
Non-negligible factors in studying the ApoM-S1P axis using EA.hy926 cells
Introduction
In 2013, Arkensteijn et al. (1) published a review exploring the relationship between the ApoM-S1P axis and lipid metabolism. The ApoM-S1P axis consists of apolipoprotein M (ApoM), sphingosine-1-phosphate (S1P), and S1P receptor subtypes (S1PRs). First identified by Xu and Dahlbäck in 1999 (2), ApoM, an important component of high-density lipoprotein (HDL), is a member of the lipocalin protein family and a physical carrier of plasma S1P (1,3). ApoM is required for the formation and maturation of preβ-HDL particles. Since the main function of preβ-HDL is to promote the outflow of intracellular cholesterol, lack of ApoM will significantly affect the reverse transport of cholesterol, and lead to the occurrence of atherosclerosis (4,5).
A lipid regulator with biological activity, S1P and its corresponding receptors have five subtypes (S1PR1 to S1PR5), which belong to the G protein-coupled receptor family and are expressed in different cells (6). ApoM delivers S1P to S1PR1 on the cell membrane, where it plays a significant role in the barrier protective effects of endothelial cells, as demonstrated in vivo and in vitro (6). Despite the importance of the ApoM-S1P axis, the role ApoM carries out in the delivery of S1P to other S1PRs remains unclear. Further research is needed to elucidate the underlying mechanism.
The human cell line EA.hy926 and human umbilical vein endothelial cells (HUVECs) are frequently used in the exploration the pathophysiological functions of endothelial cells in vitro. HUVECs were demonstrated to express S1PR1 and S1PR3 at the nucleic acid level by Dunlap et al. (7); however, the basic properties of EA.hy926 cells in relation to functional studies of the ApoM-S1P axis remain unclear.
In the present study, the expressions of coagulation factor VIII (FVIII), ApoM, C-myc-tagged and His-tagged proteins, and S1PRs was examined, and the effect of ApoM on S1PRs in EA.hy926 cells was investigated, thus providing a basis for further study into the function of the ApoM-S1P axis.
Methods
Materials
EA.hy926 cells (American Type Culture Collection, ATCC) were grown in 60-mm Petri dishes in Dulbecco’s Modified Eagle Medium (DMEM, with 4.5 g/L glucose, L-glutamine, and sodium pyruvate) with 10% FBS. Cells were cultured at 37 °C in 5% CO2 and passaged with trypsin-EDTA. Then, the cell culture medium was changed to DMEM with 1% fatty acid-free Bovine Serum Albumin (BSA) and cultured for 24 h before the experiment. ApoM antibody was purchased from GeneTex (CA, USA), and FVIII, S1PR2, S1PR4, S1PR5, and C-myc tag antibodies were obtained from Santa Cruz (CA, USA). S1PR1, S1PR3, and His tag antibodies and FITC-conjugated secondary antibodies were purchased from Abcam (Cambridge, UK). Cy3-conjugated secondary antibody was purchased from Bethyl (TX, USA). Deep-sea fish gelatin was obtained from Sigma (CA, USA). 4'6-diamidino-2-phenylindole (DAPI) dye and anti-fade solution were sourced from Boster Biological Engineering Company (Wuhan, China). Cell culture plates (6-well and 24-well) were purchased from Costar (NY, USA).
Lentivirus infection
EA.hy926 cells were infected with lentiviruses (Genechem, China), in line with the manufacturer’s instructions. Cells (2×105/well) were seeded in six-well plates and cultured for 24 h. Then, cells were infected with recombinant lentiviruses carrying the ApoM gene sequence (NM_019101, overexpression group, OE group) and lentiviruses carrying no ApoM gene sequence (negative control group, NC group), and exposed to polybrene. The transduced cells were positive for green fluorescent protein (GFP), and green fluorescence was observed by fluorescence microscopy 48 h later.
Immunofluorescence staining
Endogenous FVIII, ApoM, S1PR1-5, C-myc, and His-tagged proteins in EA.hy926 cells were detected by immunofluorescence staining. EA.hy926 cells were plated into 24-well plates at a density of 5×104/mL in 500 µL/well. When cells reached 60–70% confluence, immunofluorescence detection was performed. Samples were fixed in a 4% paraformaldehyde solution for 15 min and washed 3 times with PBS. They were then exposed to 0.3% Triton X-100 solution for 10 min and washed 3 times before the addition of 1% fish gelatin for 1 h to block nonspecific binding sites. Incubation with specific antibodies (1:200 dilution) took place at 4 °C overnight. An equal volume of 1% BSA was added to the control group instead of primary antibody, before the samples were washed 3 times, incubated with FITC-conjugated (1:1,000 dilution) or Cy3-conjugated secondary antibody (1:200 dilution) at 37 °C for 1 h in the dark, and washed with PBS 3 more times. The nucleus was stained with DAPI for 6 min, and samples were washed 3 times prior to the addition of anti-fade solution. An immunofluorescence microscope (Olympus IX73) was used to capture fluorescence images, which were then processed with CellSens software.
