30% of the total) and 41,760 deaths (15% of the total) among females in 2019 (1). Among all BC subtypes, triple negative breast cancer (TNBC) accounting for approximately 10% to 20% (2). TNBC was a special sub-group of BC which didn’t express human epidermal growth factor receptor 2 (HER2) nor estrogen receptors (ER) or progesterone receptors (PR) (3-5). According to the working principle of estrogen synthesis blocking, ER antagonists or aromatase inhibitors were widely used in the therapy of BC patients who express ER or PR. Moreover, HER2-blocking antibody such as Trastuzumab or HER2 kinase such as Trikerb can effectively treat HER2+ breast carcinomas (6,7). However, TNBC patients had the worst prognosis among all BC patients; this is partly due to endocrine therapy or therapies targeting HER2 cannot work (8). The American society of clinical oncology (ASCO) guidelines did not specify approaches for TNBC patients thus far (9). Hence, it is urgent to find one or a few accurate indicators in the genesis and development of TNBC. We hope to shed light on exploring potential therapeutic targets in TNBC by our results of data analysis.
Better understanding of TNBC pathogenesis is vital to identify diagnosis markers and novel therapeutic methods. However, it should be noted that the definite etiopathogenesis of TNBC is still unclear despite there were a great deal of researches on the mechanism in TNBC formation (10-12). It is extremely important and sorely demanded to reveal the reason and the underlying molecular mechanisms since high morbidity and mortality of TNBC, also there is an urgent needed for early diagnosis, prevention and targeted therapy biomarkers (13,14). In the present study, we want to find one or several molecular biomarkers which may eventually be applied to noninvasive diagnosis of TNBC.
Microarray was a high-throughput platform which could measure the expression of global gene. It was widely used for searching for possible genetic or epigenetic alternations, identify molecular biomarkers such as carcinomas and so on (15,16). Huge amounts of cores slice data were produced with an extensive use of microarrays, and most of them were stored and shared in public databases (17,18). Because of data redundancy and variances, researchers sometimes reached different conclusions. For getting more accurate reasons about onset and progression of TNBC, we integrated, re-analyzed and verified the data stored in public databases. Some studies were done to seek differentially expressed genes (DEGs) in TNBC though gene expression profiling microarrays (19,20). However, independent researches involving heterogeneous tissues or samples, in addition, their results were obtained from a single cohort study, so their conclusions were limited or inconsistent. Consequently, key genes and pathways were difficult to confirm according to different studies. With our study, via integrated, re-analyzed and verified available and relevant expression profiling microarray data sets that uploaded in the Gene Expression Omnibus (GEO) database by different laboratories, one-sidedness of individual researchers is overcome and statistical power increased, therefor; the screening results are more precise and reliable.
In the present study, we have downloaded three original microarray datasets GSE65194 [55 TNBC samples, 95 non-TNBC samples (36 HER+; 29 luminal A; 30 Luminal B)], GSE43358 [17 TNBC samples, 40 non-TNBC samples (14 HER+; 16 luminal A; 10 luminal B)] and GSE76275 (198 TNBC samples and 67 not triple-negative tumors) from GEO database (https://www.ncbi.nlm.nih.gov/geo). There was a total of 270 TNBC and 202 non-TNBC tissues available. Subsequently, the DEGs were screened using R language and 159 DEGs were filtered out from 21,755 genes based on 3 independent datasets which contained 472 BC cases. To better understand the molecular mechanisms of the onset and progression of TNBC, we conducted hierarchical clustering analysis, enrichment analysis for function and pathway on 159 screened DEGs. Then, by searching in protein-protein interaction (PPI) network, several important functional modules and hub genes were achieved from 159 screened DEGs.
Furthermore, we selected the top 8 DEGs with the most obvious expression difference to verify our previous results in this paper and evaluated their clinic pathological value and potential mechanism for TNBC. The expression signatures of the 8 DEGs in clinical cancer tissue were assessed by several databases. Their expressions in TNBC or non-TNBC tissues were analyzed in oncomine database. The co-expression analysis of selected 8 DEGs was performed by cBioportal reveals the co-occurrence or mutual exclusivity relationship, and gives clues to the possible underlying mechanism. The survival times of TNBC patients with high expression or low expression of DEGs were identified in the Kaplan-Meier plotter database. All in all, we hope to throw further light on the molecular mechanism of TNBC and seek out the candidate biomarkers for drug targets, diagnosis and prognosis.
