
Acidic leucine-rich nuclear phosphoprotein-32A expression contributes to adverse outcome in acute myeloid leukemia
Introduction
Acute myeloid leukemia (AML) is a clinically disparate hematopoietic malignancy, characterized by predominant fatality and high heterogeneity (1). It is the most common type of adult acute leukemia with an annual incidence of 3–8/105 (2). To date, cytogenetic analysis has been one of the most useful prognostic methods to classify AML patients into different prognostic risk groups (favorable, intermediate, unfavorable) (3). The cytogenetically normal AML (CN-AML), representing 40–50% AML patients, is the largest subset of de-novo AML cases (4,5). Although CN-AML constitutes the main body of the intermediate-risk group, yet different molecular subtypes of CN-AML diverged in transcription and DNA methylation profiles (6). Moreover, due to the wide variety of clinical outcome, the prognosis of CN-AML cannot be predicted solely by cytogenetics (7). Intriguingly, without any microscopically detectable chromosome abnormalities in its leukemic blast, CN-AML harbour aberrantly expressed genes and microRNAs, mutations, epigenetic changes that can be used as potential prognostic markers (8). It is now recognized that WT1, ASXL1, DNMT3A, FLT3-ITD and TET2 mutations presence in CN-AML represent a subgroup of patients with unfavorable prognosis, while NPM1 and CEBPA are associated with favorable prognosis (9-15). Furthermore, unfavorable prognostic factors include high expression of CPT1A, ATP1B1, RUNX1, MAPKBP1, MAP7, ERG, DNMT3B, miR-3151, miR-155, etc. (16,17). As leukemogenic mechanisms of CN-AML still remains unclear, it is vital to continue identifying new molecular biomarkers of clinical significance.
Acidic leucine-rich nuclear phosphoprotein-32A (ANP32A), also known as PP32, is an acidic leucine-rich nuclear phosphoprotein which has shown to be divergently expressed in normal tissues as well as in certain types of tumor cells (18). On one hand, ANP32A correlates with cancer progression and metastasis, demonstrated to be a candidate biomarker in patients with pancreatic tumor, colorectal cancer, hepatocellular carcinoma, oral squamous cell carcinoma, etc. (19,20). On the other hand, ANP32A is a tumor suppressor in prostate cancer, non-small cell lung cancer and breast cancer by stimulating apoptosis (18,21,22). Recent study indicates that ANP32A is a novel regulator of histone H3 acetylation and promotes leukemogenesis in AML, suggesting the expression of ANP32A might be related to the prognosis of CN-AML patients (23). Therefore, the focus on the exact functions of ANP32A in hematopoietic malignancy might lead us to the identification of ANP32A as a potential molecular marker of clinical significance in CN-AML.
Here, we presented ANP32Ahigh as an unfavorable prognostic biomarker for CN-AML. We also explored ANP32A-associated genomic and epigenomic patterns to further elucidate its function. Our study represented direct evidences for ANP32A being a prognostic biomarker in AML risk stratification and a potential therapeutic target for patients with AML.
Methods
Patients and treatment
The study was approved by the local institutional review boards. In accordance with the Declaration of Helsinki, all patients provided written informed consent. The treatment of all patients was uniformly under the protocols of Dutch-Belgian Cooperative Trial Group of Hematology-Oncology (details see in http://www.hovon.nl).
The first cohort of this study was derived from a whole AML cohort, containing 185 primarily untreated CN-AML patients, all diagnosed at Erasmus University Medical Center in Rotterdam from 1990 to 2008. The cohort age ranged from 16 to 60 years old and the median age was 47 years old. All samples were collected at the time of diagnosis and contained more than 80% blast cells after thawing (24). At least 20 metaphases from bone marrow (BM) samples were examined by conventional cytogenetic methods to diagnose normal karyotype. The presence of FLT3-ITD, FLT3-TKD and the mutations of IDH1, IDH2, NPM1, CEBPA, K-RAS, N-RAS were all assessed by reverse transcription-polymerase chain reaction (RT-PCR) assays. The clinical, cytogenetic, molecular and gene expression data of these AML cases can be downloaded from the Gene Expression Omnibus (accession number: GSE6891, details see in http://www.ncbi.nlm.nih.gov/geo) (25).
The second cohort of this study contained 162 CN-AML patients with uniform treatment was used to validate findings in the first cohort. All patients received intensive double-induction and consolidation-chemotherapy in multicenter AMLCG-1999 trial from 1999 to 2003. The cohort age ranged from 17 to 83 years old and the median age was 57.5 years old. The gene expression data can be downloaded publicly (accession number: GSE12417) (26).
Microarray analysis
Gene expression was obtained from published microarray data using Affymetrix Human Genome 133 plus 2.0 as well as U133A Gene Chips (accession number: GSE9476, GSE1159, GSE6891, GSE12417) (24,25,27,28). Seventy-three CN-AML patients with mRNA, microRNA and methylation data were derived from The Cancer Genome Atlas (TCGA) (29). Design, data quality control and normalization of microarray experiments were in accordance with the standard Affymetrix protocols. Expression of mRNA and microRNA were obtained by high throughout transcriptome sequencing (RNA-seq), while methylation data was obtained by Illumina Infinium 450K BeadChips. The expression level of ANP32A was standardized in normally distributed. Then the appropriate cut-off subdivision value was compared by the 4 quartiles of 185 CN-AML patients, which the median value showed evident distinction (Figure S1A,B,C). Therefore, median value of ANP32A expression was used to classify patients into ANP32Ahigh and ANP32Alow groups. The expression levels of ERG, DNMT3A, BAALC, WT1 and other genes were obtained using the same strategy.