RNA extraction and Duplex fluorescence RT-qPCR.
EA.hy926 cells were seeded into 6-well plates at a density of 1×105/mL in 2 mL/well. Total RNA was extracted with an RNA purification kit (Shenergy Biocolor Bioscience and Technology, China) once cells reached 80–90% confluence, according to the manufacturer’s instructions. Each group had six wells and the process was repeated twice. A spectrophotometer (at 260/280 nm) assisted with the detection of the RNA quality. Aliquots of 2 µg RNA were reverse transcribed to cDNA with a First Strand cDNA Synthesis Kit (Thermo Fisher Scientific, USA). The PCR primers and probe sets are listed in Table 1 (synthesized by Sangon Biotech Co. Ltd., China) and were designed using Primer Premier 6.00 (Premier Biosoft International, USA). The levels of target gene mRNA were quantified relative to the internal reference gene mRNA (8). RT-PCR was performed in 25 µL volumes containing 2 µL cDNA, 0.04 µL of 100 µM S1PR or ApoM primer and probe, 0.04 µL of 100 µM GAPDH primer and probe, 0.25 µL Taq DNA polymerase, 2.5 µL 10 × PCR buffer, 2.5 µL MgCl2 (25 mM), 0.5 µL 4 × dNTPs (10 mM), and 17.01 µL water. The PCR conditions were as follows: 95 °C for 3 min followed by 40 cycles of 95 °C for 5 s, and 58 °C for 12 s (S1PR2 amplification curves were analyzed at 40 and 50 PCR cycles). The relative mRNA expression was calculated by the 2−ΔΔCt method.

Full table
Western blotting
After 48 h of transfection, total cellular protein was harvested, and bicinchoninic acid assays were carried out to measure protein concentration. Each group had three wells and the process was repeated twice. Protein lysates (60, 30, or 15 µg) were separated by 10% SDS-PAGE, before transferral to polyvinylidene fluoride membranes, which were blocked with 3% BSA buffer and incubated with S1PRs and GAPDH primary antibodies. After extensive washing, membranes were incubated along with goat anti-mouse or goat anti-rabbit secondary antibodies at 37 °C for 2 h. Following incubation in ECL luminescence solution (Pierce, USA), the light signal was visualized by exposing blots to radiographic film. The radiographic film was scanned with Microtek ScanMaker i800. Quantification was performed with Quantity One software (BioRad, Germany), and GAPDH was used as a loading control to normalize target protein expressions.
Statistics
All data are expressed as mean ± SD. Statistical analysis was performed in GraphPad Prism version 6.0 (GraphPad Software, USA). Student’s t-test (two-tailed) facilitated comparisons between two groups to be performed. A value of P<0.05 was considered to be statistically significant.
Results
Identification of FVIII, ApoM, S1PR1-5, C-myc, and His-tagged proteins in EA.hy926 cells
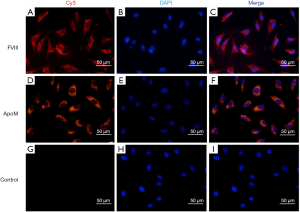
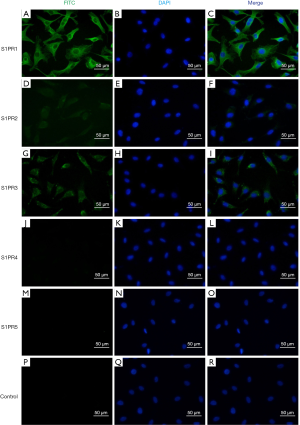
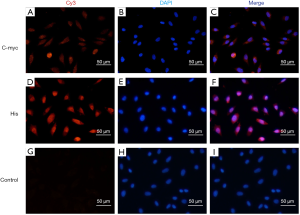
Immunofluorescence staining demonstrated that EA.hy926 cells expressed FVIII and ApoM, with FVIII evenly distributed in EA.hy926 in contrast to ApoM, which was strongly expressed and widely distributed in the cell (Figure 1). S1PR1–3 expression was detected in EA.hy926 cells, whereas S1PR4 and S1PR5 were barely detected (Figure 2). S1PR1 was highly expressed and showed a dense, patchy distribution, whereas S1PR2 and S1PR3 were expressed at low levels and showed a low-density punctate distribution. The immunofluorescence staining results also showed that C-myc and His-tagged proteins were uniformly and densely distributed in the EA.hy926 cells (Figure 3). No signals were detected when immunostaining was performed in the absence of primary antibodies.