Methods
Microarray data selection
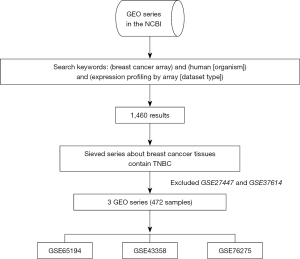
In our current study, the gene expression profiling data sets (ID: GSE65194, GSE43358, GSE76275) were obtained from National Center for Biotechnology Information-Gene Expression Omnibus (NCBI-GEO) database. We used “breast cancer array”, “human [organism]” and “expression profiling by array [dataset type]” as keywords for searching in GEO database. There were 1,460 results from this search condition. The microarray datasets were selected according to the following rules: the samples must contain human TNBC tissues; the patients did not receive special treatment, including radiotherapy and chemotherapy; and dataset tested genes were not less than 12,000. Under these conditions, we obtained 3 datasets to performing further analyze although there were 5 datasets contain TNBC tissues (GSE27447 and GSE37614 were excluded because only 10922 genes were tested in the two datasets). In other words, we collected mRNA-sequencing raw data from 3 independent original researches. We collected the following information from each individual original research: number of TNBC tissues and non-TNBC samples, GEO accession number, sequencing platforms, and the most important gene expression data. The brief information about the GEO series to be analyzed was listed in Table 1. We download the raw data of 472 specimens from 3 independent GEO series. Totally 270 TNBC and 202 non-TNBC specimens were enrolled in GSE65194, GSE43358 and GSE76275. All mRNA-sequencing data were obtained to be integrated and re-analyzed. The platform GPL570 tested 21,755 genes’ expression values. The process of obtain, sieve and filter GEO series is shown in Figure 1.

Full table
Data preprocessing before integration and re-analysis
We started to process the raw data by Affy package in R language using Robust Multiarray Average algorithm and obtained expression data of each probe. With Annotation package of Bioconductor in R program language, we converted the expression data of the probe sets to expression profile of gene according to the platform Annotation files. There is many given genes’ expression data were tested by multiple probes in each GEO series, so we averaged the multiple probes expression data for just one gene according to gene symbol. Then, the expression values file of 21,755 given genes was obtained. Of course, there were 3 tables since there were 3 independent GEO series. To integration and re-analysis of all data, we merged the gene expression value files of 472 patients from datasets of GSE65194, GSE43358 and GSE76275 into one output table according to gene symbol through using the package of sameGene in R language. Then the datasets of the output table were assigned into 2 groups: TNBC group and non-TNBC group.
We used the ComBat algorithm of the Surrogate Variable Analysis package in R language to conduct batch normalization on all expression profiling data. The function in R language could eliminate the systematic variations effectively from different independent researchers.
DEGs screening
We selected the DEGs by a Bioconductor software package called LIMMA in R program language using linear models from the normalized data of TNBC and Non-TNBC tissues. Expression values of DEGs with |Fold Change| >3, also known as |PP2FoldChange| (|log2FC|) >1.585 and adjusted P value <0.05 were the focus we pay attention to.
We draw the Volcano plot to show the distribution of all 21,755 genes for P value and fold change. Heat map of expression hierarchical clustering analysis for top 50 genes was performed to investigate probable discrepancies between TNBC and non-TNBC tissues.
Function and pathway enrichment analysis of 159 screened DEGs
To gain biological function sights of involved DEGs, we did functional enrichment analysis with FunRich. The FunRich software is a standalone functional enrichment and network analysis tool. It can be utilized to perform biological process, cellular component, and biological pathway enrichment analysis for a batch of genes with a filtering condition of P value <0.05.
PPI network and module analysis
The PPI network of DEGs can help us better understanding the molecular mechanisms by highlight the core signal pathways and core genes in carcinogenesis. It was constructed based on Search Tool for the Retrieval of Interacting Genes/Proteins (STRING) database (21) and clarified which gene encoded protein had most interactions with other DEGs. Our study constructed and visualized the PPI network of 159 screened DEGs with a cut-off criterion of interaction score >0.4 and p value <0.05.
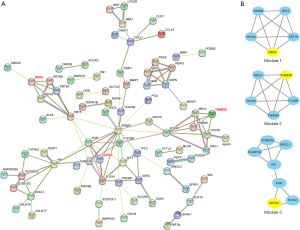
Moreover, to screen out ranked top proteins (these proteins corresponded genes called hub genes) which had most interactions with other DEGs, and correspondent modules of hub genes within PPI network, we conducted MCODE analysis by Cytoscape software with filter criteria of a number of nodes >4 and MCODE score >3. We selected and listed the top 3 hub genes with the threshold of connection degree >10.
Oncomine database verify analysis of top 8 DEGs selected from GEO database
Oncomine is a web-based platform for data store and re-analysis. It contained about 4700 genome-wide expression analyses of over 50 different tumors (22). The expression levels of top 8 DEGs with most obvious expression changes were analyzed using Oncomine Database (https://www.oncomine.org). We obtained and compared the mRNA expression fold change of screened DEGs in TNBC tissues and non-TNBC tissues through Oncomine. Co-expression analysis in Oncomine was used to find out which genes had synchronous expression patterns with our concerned DEGs. Based on heatmap and expression value fold change, we analyzed the co-expression profiles of N-acetyltransferase 1 (NAT1) in TNBC using Oncomine.