Statistical analysis
Overall survival (OS) was defined as the time from diagnosis date to death by any causes. Event free survival (EFS) was defined as the time from diagnosis date to the removal from the study because of the end of complete remission (CR), relapse or death (the censorship equals to 1 when death event was observed). The Kaplan-Meier method and the log-rank test were used to evaluate and validate the association between ANP32A expression and OS, EFS. The fisher exact and the Wilcoxon rank-sum test, analyzing categorical and continuous variables respectively, were used to estimate the association between ANP32A expression levels and the patients’ clinical, molecular characteristics. Multivariate Cox proportional hazard models were used to assess the effect of ANP32A expression on OS and EFS with other known risk factors in presence. Except ANP32A and other 3 clinical & demographical variables were chosen, two gene mutations were further brought into our investigation which were shown to have a strong association with AML (30,31). Student’s t-test as well as multiple hypothesis correction (False Discovery Rate, FDR) were used to identify discrepancy in gene expression, microRNA expression, and DNA methylation profiles between ANP32Ahigh and ANP32Alow groups. All analyses were performed using R3.2.3 and its related software packages.
Results
Overexpression of ANP32A in AML patients
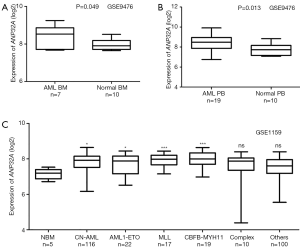
ANP32A expression were analyzed in two microarray assays, including BM and peripheral blood samples (PB) in AML patients and healthy donors. The expression of ANP32A was remarkably high in AML BM than in Normal BM (Figure 1A, P=0.049, 7 AML BM samples vs. 10 normal BM samples, GEO accession number GSE9476). Overexpression of ANP32A was further validated by PB samples in the same microarray assay (Figure 1B, P=0.013, 19 AML PB samples vs. 10 normal PB samples). Furthermore, overexpression of ANP32A was found in different subgroups in AML patients than normal BM, including 116 CN-AML, 22 AML1-ETO, 17 MLL-translocation, and 19 CBFβ-MYH11 (Figure 1C, GEO accession number GSE1159). These results indicated that ANP32A was evidently overexpressed in AML patients.
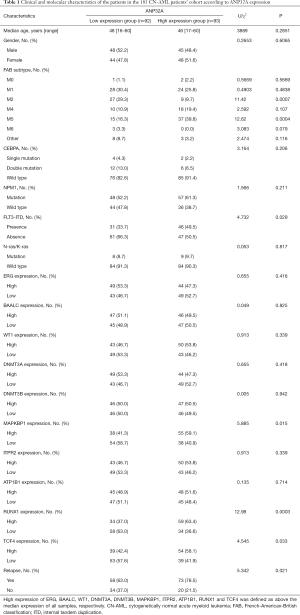
Clinical and molecular characteristics between ANP32Alow and ANP32Ahigh group in AML patients
In a cohort of 185 CN-AML patients derived from GSE6891 assay (435 AML patients, no M3), ANP32Ahigh group tends to have less FAB-M2 subtype patients (P=0.0007) and more FAB-M5 patients (P=0.0004). ANP32A was significantly overexpressed in patients with FLT3-ITD (P=0.029), while no significant differences between ANP32Alow and ANP32Ahigh group were observed in CEBPA, NPM1 and N-ras/K-ras mutation groups (P=0.206, P=0.211, and P=0.817, respectively). Besides, ANP32Ahigh group was more likely to have a higher expression of MAPKBP1, RUNX1, and TCF4, which were known adverse prognostic biomarkers in AML. Furthermore, patients in ANP32Ahigh group was more likely to relapse than those in ANP32Alow group (Table 1).