S1PRs mRNA and protein expression in EA.hy926 cells
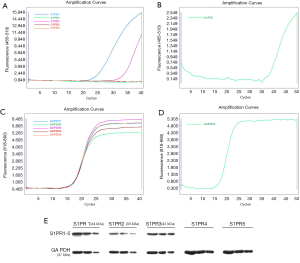
The generation of smooth amplification curves of S1PRs genes took place in the FAM channel (465–510 nm, Figure 4A,B). The GAPDH gene amplification curve was produced from the same cDNA amplification that appeared in the CY5 channel (618–660 nm, Figure 4C,D). The expression of S1PR1 and S1PR3 mRNA was detected in EA.hy926 cells in 40 cycles of PCR, whereas S1PR2, S1PR4, and S1PR5 were not detected. When the number of PCR cycles increased to 50, S1PR2 was detected, and the CT value was 40.3. Western blotting detection of S1PR1–5 in the EA.hy926 cells indicated that S1PR1, S1PR2, and S1PR3 were expressed in cells, whereas S1PR4 and S1PR5 were not (Figure 4E).
ApoM overexpression in EA.hy926 cells
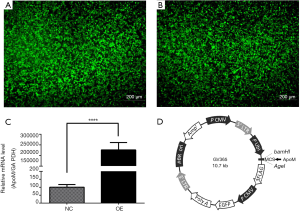
Cells infected with GFP-tagged overexpression lentiviruses and mock-control lentiviruses were labeled as the OE and NC groups, respectively. In the OE group, ApoM mRNA levels were significantly increased (P<0.0001, Figure 5; ~2,000-fold). The vector map used to construct recombinant lentiviruses is shown in Figure 5D. The results demonstrated the successful establishment of a high-expression ApoM cell model.
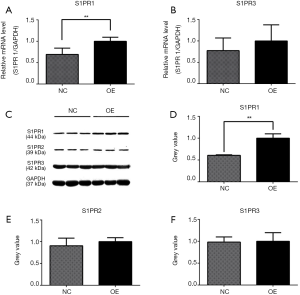
The effects of ApoM on S1PR1-3 mRNA and protein expression
The assessment of S1PR1–3 mRNA levels in the OE and NC cells showed that the expression of S1PR1 mRNA was 1.44-fold higher (P=0.0015) in the OE group than in the NC group (Figure 6A). There was no significant difference between the S1PR3 mRNA levels of the two groups (Figure 6B). S1PR2 mRNA was not detected in the two groups, even when the number of PCR cycles was increased to 50. To determine whether ApoM affected S1PR1–3 protein expression, the gray scale value of the protein bands from the Western blotting was analyzed (Figure 6C). The overexpression of ApoM increased S1PR1 protein expression 1.65–fold (P=0.0025, Figure 6D), whereas there were no significant differences observed between the S1PR2 and S1PR3 protein levels of the two groups (Figure 6E,F).
Discussion
HUVECs, which are derived from the maternal umbilical cord, are commonly used in the study of vascular endothelial cell function in in vitro experiments. However, nutrient-rich conditions are needed for their growth. Even so, they grow slowly after 6–8 passages. EA.hy926 cells have been fused with HUVECs and thioguanine-resistant A549 lung cancer cells, which are induced by polyethylene glycol. The resulting cell line shares the characteristics of endothelial cells and is able to grow and proliferate indefinitely (9). Because the culturing of EA.hy926 cells can be carried out in vitro quickly and easily, EA.hy926 cells served as a model in this study. The results indicated that FVIII and endogenous ApoM were expressed by EA.hy926 cells. FVIII expression and secretion are characteristics of endothelial cells which point towards the source of EA.hy926 cells. As a secretory protein, ApoM primarily localizes to liver and kidney tissues, and it is secreted by hepatocytes and renal tubular epithelial cells (10). The results of this study showed EA.hy926 cells to express ApoM, and this could be applied to future studies of the ApoM-S1P axis. However, only S1PR1 has thus far been reported to be affected by ApoM in EA.hy926 cells (11), and it is still not clear whether expression of other S1PRs is affected by ApoM; we therefore examined the basal expression of S1PRs and the impact of ApoM on S1PRs in EA.hy926 cells.