Kaplan-Meier plotter analysis of DEGs co-expression with NAT1
The Kaplan-Meier plotter is a web-based platform which was used to assess the effect of over 50,000 genes on survival in 6,234 BC samples (23). The prognostic value of NAT1 and its co-expression genes GATA binding protein 3 (GATA3) and Signal peptide, CUB domain and EGF like domain containing 2 (SCUBE2) in BC and TNBC were analyzed by Kaplan-Meier Plotter (http://kmplot.com/analysis/), and the log-rank test was used to test for significance (24).
Genetic alteration and co-expression analysis of interested DEGs
The cBioPortal (http://www.cbioportal.org) (25) is a web-based platform used for gene-based data interactive exploration. The alterations such as deep deletion, mRNA/protein upregulation and downregulation, missense mutation and so on of NAT1 and its coexpression genes GATA3, trefoil factor 3 (TFF3), forkhead box A1 (FOXA1), G protein-coupled receptor 160 (GPR160) and SCUBE2 in Breast Invasive Carcinoma (TCGA, provisional) was analyzed using cBioportal. The cBioPortal is also used for co-occurrence or mutual exclusivity and customizable correlation plot’s analysis.
Results
Normalization of mRNA expression data
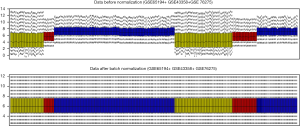
We normalized mRNA expression data of 21,755 genes by batch normalization according to a median method of 472 samples (270 TNBC and 202 non-TNBC specimens). The mRNA expression values of 472 TNBC and non-TNBC specimens before and after batch normalizations were shown in Figure 2. X-axis represents independent samples. Y-axis represents expression value of genes. Black bold horizontal line displayed the medium expression value of 472 samples, after batch normalization, the medium expression value of 472 samples was almost consistent means the normalized data was eligible, so that we can do analysis further.
Selection of DEGs and expression hierarchical clustering analysis
The Limma package in R language was utilized to compare gene expression between TNBC tissues and Non-TNBC tissues and screened out DEGs with the criterion of |log2 FC| >1.585 and P<0.05. The Limma package identified all DEGs through t tests statistic algorithm. We ranked significant genes based on fold change of genes’ expression values.
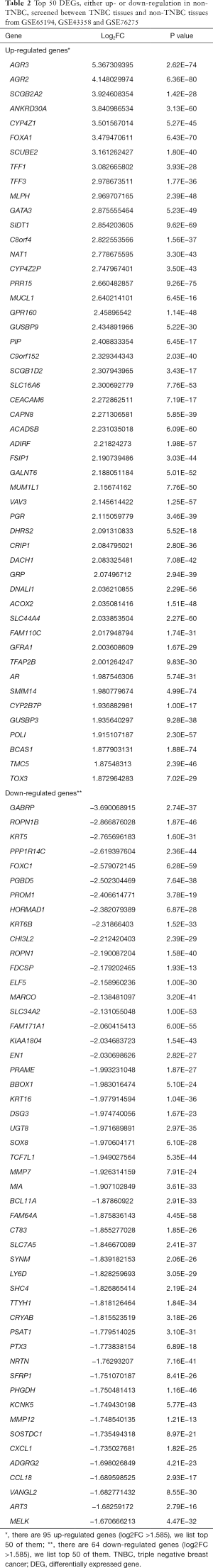
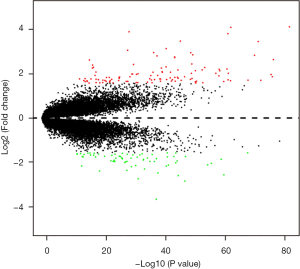
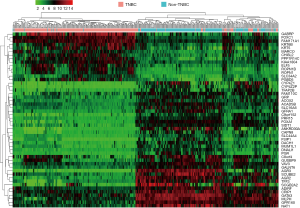
In total, 159 DEGs (95 up-regulated and 64 down-regulated) obtained based on genes’ expression data of 472 patients (270 TNBC and 202 non-TNBC from 3 GEO series). We list top 50 DEGs according to fold change of genes’ expression values in Table 2. The volcano plot (Figure 3) showed the distribution of all genes, the abscissa axis represents P values [−log10 (P value)] and axis of ordinates stands for fold change (log2 FC). The up-regulated DEGs displayed by red dots, and green dots stand for down-regulated DEGs.
Full table
In Figure 4, To display and compare gene expression differences between TNBC and non-TNBC more intuitive, we draw the heat map with R software and select fold change patterns of top 50 highly DEGs to perform hierarchical clustering.
Function and pathway enrichment analysis of 159 screened DEGs
To further investigate the biologic effects of aberrantly-expressed DEGs in TNBC, we performed biological process enrichment analysis of 159 screened DEGs using Funrich software. The top 10 enriched biological processes are shown in Figure 5A. The functions of 159 DEGs in the biological process were largely focused on signal transduction, cell communication, regulation of nucleic acid metabolism, and so on.