Full table
Overexpression of ANP32A was associated with unfavorable outcome in AML patients
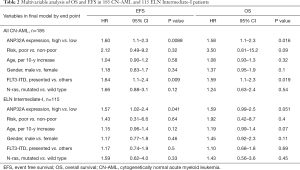
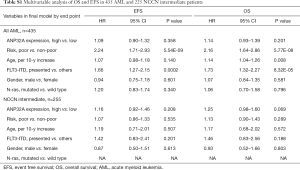
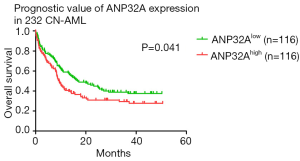
As patients in ANP32Ahigh group was more likely to relapse than those in ANP32Alow group in the cohort of 185 CN-AML patients (Table 1, P=0.021), the survival analysis was further carried out in a whole cohort of CN-AML patients (n=185) and ELN Intermediate-I patients (n=115). ANP32Ahigh group showed significantly shorter OS and EFS in CN-AML patients’ cohort (Figure 2A,B, ANP32Ahigh vs. ANP32Alow: median OS: 11.9 vs. 37.4 months, P=0.012; median EFS: 8.4 vs. 18.3 months, P=0.005) as well as in ELN Intermediate-I patients’ cohort (Figure 2C,D, ANP32Ahigh vs. ANP32Alow: median OS: 8.5 vs. 30.4 months, P=0.018; median EFS: 6.9 vs. 13.0 months, P=0.045). To further validate the prognostic significance of ANP32A expression, multivariable analysis was carried out in these two cohorts. It showed that ANP32Ahigh group had 0.47 times higher risks on EFS and 0.46 times higher risks on OS in CN-AML patients’ cohort (Table 2, P=0.0088 and P=0.016, respectively). The presence of FLT3-ITD was another adverse factor (P=0.009 in EFS and P=0.019 in OS). As for the ELN Intermediate-I patients’ cohort, ANP32Ahigh group had 0.45 times higher risks on EFS and 0.48 times higher risks on OS (Table 2, P=0.041 and P=0.051, respectively), indicating ANP32A expression had more prognostic significance in CN-AML cohort. In order to further investigate the prognostic value of ANP32A expression, survival analysis was also performed in 435 non-M3 AML patients and 255 NCCN Intermediate Risk AML patients. The results indicated a remarkable shorter OS and EFS of ANP32Ahigh group in 435 non-M3 AML patients’ cohort (Figure 2E,F, ANP32Ahigh vs. ANP32Alow: median OS: 16.9 vs. 24.2 months, P=0.034; median EFS: 9.0 vs. 13.1 months, P=0.011) and 255 NCCN Intermediate Risk AML patients’ cohort (Figure 2G,H, ANP32Ahigh vs. ANP32Alow: median OS: 16.5 vs. 24.7 months, P=0.048; median EFS: 9.0 vs. 14.1 months, P=0.039). However, the multivariate analysis in these two cohorts didn’t confirm the independent prognostic value of ANP32A (see Table S1), further indicating the prognostic significance of ANP32A in CN-AML. To validate the unfavorable outcome associated with ANP32Ahigh in AML patients, another survival analysis was conducted in another independent cohort of 232 CN-AML patients derived from GSE12417 assay. ANP32Ahigh group showed significantly shorter OS in CN-AML patient cohort (Figure S2, ANP32Ahigh vs. ANP32Alow: median OS: 9.4 vs. 16.7 months, P=0.041).

Full table
Full table
Genome-wide gene-expression profiles associated with ANP32A expression
In order to further investigate the biological value of ANP32A in leukemogenesis, we derived ANP32A-associated gene expression profiles from genome-wide microarray analysis in 73 CN-AML patients from TCGA dataset; 1,401 up-regulated and 543 down-regulated genes were found to be significantly associated with ANP32Ahigh group (FDR-adjusted P<0.001, Figure 3A,B). The up-regulated genes included: (I) genes encoded transcription factor proteins: MYB, PHF10 (32); (II) independent unfavorable prognostic factor in CN-AML: ERG (33); (III) gene encoded cohesion complex: SMC3 (34); (IV) gene involved in phosphatases: PTPN11 (34); (V) genes regulated cell growth and signal transduction pathways: ANXA family genes (35); (VI) genes promoted tumorigenesis: CDK4 (36); ZPF91, involved in leukemogenesis and could activate HIF1a via NF-kB to promote tumorigenesis (37); SRSF family genes (SRSF1, SRSF3, SRSF10), which were oncogene and overexpressed in multiple cancers to promote cell proliferation and transformation (38); (VII) gene related to ribosome biogenesis: NPM1 (39); (VIII) gene involved in fine-tuning hematopoietic stem and progenitor cell homeostasis: PHF6 (40). The down-regulated genes included: (I) gene inactivated in solid tumor and acute lymphoblastic leukemia (ALL): CREBBP (41); (II) gene improved tumor immunogenicity and enhanced Fas-induced apoptosis: TNFRSF14 (42); (III) genes interacted with NAD metabolism and p53 signaling: SIRT family genes (SIRT2, SIRT7) (43); (IV) genes interacted with WNT-pathway and B-cell receptor signaling mediators: IRF9, BCL9L, SIT1 (44).
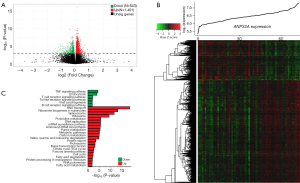
There is evidence that the uncovering of unique gene expression profiles with the identification of dysregulated signaling pathways is now providing new insights into the leukemogenesis of CN-AML (45). Therefore, data from MSigDB was used to investigate cell signaling pathways associated with ANP32A (46). A total of 6 down-regulated and 22 up-regulated pathways were found to be significantly associated with ANP32Ahigh group (P<0.05, Figure 3C). The up-regulated pathways included: (I) important molecular and biological pathways, such as RNA transport, ribosome biogenesis in eukaryotes, spliceosome, ribosome, DNA replication, protein export, proteasome, cell cycle, and RNA polymerase; (II) metabolic pathways of proteins, amino acid and nucleic acid, such as basal transcription factors, carbon metabolism, pyrimidine metabolism, purine metabolism, and valine, leucine, isoleucine degradation; (III) lipid metabolic pathways, such as fatty acid degradation and fatty acid metabolism. On the contrary, the down-regulated pathways were mostly immune-related ones, such as T and B cell receptor signaling pathway, TNF signaling pathway, Toll-like receptor signaling pathway. These dysregulated signaling pathways might explain the involvement between ANP32A and the leukemogenesis of CN-AML.