In the present study, cell immunofluorescence staining, Western blotting, and RT-qPCR were used to demonstrate that EA.hy926 cells highly expressed S1PR1, slightly expressed S1PR3, weakly expressed S1PR2, and did not express S1PR4 and S1PR5. We also showed that the levels of S1PR1 were significantly higher in EA.hy926 cells infected with lentivirus carrying the ApoM gene than in control cells, which is consistent with the findings of our previous research (11). In contrast, S1PR3 expression was not affected, indicating that ApoM upregulates S1PR1 mRNA and protein expression. Our results indicated that ApoM carried S1P and played a physiological role in endothelial cells, mainly through S1PR1. However, S1PR1 expression was affected by ApoM itself and needed to be considered. Since S1PR4 and S1PR5 are not expressed by EA.hy926 cells, it is suggested that it is not necessary for researchers to detect them in future experiments. S1P is transported by ApoM or albumin to the membrane in effector cells, where it binds different receptor subtypes and activates multiple intracellular signaling pathways in target cells. S1PR1 is widely distributed and highly expressed in endothelial cells, lymphocytes, and astrocytes (12). Its main physiological functions include neuronal migration, immune regulation, cardiovascular and nervous system embryonic development, angiogenesis, and endothelial barrier function (13-17). ApoM may promote S1PR1 expression and thus play a part in vascular protection, in turn providing a basis for further research on the physiological role of S1P in S1PR1 activation in the cell membrane.
The results of this study showed S1PR2 to be weakly expressed in EA.hy926-untreated cells; however, the lentiviral-infected cells, including those in the OE and NC groups, showed undetectable levels of S1PR2 mRNA. There are three possible explanations for these findings: (I) the expression of S1PR2 mRNA may be extremely low and could not be detected, despite an increase in the number of PCR amplification cycles to 50; (II) the S1PR2 protein band may be non-specific or produced by cross-reaction with other receptors; and (III) lentivirus infection may affect growth and S1PR2 expression in EA.hy926 cells. Therefore, other cell lines and detection methods could assist with further verifying our results.
C-myc and His tags are widely used in cell experiments (18-20), such as for the localization of proteins in cellular immunofluorescence assays, and as indicator proteins in Western blots. Exogenous recombinant ApoM with tag proteins could be applied in in vitro studies to ascertain whether ApoM acts as a mediator for the delivery of S1P to S1PRs on the cell membrane, as exogenous ApoM can be directly localized by using antibodies against tag proteins, thereby distinguishing it from endogenous ApoM. The present results showed that EA.hy926 cells expressed C-myc and His-tag proteins, which were therefore not suitable as exogenous tag proteins for recombinant ApoM. His-tag is currently the most commonly used tag for prokaryotic protein expression. We detected His-tag expression in EA.hy926 cells by immunofluorescence and analyzed the possible causes. When the EA.hy926 cell line was established, the primary cultured human umbilical vein cells were fused with a thioguanine-resistant A549 clone inserting the His-tag, but this was not specifically mentioned in the article on establishing EA.hy926 cell line (21).
In summary, the EA.hy926 cells with the best macro-vascular characteristics expressed ApoM and S1PRs, which points towards their potential value for in vitro studies of the ApoM-S1P axis. ApoM overexpression would carry more S1P, promoting the expression of S1PR1, and thereby transferring more S1P to bind to S1PR1, which plays a role in vascular endothelial cell protection. However, the specific underlying mechanism is yet to be determined.
Acknowledgments
We thank International Science Editing for language editing.
Funding: This research was supported by the National Natural Science Foundation of China (No. 81370372), the Natural Science Foundation of Jiangsu Province (No. BK20151179 and No. BK20191158), Jiangsu Provincial Youth Medicine Key Talent Project (No. QNRC2016282), the International Cooperation Foundation of Changzhou (No. CZ20160013 and No. CZ20190022), and Major Science and Technology Project of Changzhou Health Commission (No. ZD201804).