In order to master which biologic pathways the DEGs involved in, biological pathway enrichment analysis was done, and we found that DEGs mainly enriched in Mesenchymal-to-epithelial transition (MET), mTOR signaling pathway, IL-3 mediated signaling pathway and so on (Figure 5B).
PPI network and module analysis
We constructed the PPI network for 159 screened DEGs using the open-access database STRING (Figure 6A). To find out the hub genes among 159 DEGs, we did module analysis through MODE function in Cytoscape software and top 3 hub genes and modules were shown in Figure 6B. Desmoglein 3 (DSG3), family with sequence similarity 83 member D (FAM83D) and GATA3 were the top 3 hub genes.
Validation of the expression of top 8 aberrantly expressed DEGs based on heat map in Oncomine Database
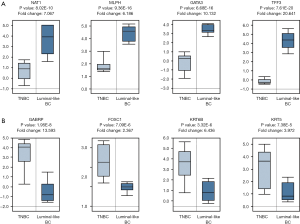
To further elucidate the expression patterns of the DEGs between TNBC and non-TNBC tissues was consistent with our analysis result based on GEO data; a clinical study was performed in the light of previous results in Oncomine database. We screened out 159 DEGs from 3 GEO series, 472 samples, including 64 down-regulated DEGs and 95 up-regulated DEGs. We selected 4 down-regulated DEGs and 4 up-regulated DEGs which have most obvious expression changes according to heat map clustering analysis result for further analysis. The top 8 aberrantly expressed DEGs were NAT1, MLPH, GATA3, TFF3, GABRP, FOXC1, KRT6B and KRT5. The results obtained according to Oncomine database confirmed that the expressions of the top 8 DEGs were consistent with our previous studies in accordance with data from GEO series (P<0.001) (Figure 7).
Potential molecular mechanism associated with NAT1 in TNBC
To clarify which genes set had synchronous expression patterns, we performed Co-expression analysis using Oncomine. Based on previous results of difference analysis and clustering analysis, NAT1 is most obvious of expression change’s gene. A set of genes had a tendency toward co-expressed with NAT1 in BC was picked up and presented in Figure 8A. The co-expression profiles for NAT1 were listed with a strong cluster of top 10% genes based on 167 breast carcinoma tissues. On the bright side, we found that there were 5 genes (GATA3, TFF3, FOXA1, SCUBE2, GPR160) which co-expression with NAT1 were DEGs that screened out from TNBC based on our previous results.
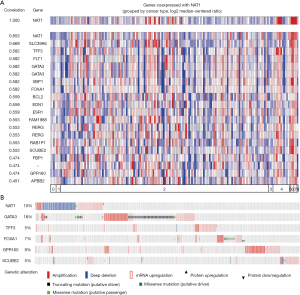
The OncoPrint from cBioPortal can provide a graphical summary of genomic/mRNA expression/protein expression alterations of sets of genes in different cancer samples. It summarized copy number alterations such as amplifications and deletions, genomic alterations such as mutations, and mRNA or protein expression changes. We analyzed genomic alterations of NAT1 and its co-expression genes using cBioPortal and 52% cases with genetic alterations had been obtained (Figure 8B). The mutual exclusivity from cBioPortal can provide information of genes’ potential relationship in different cancer, so we used cBioPortal to explore the probable co-occurrence of NAT1 and its co-expression genes. As Table 3 showed, same as DEGs, SCUBE2 and GATA3 had a tendency towards co-occurrence with NAT1 in TNBC.
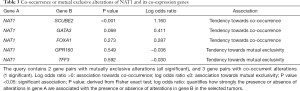
Full table
Key DEGs expression in TNBC was related to survival outcome based on Kaplan-Meier plotter
According to our previous bioinformatics analyses and validation, the expression of NAT1 was down-regulated obviously in TNBC. Same as DEGs, GATA3 and SCUBE2 co-expressed with NAT1, and had a tendency towards a co-occurrence with NAT1 in TNBC. To explore the association of NAT1, GATA3 and SCUBE2 expression with survival outcome of TNBC patients, we draw survival curves using Kaplan-Meier plotter database. As Figure 9A,B,C showed, the high expression of NAT1, GATA3 and SCUBE2 were positive correlated with better prognosis (P<0.001). However, in TNBC patients, the effect of mRNA expression of NAT1, GATA3 and SCUBE2 on cancer progression wasn’t statistically significant (P>0.05). But based on the figures, differentially expressed NAT1 and GATA3 had diverse survival times; no statistical differences maybe on account of insufficient samples.
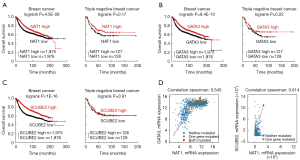
The correlation plots from cBioPortal were used to visualizing the relationship between mRNA expression of NAT1 and GATA3 or SCUBE2 (Figure 9D). Their mRNA expression had positive correlation.