Genome-wide microRNA profiles associated with ANP32A expression
MicroRNAs have been increasingly used to diagnose and assess prognosis in hematopoietic malignancy. Dysregulated microRNAs could promote aggressive tumor phenotype, identify risk classification, and reflect therapeutic effects (47). The genome-wide microRNA profiles were analyzed and 61 microRNAs were identified to be significantly correlated to ANP32A expression, including 51 positive and 10 negative microRNAs (P<0.05, Figure 4A,B). Positively correlated microRNAs included miR-17, miR-19b-1, miR-20a, miR-25, miR-106a, miR-106b, miR-125b-1, miR-99a, miR-301b, miR-199b, miR-191, miR-629, etc. MiR-17, miR-19b-1 and miR-20a, the members of miR-17-92 cluster, were found to be upregulated in AML and helped apoptotic resistance in K562 cells (48). MiR-25 had statistically higher expression in c-kit subgroup and might be involved in leukemogenesis in pediatric AML (49). MiR-106a and miR-106b, paralogs of miR-17-92 cluster, were found to enhance proliferation of T-ALL cells and significantly upregulated in relapsed pediatric MLL-AML, respectively (50). By targeting genes associated with tumorigenesis, miR-125b-1 might act as an onco-microRNA and its overexpression alone in mice could induce tumors in multiple hematopoietic lineages (51). MiR-99a upregulation was proven to be associated with poor prognosis of AML, leading to cell expansion and progression of AML (52). MiR-301b and miR-199b were found upregulated in AML patients at diagnosis and trended to normalization after chemotherapy (53). Overexpression of miR-191 had significantly worse OS and EFS than those with low expression (54). MiR-629 was found upregulated in pediatric T-ALL, and might be used for distinguishing pediatric ALL subtypes (55). Negatively correlated microRNAs included miR-24-2, miR-26b, miR-197. MiR-24-2 was proved to be down-regulated in acute leukemia cell lines compared to hematopoietic stem progenitor cells (56). Decreased expression of miR-26b was found in T-ALL cells and it functioned as a tumor suppressor in T-ALL. Furthermore, it could inhibit the PI3K/AKT pathway, reduce proliferation and promote apoptosis of T-ALL cells (57). Overexpression of miR-197 inhibited tumor growth and prolonged OS in subcutaneous mouse xenograft model in multiple myeloma, revealing its novel role as tumor suppressor (58). These results indicated that microRNA expression might link to unfavorable outcome in ANP32Ahigh group.
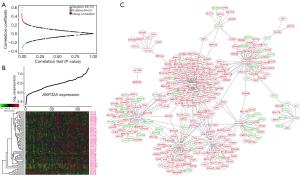
To further validate the results of the microarray platform and microRNAs, interaction network analysis was used to explore the interaction and correlation between microRNAs and their target genes (Figure 4C). The results showed the tumor suppressors repressed by the up-regulated microRNAs, led to worse outcome. For example, SIRT7, targeted by miR-629, possessed tumor-suppressing properties in breast cancer, pancreatic cancer, head and neck squamous cell carcinoma, etc. (59). KLF13, targeted by miR-106b, inhibited the transcription of MYB and BCL2 in ALL (60). Additionally, many oncogenes were the targets of the down-regulated microRNAs. SUGT1, targeted by miR-24-2, contributed to cancer development by stabilizing oncoproteins (61). FBXO45, targeted by miR-26b, exerted anti-apoptotic effect by interfering the proteasome-dependent degradation of p73, a known pro-apoptotic factor (62). These results suggested why ANP32A acted as an unfavorable prognosticator and could help provide a comprehensive view of the molecular mechanisms of ANP32A.
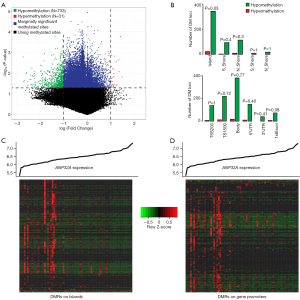
Genome-wide methylation profiles associated with ANP32A expression
Differential methylated regions (DMR) were further analyzed to uncover the different methylation patterns between ANP32Ahigh group and ANP32Alow group of CN-AML. A total of 733 hypomethylation and 31 hypermethylation DMRs were discovered from the comparison between ANP32Ahigh group and ANP32Alow group (P<0.05, |Log2(FC)| >1, Figure 5A). In addition, position distributions around CpG islands were compared in these aberrant DMRs. It showed that significantly more hypomethylation DMRs were found in the CpG islands (P=0.03, Figure 5B). Furthermore, position distributions of different structural fragments of genes were analyzed. The results indicated relatively more hypomethylation DMRs lied in 1st Exon regions (P=0.08, Figure 5B). Finally, the heatmaps showed ANP32A-associated DMRs enriched on islands and gene promoter regions (Figure 5C,D).
Discussion
Recent study provided strong evidence that ANP32A was a key factor to deregulate histone H3 acetylation, and played a role as an oncogene in leukemogenesis (23). It could interact with DNA-binding transcription factors, thus influencing gene expression (63). ANP32A acted as an oncogene in certain solid tumors, such as colorectal cancer, liver cancer, ovarian cancer, etc. (19,64). Besides, study showed ANP32A might repress ERK and subsequently inhibit RUNX1 and FLI1 to promote megakaryocyte differentiation in acute megakaryoblastic leukemia (65). In our study, we provided direct evidence that high expression of ANP32A predicted unfavorable outcomes for CN-AML. Firstly, high expression of ANP32A was shown in AML BM and PB than in normal BM and PB samples, as well as in CN-AML BM than in normal BM samples (Figure 1A,B,C). Secondly, survival prognosis with gene expression were well studied previously (66,67) as well as the various functionality of omics data (68-73). After validating in relatively large independent CN-AML cohort, high expression of ANP32A was shown to associate with unfavorable outcomes. Similar results were also shown in ELN Intermediate-I cohort, NCCN-Intermediate Risk cohort, and non-M3 AML cohort, all containing a variety of karyotypes of AML. These results were consistent with earlier findings of ANP32A, suggesting its potential role of a prognosticator and therapeutic target of AML. Furthermore, our results could provide evidence for further fine stratification of AML, especially in CN-AML, ELN Intermediate-I, NCCN-Intermediate Risk patients.