Footnote
Conflicts of Interest: The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Arkensteijn BW, Berbee JF, Rensen PC, et al. The apolipoprotein m-sphingosine-1-phosphate axis: biological relevance in lipoprotein metabolism, lipid disorders and atherosclerosis. Int J Mol Sci 2013;14:4419-31. [Crossref] [PubMed]
- Xu N, Dahlback B. A novel human apolipoprotein (apoM). J Biol Chem 1999;274:31286-90. [Crossref] [PubMed]
- Christoffersen C, Obinata H, Kumaraswamy SB, et al. Endothelium-protective sphingosine-1-phosphate provided by HDL-associated apolipoprotein M. Proc Natl Acad Sci U S A 2011;108:9613-8. [Crossref] [PubMed]
- Wolfrum C, Poy MN, Stoffel M. Apolipoprotein M is required for prebeta-HDL formation and cholesterol efflux to HDL and protects against atherosclerosis. Nat Med 2005;11:418-22. [Crossref] [PubMed]
- Nádró B, Juhasz L, Szentpeteri A, et al. The role of apolipoprotein M and sphingosine 1-phosphate axis in the prevention of atherosclerosis. Orv Hetil 2018;159:168-75. [PubMed]
- Maceyka M, Harikumar KB, Milstien S, et al. Sphingosine-1-phosphate signaling and its role in disease. Trends Cell Biol 2012;22:50-60. [Crossref] [PubMed]
- Dunlap KA, Kwak HI, Burghardt RC, et al. The sphingosine 1-phosphate (S1P) signaling pathway is regulated during pregnancy in sheep. Biol Reprod 2010;82:876-87. [Crossref] [PubMed]
- Yu MM, Yao S, Luo KM, et al. Apolipoprotein M increases the expression of vitamin D receptor mRNA in colorectal cancer cells detected with duplex fluorescence reverse transcription-quantitative polymerase chain reaction. Mol Med Rep 2017;16:1167-72. [Crossref] [PubMed]
- Bouïs D, Hospers GA, Meijer C, et al. Endothelium in vitro: a review of human vascular endothelial cell lines for blood vessel-related research. Angiogenesis 2001;4:91-102. [Crossref] [PubMed]
- Zhang XY, Dong X, Zheng L, et al. Specific tissue expression and cellular localization of human apolipoprotein M as determined by in situ hybridization. Acta Histochem 2003;105:67-72. [Crossref] [PubMed]
- Wang M, Luo GH, Liu H, et al. Apolipoprotein M induces inhibition of inflammatory responses via the S1PR1 and DHCR24 pathways. Mol Med Rep 2019;19:1272-83. [PubMed]
- Chae SS, Proia RL, Hla T. Constitutive expression of the S1P1 receptor in adult tissues. Prostaglandins Other Lipid Mediat 2004;73:141-50. [Crossref] [PubMed]
- Li C, Li JN, Kays J, et al. Sphingosine 1-phosphate enhances the excitability of rat sensory neurons through activation of sphingosine 1-phosphate receptors 1 and/or 3. J Neuroinflammation 2015;12:70. [Crossref] [PubMed]
- Baeyens A, Fang V, Chen C, et al. Exit Strategies: S1P Signaling and T Cell Migration. Trends Immunol 2015;36:778-87. [Crossref] [PubMed]
- Clay H, Wilsbacher LD, Wilson SJ, et al. Sphingosine 1-phosphate receptor-1 in cardiomyocytes is required for normal cardiac development. Dev Biol 2016;418:157-65. [Crossref] [PubMed]
- Pi J, Tao T, Zhuang T, et al. A MicroRNA302-367-Erk1/2-Klf2-S1pr1 Pathway Prevents Tumor Growth via Restricting Angiogenesis and Improving Vascular Stability. Circ Res 2017;120:85-98. [Crossref] [PubMed]
- Sun X, Mathew B, Sammani S, et al. Simvastatin-induced sphingosine 1-phosphate receptor 1 expression is KLF2-dependent in human lung endothelial cells. Pulm Circ 2017;7:117-25. [Crossref] [PubMed]
- Jaldin-Fincati JR, Bilan PJ, Klip A. GLUT4 Translocation in Single Muscle Cells in Culture: Epitope Detection by Immunofluorescence. Methods Mol Biol 2018;1713:175-92.
- Zhang Y, Wu S, Song S, et al. Generation and characterization of a potentially applicable Vero cell line constitutively expressing the Schmallenberg virus nucleocapsid protein. Cytotechnology 2017;69:145-56. [Crossref] [PubMed]
- Huang M, Chen R, Ren G. Secretory expression and purification of Bacillus licheniformis keratinase in insect cells. PLoS One 2017;12:e0183764. [Crossref] [PubMed]
- Edgell CJ, McDonald CC, Graham JB. Permanent cell line expressing human factor VIII-related antigen established by hybridization. Proc Natl Acad Sci U S A 1983;80:3734-7. [Crossref] [PubMed]