Discussion
TNBC was a distinctive sub-group of BC which didn’t express HER2 nor ER or PR (26-28) and accounted for 10–20% of BC patients (29). TNBC patients always had poor prognosis, higher probability to metastasis, and the lower five-year survival rates. Fortunately, scientific advancements of BC improved the survival outcome of BC patients greatly. Molecular markers of HER2, ER and PR are widely applied in clinical for specific treatments, with excellent predictive value and prognostic value (3,30,31). However, TNBC remains the most intractable problem among all BC subtypes since lack of molecular therapeutic targets. TNBC patients had only limited treatment options because of hormone receptor and HER2 receptor expressed low and researchers are keenly attempting to identify new treatments (32,33).
A field which has recently contributed significantly to improved diagnostics, classification and prognostics are BC transcriptomics, a whole transcriptome high throughput sequencing and analysis technique, which identifies changes in the RNA expression, is now being used to obtain more effective information of the molecular mechanism of BC (34,35). With our study, via integrated, re-analyzed and verified available and relevant expression profiling microarray data sets that uploaded in GEO database by different laboratories, one-sidedness of individual researchers is overcome and statistical power increased, therefor; the screening results are more precise and reliable.
In the current research, we focused on the differentially expressed mRNAs between TNBC and non-TNBC tissues according to GEO microarray data. From 472 samples from independent labs and 21,755 genes, we screened out 64 down-regulated and 95 up-regulated DEGs under condition of |Fold Change of genes expression values| >3 and P<0.05.
Biological process analysis of all DEGs demonstrates that the DEGs were mainly involved signal transduction, cell communication and nucleic acid metabolism. Qiu et al. reported that inhibition of the Notch1 signal pathway can induce apoptosis, inhibit angiogenesis, and influence cell cycle distribution in TNBC (36). The RAF/MEK/ERK signal transduction pathway also plays an important role in TNBC, De Luca et al. reported that it regulates cell growth and proliferation, differentiation and migration of BC cells (37). Wnt/β-catenin pathway had also been well-documented of taking a vital role in the development of TNBC (38). Nevertheless, researches had continued to explore a more definite relationship between signal transduction molecules and TNBC patients’ survival rate since their conclusion still controversial. Furthermore, the enriched biological pathways of the DEGs we selected included MET, mTOR signaling pathway and IL-3 mediated signaling pathway. Numerous studies had reported epithelial-to-mesenchymal transition (EMT) can be induced by receptor tyrosine kinases (RTKs) including c-Met, epithelial growth factor-receptors and IGF-1R, and EMT always promoted tumor progression, in development of carcinoma, RAF/MEK/ERK1/2 axis, PI3k/Akt/mTOR pathways, and phosphorylation of β-catenin were play a critical role (39-41). Function analysis helps us better to understand the mechanism of TNBC and provides a guide for TNBC prevention and treatment; however, further laboratory and clinical researchers are required to verify this.
PPI network clearly revealed the function relationship of all 159 DEGs, further analysis identified top 3 hub genes: DSG3, FAM83D and GATA3. DSG3 reflects a strong relationship with Keratins in functional connections. Coincidentally, we assessed the expression difference of KRT5 and KRT6B in TNBC tissues and control specimens using oncomine database and the result consistent with our analysis result based on GEO database. Zinc-finger binding transcription factor GATA3 which belonged to GATA family had been reported many times closely associated with breast carcinomas. It was a specific marker based on our existing knowledge (42). In the current study, we verified that as an aberrantly expressed mRNA, GATA3 is down-regulated in TNBC and survival outcome of TNBC patients with low GATA3 was worse.
To verify our previous results in this paper, we assessed the expression levels of top 8 DEGs with most obvious fold changes. The expression levels of NAT1, MLPH, GATA3, TFF3, GABRP, FOXC1, KRT6B and KRT5 were analyzed in Oncomine database, respectively. And the results are consistent with the previous conclusion obtained from GEO series. It proved that the DEGs screened from GEO database are reliable. There were a few researches about the relationship with MLPH (43), GATA3 (44), TFF3 (45), GABRP (46), FOXC1 (47), KRT6B, KRT5 (48) and TNBC. However, few reports have revealed the association between NAT1 and TNBC. Clustering analysis which performed to investigate probable discrepancies between TNBC and Non-TNBC tissues also revealed that NAT1 is most obvious of expression change’s gene.