It is at present unclear about the pathogenesis of AML. However, previous studies showed the molecular biology of leukemia and identified favorable or unfavorable prognosticators, facilitating the understanding of leukemogenesis. In our study, associations between ANP32A expression and previously known prognosticators were explored in a cohort of 185 CN-AML patients. We found that ANP32Ahigh group contained significantly more M5 (P=0.004) and less M2 (P=0.007) FAB subtype, suggesting that ANP32Ahigh expressers carried more mature cells. Furthermore, ANP32Ahigh group had significantly more FLT3-ITD mutation (P=0.029), and remarkably high expression of MAPKBP1 (P=0.015), RUNX1 (P=0.0003) and TCF4 (P=0.033). all of which were known independently adverse prognosticators in AML (25). Besides, ANP32Ahigh group had more relapse patients than ANP32Alow group (P=0.021). These results indicated that ANP32A might be another independent biomarker of unfavorable outcome for CN-AML.
There is a growing number of evidences demonstrated that aberrant gene expression, microRNA post-transcriptional regulation and DNA methylation played significant role in leukemogenesis (74-77). Our study further discussed how overexpression of ANP32A affected prognosis from these three aspects. Firstly, we investigated genome-wide genes and cell signaling pathways that associated with ANP32A. Genes promoted tumorigenesis were up-regulated, such as CDK4, ZPF91, SRSF family genes, as well as ERG, an independent unfavorable prognosticator in CN-AML. On the contrary, genes inactivated in tumor and immunogenicity were down-regulated, such as CREBBP and TNFRSF14, respectively. Secondly, cell signaling pathway analysis results were in accordance with ANP32A’s unfavorable prognosticator role. Results showed up-regulation of cell cycle pathway, lipid metabolic pathway, metabolic pathways of proteins, amino acid and nucleic acid as well as down-regulation of immune-related pathways of T cell, B cell and Toll-like receptor signaling pathway and TNF signaling pathway. All these results further validated the previous study that ANP32A was a member in nuclear protein family implicated in multiple cellular pathways (78).
Furthermore, in our study, ANP32A was found to correlate positively or negatively to microRNAs, such as members of miR-17-92 cluster and miR-24-2, which played novel role in leukemogenesis or tumor suppression, respectively. Also, synchronized changes of microRNA-mRNA pairs along with ANP32A expression might contribute to the aggravation of CN-AML malignancy. For instance, the pair of positive correlated miR-629 and down regulated SIRT7, the pair of negative correlated miR-26b and up regulated FBXO45, etc. These genes, microRNAs, pathways, microRNA-mRNA pairs might be attributable to the adverse outcome of CN-AML.
Finally, genome-wide methylation profiles associated with ANP32A expression were analyzed. Results showed that more hypomethylation DMRs lied around CpG islands and in 1st Exon regions. Hypomethylation in different genic regions had been proven to exhibit a significant adverse effect on gene expression, resulting in malignant transformation and tumor progression (79). For example, in chronic lymphocytic leukemia (CLL), epigenomic analysis detected widespread DNA hypomethylation in the gene body, implicating functional and clinical roles of DNA hypomethylation in leukemogenesis (80). The hypomethylation DMRs detected in our study might possibly contribute to the unfavorable prognosis and provide deeper understanding in leukemogenesis.
In summary, we had performed a large-scale analysis of ANP32A in CN-AML. Combined with recent study that ANP32A could promote leukemogenesis, we further validated that overexpression of ANP32A was an unfavorable biomarker for CN-AML. We provided evidence that ANP32Ahigh patients had different kinds of adverse molecular characteristics. Furthermore, we highlighted the power of multi-omics analysis to provide new insights into the leukemogenic mechanisms of CN-AML and define their clinical implications.
Conclusions
We proved ANP32A is a novel, potential unfavorable prognosticator and therapeutic target for AML.
Acknowledgments
Funding: The present study was supported by grants from PLA General Hospital Science and technology Project (18KMM01) and Beijing Natural Science Foundation (7204305).