To explore further the potential role of NAT1 in TNBC, we did co-expression analysis for NAT1 in BC tissues, and we found that there were 5 genes (GATA3, TFF3, FOXA1, SCUBE2, GPR160) which co-expression with NAT1 also were DEGs that we screened out from TNBC. We analyzed genomic alterations, co-occurrence or mutual exclusivity relationship between NAT1 and its co-expression genes using cBioPortal. There was a tendency towards co-occurrence between NAT1 and SCUBE2 or GATA3 in TNBC. That is to say, same as DEGs, GATA3 and SCUBE2 co-expressed with NAT1, and had a tendency towards a co-occurrence with NAT1 in TNBC. From all above results, we speculate that NAT1, SCUBE2 and GATA3 play an important role in TNBC. The survival curves show that the probability of BC progression was found to be statistically significant with high NAT1, GATA3 and SCUBE2 expression as compared to that of low NAT1, GATA3 and SCUBE2 expression (P<0.01). The mRNA of GATA3 and SCUBE2 had positive correlation with NAT1.
This study had several limitations. Firstly, the survival curves of NAT1, GATA3 and SCUBE2 between TNBC and Non-TNBC wasn’t statistically significant (P>0.05), but on the basis of the figure, differentially expressed NAT1 and GATA3 have diverse survival times, statistical nonsense may on account of insufficient samples. Secondly, although we performed data integration, normalization and reanalysis for sequencing results from 3 independent labs, and preliminary validated our results, more experiments are needed to make further research. Of course, we will integrate existing results into our future experiments and continue to explore the molecular mechanisms of TNBC.
Through our research, via integrated, re-analyzed and verified available and relevant expression profiling microarray data sets that uploaded in GEO database by different laboratories, one-sidedness of individual researchers is overcome and statistical power increased, therefor; the screening results are more precise and reliable. Our study provides information for researchers to identify potential candidate molecular biomarkers and pathways, which play an important role in TNBC may be utilized for drug targets, diagnosis and prognosis. It was obvious that our results give more insight of TNBC carcinogenesis and were on behalf of a valuable resource for future studies.
Conclusions
Our research integrated, re-analyzed and verified whole genome sequencing microarray data from 3 individual labs, there are 159 DEGs match our screening condition. Biological process analysis, biological pathway analysis, and PPI network helped us identify some genes and pathways significantly correlated with TNBC progression. Validation experiments verified that expression patterns of top 8 DEGs consisted of NAT1, MLPH, GATA3, TFF3, GABRP, FOXC1, KRT6B and KRT5 in oncomine database are consistent with their expression level in GEO series. Clustering analysis which performed to investigate probable discrepancies between TNBC and Non-TNBC tissues revealed that NAT1 is most obvious of expression change’s gene. Moreover, same as DEGs, GATA3 and SCUBE2 co-expressed with NAT1, and had a tendency towards a co-occurrence with NAT1 in TNBC. As a hub gene of PPI network, the mRNA of GATA3 had positive correlation with NAT1. The survival curves show that the probability of BC progression was found to be statistically significant with high NAT1, GATA3 and SCUBE2 expression as compared to that of low NAT1, GATA3 and SCUBE2 expression (P<0.01). From all above results, we speculate that NAT1, SCUBE2 and GATA3 play an important role in TNBC.
Acknowledgments
Funding: The study was supported by the Nosocomial Scientific Research Fund Projects from International Peace Maternity and Child Health Hospital of Shanghai Jiao Tong University School of Medicine (No. GFY5801) and Shanghai Sailing Program (No. 19YF1452200).
Footnote
Conflicts of Interest: The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. This article does not contain any studies with human participants or animals performed by any of the authors. The human data we filtered, verified and deeply analyzed were using a combined bioinformatics approach based on multiple databases.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Siegel RL, Miller KD, Jemal A. Cancer statistics, 2019. CA Cancer J Clin 2019;69:7-34. [Crossref] [PubMed]
- Al-Mahmood S, Sapiezynski J, Garbuzenko OB, et al. Metastatic and triple-negative breast cancer: challenges and treatment options. Drug Deliv Transl Res 2018;8:1483-507. [Crossref] [PubMed]
- Wang C, Kar S, Lai X, et al. Triple negative breast cancer in Asia: An insider's view. Cancer Treat Rev 2018;62:29-38. [Crossref] [PubMed]
- Kumar RV, Panwar D, Amirtham U, et al. Estrogen receptor, Progesterone receptor, and human epidermal growth factor receptor-2 status in breast cancer: A retrospective study of 5436 women from a regional cancer center in South India. South Asian J Cancer 2018;7:7-10. [Crossref] [PubMed]
- da Silva JL, Cardoso Nunes NC, Izetti P, et al. Triple negative breast cancer: A thorough review of biomarkers. Crit Rev Oncol Hematol 2020;145:102855. [Crossref] [PubMed]
- DeSantis CE, Ma J, Goding Sauer A, et al. Breast cancer statistics, 2017, racial disparity in mortality by state. CA Cancer J Clin 2017;67:439-48. [Crossref] [PubMed]
- Lima ZS, Ghadamzadeh M, Arashloo FT, et al. Recent advances of therapeutic targets based on the molecular signature in breast cancer: genetic mutations and implications for current treatment paradigms. J Hematol Oncol 2019;12:38. [Crossref] [PubMed]
- El Hachem G, Gombos A, Awada A. Recent advances in understanding breast cancer and emerging therapies with a focus on luminal and triple-negative breast cancer. F1000Res 2019. [Crossref] [PubMed]
- Costa RLB, Gradishar WJ. Triple-Negative Breast Cancer: Current Practice and Future Directions. J Oncol Pract 2017;13:301-3. [Crossref] [PubMed]
- Kim C, Gao R, Sei E, et al. Chemoresistance Evolution in Triple-Negative Breast Cancer Delineated by Single-Cell Sequencing. Cell 2018;173:879-93.e13. [Crossref] [PubMed]
- Staaf J, Glodzik D, Bosch A, et al. Whole-genome sequencing of triple-negative breast cancers in a population-based clinical study. Nat Med 2019;25:1526-33. [Crossref] [PubMed]
- Kim IS, Gao Y, Welte T, et al. Immuno-subtyping of breast cancer reveals distinct myeloid cell profiles and immunotherapy resistance mechanisms. Nat Cell Biol 2019;21:1113-26. [Crossref] [PubMed]
- Sharma P, Barlow WE, Godwin AK, et al. Validation of the DNA Damage Immune Response Signature in Patients With Triple-Negative Breast Cancer From the SWOG 9313c Trial. J Clin Oncol 2019;37:3484-92. [Crossref] [PubMed]
- Jiang YZ, Ma D, Suo C, et al. Genomic and Transcriptomic Landscape of Triple-Negative Breast Cancers: Subtypes and Treatment Strategies. Cancer Cell 2019;35:428-40.e5. [Crossref] [PubMed]
- Li H, Wang X, Fang Y, et al. Integrated expression profiles analysis reveals novel predictive biomarker in pancreatic ductal adenocarcinoma. Oncotarget 2017;8:52571-83. [PubMed]
- Wang Z, Yang B, Zhang M, et al. lncRNA Epigenetic Landscape Analysis Identifies EPIC1 as an Oncogenic lncRNA that Interacts with MYC and Promotes Cell-Cycle Progression in Cancer. Cancer Cell 2018;33:706-20.e9. [Crossref] [PubMed]
- Zhao W, Ajani JA, Sushovan G, et al. Galectin-3 Mediates Tumor Cell-Stroma Interactions by Activating Pancreatic Stellate Cells to Produce Cytokines via Integrin Signaling. Gastroenterology 2018;154:1524-37.e6. [Crossref] [PubMed]
- Sanchez A, Furberg H, Kuo F, et al. Transcriptomic signatures related to the obesity paradox in patients with clear cell renal cell carcinoma: a cohort study. Lancet Oncol 2020;21:283-93. [Crossref] [PubMed]
- Shaheen S, Fawaz F, Shah S, et al. Differential Expression and Pathway Analysis in Drug-Resistant Triple-Negative Breast Cancer Cell Lines Using RNASeq Analysis. Int J Mol Sci 2018. [Crossref] [PubMed]
- Peng C, Ma W, Xia W, et al. Integrated analysis of differentially expressed genes and pathways in triplenegative breast cancer. Mol Med Rep 2017;15:1087-94. [Crossref] [PubMed]
- Szklarczyk D, Morris JH, Cook H, et al. The STRING database in 2017: quality-controlled protein-protein association networks, made broadly accessible. Nucleic Acids Res 2017;45:D362-8. [Crossref] [PubMed]
- Liu W, Ouyang S, Zhou Z, et al. Identification of genes associated with cancer progression and prognosis in lung adenocarcinoma: Analyses based on microarray from Oncomine and The Cancer Genome Atlas databases. Mol Genet Genomic Med 2019;7:e00528. [PubMed]
- Hou GX, Liu P, Yang J, et al. Mining expression and prognosis of topoisomerase isoforms in non-small-cell lung cancer by using Oncomine and Kaplan-Meier plotter. PLoS One 2017;12:e0174515. [Crossref] [PubMed]
- Xiong J, Zhang X, Chen X, et al. Prognostic roles of mRNA expression of notch receptors in non-small cell lung cancer. Oncotarget 2017;8:13157-65. [PubMed]