Footnote
Conflicts of Interest: The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. The study was approved by the local institutional review boards. In accordance with the Declaration of Helsinki, all patients provided written informed consent.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Papaemmanuil E, Gerstung M, Bullinger L, et al. Genomic Classification and Prognosis in Acute Myeloid Leukemia. N Engl J Med 2016;374:2209-21. [Crossref] [PubMed]
- Sanz MA, Iacoboni G, Montesinos P, et al. Emerging strategies for the treatment of older patients with acute myeloid leukemia. Ann Hematol 2016;95:1583-93. [Crossref] [PubMed]
- Shimizu H, Yokohama A, Ishizaki T, et al. Clonal evolution detected with conventional cytogenetic analysis is a potent prognostic factor in adult patients with relapsed AML. Hematol Oncol 2018;36:252-7. [Crossref] [PubMed]
- Arber DA, Orazi A, Hasserjian R, et al. The 2016 revision to the World Health Organization classification of myeloid neoplasms and acute leukemia. Blood 2016;127:2391-405. [Crossref] [PubMed]
- Qu X, Othus M, Davison J, et al. Prognostic methylation markers for overall survival in cytogenetically normal patients with acute myeloid leukemia treated on SWOG trials. Cancer 2017;123:2472-81. [Crossref] [PubMed]
- Woods BA, Levine RL. The role of mutations in epigenetic regulators in myeloid malignancies. Immunol Rev 2015;263:22-35. [Crossref] [PubMed]
- Wang M, Yang C, Zhang L, et al. Molecular Mutations and Their Cooccurrences in Cytogenetically Normal Acute Myeloid Leukemia. Stem Cells Int 2017;2017:6962379. [Crossref] [PubMed]
- Marcucci G, Yan P, Maharry K, et al. Epigenetics meets genetics in acute myeloid leukemia: clinical impact of a novel seven-gene score. J Clin Oncol 2014;32:548-56. [Crossref] [PubMed]
- Toogeh G, Ramzi M, Faranoush M, et al. Prevalence and Prognostic Impact of Wilms' Tumor 1 (WT1) Gene, Including SNP rs16754 in Cytogenetically Normal Acute Myeloblastic Leukemia (CN-AML): An Iranian Experience. Clin Lymphoma Myeloma Leuk 2016;16:e21-6. [Crossref] [PubMed]
- Azari-Yam A, Tavakkoly-Bazzaz J, Semnani Y, et al. FLT3 Gene Mutation Profile and Prognosis in Adult Acute Myeloid Leukemia. Clin Lab 2016;62:2011-7. [Crossref] [PubMed]
- Ahn JS, Kim HJ, Kim YK, et al. Adverse prognostic effect of homozygous TET2 mutation on the relapse risk of acute myeloid leukemia in patients of normal karyotype. Haematologica 2015;100:e351-3. [Crossref] [PubMed]
- Balsat M, Renneville A, Thomas X, et al. Postinduction Minimal Residual Disease Predicts Outcome and Benefit From Allogeneic Stem Cell Transplantation in Acute Myeloid Leukemia With NPM1 Mutation: A Study by the Acute Leukemia French Association Group. J Clin Oncol 2017;35:185-93. [Crossref] [PubMed]
- Sarojam S, Raveendran S, Vijay S, et al. Characterization of CEBPA Mutations and Polymorphisms and their Prognostic Relevance in De Novo Acute Myeloid Leukemia Patients. Asian Pac J Cancer Prev 2015;16:3785-92. [Crossref] [PubMed]
- Huang S, Jiang MM, Chen GF, et al. Epigenetic Silencing of Eyes Absent 4 Gene by Acute Myeloid Leukemia 1-Eight-twenty-one Oncoprotein Contributes to Leukemogenesis in t(8;21) Acute Myeloid Leukemia. Chin Med J (Engl) 2016;129:1355-62. [Crossref] [PubMed]
- Huang S, Yang H, Li Y, et al. Prognostic Significance of Mixed-Lineage Leukemia (MLL) Gene Detected by Real-Time Fluorescence Quantitative PCR Assay in Acute Myeloid Leukemia. Med Sci Monit 2016;22:3009-17. [Crossref] [PubMed]
- Díaz-Beyá M, Brunet S, Nomdedeu J, et al. The expression level of BAALC-associated microRNA miR-3151 is an independent prognostic factor in younger patients with cytogenetic intermediate-risk acute myeloid leukemia. Blood Cancer J 2015;5:e352. [Crossref] [PubMed]
- Zhou G, Cao Y, Dong W, et al. The clinical characteristics and prognostic significance of AID, miR-181b, and miR-155 expression in adult patients with de novo B-cell acute lymphoblastic leukemia. Leuk Lymphoma 2017;58:1-9. [Crossref] [PubMed]
- Schramedei K, Morbt N, Pfeifer G, et al. MicroRNA-21 targets tumor suppressor genes ANP32A and SMARCA4. Oncogene 2011;30:2975-85. [Crossref] [PubMed]
- Li C, Ruan HQ, Liu YS, et al. Quantitative proteomics reveal up-regulated protein expression of the SET complex associated with hepatocellular carcinoma. J Proteome Res 2012;11:871-85. [Crossref] [PubMed]
- Velmurugan BK, Yeh KT, Lee CH, et al. Acidic leucine-rich nuclear phosphoprotein-32A (ANP32A) association with lymph node metastasis predicts poor survival in oral squamous cell carcinoma patients. Oncotarget 2016;7:10879-90. [Crossref] [PubMed]
- Bai J, Brody JR, Kadkol SS, et al. Tumor suppression and potentiation by manipulation of pp32 expression. Oncogene 2001;20:2153-60. [Crossref] [PubMed]
- Pan W, da Graca LS, Shao Y, et al. PHAPI/pp32 suppresses tumorigenesis by stimulating apoptosis. J Biol Chem 2009;284:6946-54. [Crossref] [PubMed]
- Yang X, Lu B, Sun X, et al. ANP32A regulates histone H3 acetylation and promotes leukemogenesis. Leukemia 2018;32:1587-97. [Crossref] [PubMed]
- Valk PJ, Verhaak RG, Beijen MA, et al. Prognostically useful gene-expression profiles in acute myeloid leukemia. N Engl J Med 2004;350:1617-28. [Crossref] [PubMed]
- Verhaak RG, Wouters BJ, Erpelinck CA, et al. Prediction of molecular subtypes in acute myeloid leukemia based on gene expression profiling. Haematologica 2009;94:131-4. [Crossref] [PubMed]
- Metzeler KH, Hummel M, Bloomfield CD, et al. An 86-probe-set gene-expression signature predicts survival in cytogenetically normal acute myeloid leukemia. Blood 2008;112:4193-201. [Crossref] [PubMed]
- Stirewalt DL, Meshinchi S, Kopecky KJ, et al. Identification of genes with abnormal expression changes in acute myeloid leukemia. Genes Chromosomes Cancer 2008;47:8-20. [Crossref] [PubMed]
- de Jonge HJ, Woolthuis CM, Vos AZ, et al. Gene expression profiling in the leukemic stem cell-enriched CD34+ fraction identifies target genes that predict prognosis in normal karyotype AML. Leukemia 2011;25:1825-33. [Crossref] [PubMed]
- Cancer Genome Atlas Research Network, Ley TJ, Miller C, et al. Genomic and epigenomic landscapes of adult de novo acute myeloid leukemia. N Engl J Med 2013;368:2059-74. [Crossref] [PubMed]
- Bacher U, Haferlach T, Schoch C, et al. Implications of NRAS mutations in AML: a study of 2502 patients. Blood 2006;107:3847-53. [Crossref] [PubMed]
- Daver N, Schlenk RF, Russell NH, et al. Targeting FLT3 mutations in AML: review of current knowledge and evidence. Leukemia 2019;33:299-312. [Crossref] [PubMed]
- Li C, Nie H, Wang M, et al. MicroRNA-409-3p regulates cell proliferation and apoptosis by targeting PHF10 in gastric cancer. Cancer Lett 2012;320:189-97. [Crossref] [PubMed]
- Mandoli A, Singh AA, Prange KHM, et al. The Hematopoietic Transcription Factors RUNX1 and ERG Prevent AML1-ETO Oncogene Overexpression and Onset of the Apoptosis Program in t(8;21) AMLs. Cell Rep 2016;17:2087-100. [Crossref] [PubMed]
- Bullinger L, Dohner K, Dohner H. Genomics of Acute Myeloid Leukemia Diagnosis and Pathways. J Clin Oncol 2017;35:934-46. [Crossref] [PubMed]
- Gerke V, Creutz CE, Moss SE. Annexins: linking Ca2+ signalling to membrane dynamics. Nat Rev Mol Cell Biol 2005;6:449-61. [Crossref] [PubMed]
- Pikman Y, Alexe G, Roti G, et al. Synergistic Drug Combinations with a CDK4/6 Inhibitor in T-cell Acute Lymphoblastic Leukemia. Clin Cancer Res 2017;23:1012-24. [Crossref] [PubMed]
- Ma J, Mi C, Wang KS, et al. Zinc finger protein 91 (ZFP91) activates HIF-1alpha via NF-kappaB/p65 to promote proliferation and tumorigenesis of colon cancer. Oncotarget 2016;7:36551-62. [PubMed]
- Zhou L, Guo J, Jia R. Oncogene SRSF3 suppresses autophagy via inhibiting BECN1 expression. Biochem Biophys Res Commun 2019;509:966-72. [Crossref] [PubMed]
- Pozzo F, Bittolo T, Vendramini E, et al. NOTCH1-mutated chronic lymphocytic leukemia cells are characterized by a MYC-related overexpression of nucleophosmin 1 and ribosome-associated components. Leukemia 2017;31:2407-15. [Crossref] [PubMed]
- McRae HM, Garnham AL, Hu Y, et al. PHF6 regulates hematopoietic stem and progenitor cells and its loss synergizes with expression of TLX3 to cause leukemia. Blood 2019;133:1729-41. [Crossref] [PubMed]
- Dixon ZA, Nicholson L, Zeppetzauer M, et al. CREBBP knockdown enhances RAS/RAF/MEK/ERK signaling in Ras pathway mutated acute lymphoblastic leukemia but does not modulate chemotherapeutic response. Haematologica 2017;102:736-45. [Crossref] [PubMed]
- Launay E, Pangault C, Bertrand P, et al. High rate of TNFRSF14 gene alterations related to 1p36 region in de novo follicular lymphoma and impact on prognosis. Leukemia 2012;26:559-62. [Crossref] [PubMed]
- Grohmann T, Penke M, Petzold-Quinque S, et al. Inhibition of NAMPT sensitizes MOLT4 leukemia cells for etoposide treatment through the SIRT2-p53 pathway. Leuk Res 2018;69:39-46. [Crossref] [PubMed]
- Hansen MC, Nyvold CG, Roug AS, et al. Nature and nurture: a case of transcending haematological pre-malignancies in a pair of monozygotic twins adding possible clues on the pathogenesis of B-cell proliferations. Br J Haematol 2015;169:391-400. [Crossref] [PubMed]