- Wu P, Heins ZJ, Muller JT, et al. Integration and Analysis of CPTAC Proteomics Data in the Context of Cancer Genomics in the cBioPortal. Mol Cell Proteomics 2019;18:1893-8. [Crossref] [PubMed]
- Fabbri F, Salvi S, Bravaccini S. Know your enemy: Genetics, aging, exposomic and inflammation in the war against triple negative breast cancer. Semin Cancer Biol 2020;60:285-93. [PubMed]
- Tajbakhsh A, Rivandi M, Abedini S, et al. Regulators and mechanisms of anoikis in triple-negative breast cancer (TNBC): A review. Crit Rev Oncol Hematol 2019;140:17-27. [Crossref] [PubMed]
- Vojtek M, Marques MPM, Ferreira I, et al. Anticancer activity of palladium-based complexes against triple-negative breast cancer. Drug Discov Today 2019;24:1044-58. [Crossref] [PubMed]
- He MY, Rancoule C, Rehailia-Blanchard A, et al. Radiotherapy in triple-negative breast cancer: Current situation and upcoming strategies. Crit Rev Oncol Hematol 2018;131:96-101. [Crossref] [PubMed]
- Sporikova Z, Koudelakova V, Trojanec R, et al. Genetic Markers in Triple-Negative Breast Cancer. Clin Breast Cancer 2018;18:e841-50. [Crossref] [PubMed]
- Damaskos C, Garmpi A, Nikolettos K, et al. Triple-Negative Breast Cancer: The Progress of Targeted Therapies and Future Tendencies. Anticancer Res 2019;39:5285-96. [Crossref] [PubMed]
- Shi Y, Jin J, Ji W, et al. Therapeutic landscape in mutational triple negative breast cancer. Mol Cancer 2018;17:99. [Crossref] [PubMed]
- Papadimitriou M, Mountzios G, Papadimitriou CA. The role of PARP inhibition in triple-negative breast cancer: Unraveling the wide spectrum of synthetic lethality. Cancer Treat Rev 2018;67:34-44. [Crossref] [PubMed]
- Lederer AR, La Manno G. The emergence and promise of single-cell temporal-omics approaches. Curr Opin Biotechnol 2020;63:70-8. [Crossref] [PubMed]
- Pérez-Pena J, Tibor Fekete J, Paez R, et al. A Transcriptomic Immunologic Signature Predicts Favorable Outcome in Neoadjuvant Chemotherapy Treated Triple Negative Breast Tumors. Front Immunol 2019;10:2802. [Crossref] [PubMed]
- Qiu M, Peng Q, Jiang I, et al. Specific inhibition of Notch1 signaling enhances the antitumor efficacy of chemotherapy in triple negative breast cancer through reduction of cancer stem cells. Cancer Lett 2013;328:261-70. [Crossref] [PubMed]
- De Luca A, Maiello MR, D'Alessio A, et al. The RAS/RAF/MEK/ERK and the PI3K/AKT signalling pathways: role in cancer pathogenesis and implications for therapeutic approaches. Expert Opin Ther Targets 2012;16 Suppl 2:S17-27. [Crossref] [PubMed]
- Pohl SG, Brook N, Agostino M, et al. Wnt signaling in triple-negative breast cancer. Oncogenesis 2017;6:e310. [Crossref] [PubMed]
- Rajput S, Guo Z, Li S, et al. PI3K inhibition enhances the anti-tumor effect of eribulin in triple negative breast cancer. Oncotarget 2019;10:3667-80. [Crossref] [PubMed]
- Xie W, Zhang Y, Zhang S, et al. Oxymatrine enhanced anti-tumor effects of Bevacizumab against triple-negative breast cancer via abating Wnt/beta-Catenin signaling pathway. Am J Cancer Res 2019;9:1796-814. [PubMed]
- Wright TD, Raybuck C, Bhatt A, et al. Pharmacological inhibition of the MEK5/ERK5 and PI3K/Akt signaling pathways synergistically reduces viability in triple-negative breast cancer. J Cell Biochem 2020;121:1156-68. [Crossref] [PubMed]
- Byrne DJ, Deb S, Takano EA, et al. GATA3 expression in triple-negative breast cancers. Histopathology 2017;71:63-71. [Crossref] [PubMed]
- Gromov P, Espinoza JA, Talman ML, et al. FABP7 and HMGCS2 are novel protein markers for apocrine differentiation categorizing apocrine carcinoma of the breast. PLoS One 2014;9:e112024. [Crossref] [PubMed]
- Kim S, Moon BI, Lim W, et al. Expression patterns of GATA3 and the androgen receptor are strongly correlated in patients with triple-negative breast cancer. Hum Pathol 2016;55:190-5. [Crossref] [PubMed]
- Yao Y, Chu Y, Xu B, et al. Risk factors for distant metastasis of patients with primary triple-negative breast cancer. Biosci Rep 2019. [Crossref] [PubMed]
- Wali VB, Patwardhan GA, Pelekanou V, et al. Identification and Validation of a Novel Biologics Target in Triple Negative Breast Cancer. Sci Rep 2019;9:14934. [Crossref] [PubMed]
- Han B, Zhou B, Qu Y, et al. FOXC1-induced non-canonical WNT5A-MMP7 signaling regulates invasiveness in triple-negative breast cancer. Oncogene 2018;37:1399-408. [Crossref] [PubMed]
- Zhang X, Li Q, Zhao H, et al. Pathological expression of tissue factor confers promising antitumor response to a novel therapeutic antibody SC1 in triple negative breast cancer and pancreatic adenocarcinoma. Oncotarget 2017;8:59086-102. [PubMed]