- Pölönen P, Mehtonen J, Lin J, et al. Hemap: An interactive online resource for characterizing molecular phenotypes across hematologic malignancies. Cancer Res 2019;79:2466-79. [Crossref] [PubMed]
- Liberzon A, Birger C, Thorvaldsdottir H, et al. The Molecular Signatures Database (MSigDB) hallmark gene set collection. Cell Syst 2015;1:417-25. [Crossref] [PubMed]
- Tian XP, Huang WJ, Huang HQ, et al. Prognostic and predictive value of a microRNA signature in adults with T-cell lymphoblastic lymphoma. Leukemia 2019;33:2454-65. [Crossref] [PubMed]
- Wu Y, Heinrichs J, Bastian D, et al. MicroRNA-17-92 controls T-cell responses in graft-versus-host disease and leukemia relapse in mice. Blood 2015;126:1314-23. [Crossref] [PubMed]
- Xu LH, Guo Y, Cen JN, et al. Overexpressed miR-155 is associated with initial presentation and poor outcome in Chinese pediatric acute myeloid leukemia. Eur Rev Med Pharmacol Sci 2015;19:4841-50. [PubMed]
- Verboon LJ, Obulkasim A, de Rooij JD, et al. MicroRNA-106b~25 cluster is upregulated in relapsed MLL-rearranged pediatric acute myeloid leukemia. Oncotarget 2016;7:48412-22. [Crossref] [PubMed]
- Li G, So AY, Sookram R, et al. Epigenetic silencing of miR-125b is required for normal B-cell development. Blood 2018;131:1920-30. [Crossref] [PubMed]
- Si X, Zhang X, Hao X, et al. Upregulation of miR-99a is associated with poor prognosis of acute myeloid leukemia and promotes myeloid leukemia cell expansion. Oncotarget 2016;7:78095-109. [Crossref] [PubMed]
- Koutova L, Sterbova M, Pazourkova E, et al. The impact of standard chemotherapy on miRNA signature in plasma in AML patients. Leuk Res 2015;39:1389-95. [Crossref] [PubMed]
- Garzon R, Volinia S, Liu CG, et al. MicroRNA signatures associated with cytogenetics and prognosis in acute myeloid leukemia. Blood 2008;111:3183-9. [Crossref] [PubMed]
- Almeida RS, Costa ESM, Coutinho LL, et al. MicroRNA expression profiles discriminate childhood T- from B-acute lymphoblastic leukemia. Hematol Oncol 2019;37:103-12. [Crossref] [PubMed]
- Scheibner KA, Teaboldt B, Hauer MC, et al. MiR-27a functions as a tumor suppressor in acute leukemia by regulating 14-3-3theta. PLoS One 2012;7:e50895. [Crossref] [PubMed]
- Yuan T, Yang Y, Chen J, et al. Regulation of PI3K signaling in T-cell acute lymphoblastic leukemia: a novel PTEN/Ikaros/miR-26b mechanism reveals a critical targetable role for PIK3CD. Leukemia 2017;31:2355-64. [Crossref] [PubMed]
- Yang Y, Li F, Saha MN, et al. miR-137 and miR-197 Induce Apoptosis and Suppress Tumorigenicity by Targeting MCL-1 in Multiple Myeloma. Clin Cancer Res 2015;21:2399-411. [Crossref] [PubMed]
- Li W, Zhu D, Qin S. SIRT7 suppresses the epithelial-to-mesenchymal transition in oral squamous cell carcinoma metastasis by promoting SMAD4 deacetylation. J Exp Clin Cancer Res 2018;37:148. [Crossref] [PubMed]
- Jing D, Bhadri VA, Beck D, et al. Opposing regulation of BIM and BCL2 controls glucocorticoid-induced apoptosis of pediatric acute lymphoblastic leukemia cells. Blood 2015;125:273-83. [Crossref] [PubMed]
- Ogi H, Sakuraba Y, Kitagawa R, et al. The oncogenic role of the cochaperone Sgt1. Oncogenesis 2015;4:e149. [Crossref] [PubMed]
- Wang K, Qu X, Liu S, et al. Identification of aberrantly expressed F-box proteins in squamous-cell lung carcinoma. J Cancer Res Clin Oncol 2018;144:1509-21. [Crossref] [PubMed]
- Kadota S, Nagata K. pp32, an INHAT component, is a transcription machinery recruiter for maximal induction of IFN-stimulated genes. J Cell Sci 2011;124:892-9. [Crossref] [PubMed]
- Ouellet V, Le Page C, Guyot MC, et al. SET complex in serous epithelial ovarian cancer. Int J Cancer 2006;119:2119-26. [Crossref] [PubMed]
- Sun X, Lu B, Han C, et al. ANP32A dysregulation contributes to abnormal megakaryopoiesis in acute megakaryoblastic leukemia. Blood Cancer J 2017;7:661. [Crossref] [PubMed]
- Huang Z, Zhan X, Xiang S, et al. SALMON: Survival Analysis Learning With Multi-Omics Neural Networks on Breast Cancer. Front Genet 2019;10:166. [Crossref] [PubMed]
- Shao W, Wang T, Huang Z, et al. editors. Diagnosis-Guided Multi-modal Feature Selection for Prognosis Prediction of Lung Squamous Cell Carcinoma. International Conference on Medical Image Computing and Computer-Assisted Intervention; 2019: Springer.
- Johnson TS, Li S, Franz E, et al. PseudoFuN: Deriving functional potentials of pseudogenes from integrative relationships with genes and microRNAs across 32 cancers. GigaScience 2019;8:giz046. [Crossref] [PubMed]
- Johnson TS, Wang T, Huang Z, et al. LAmbDA: label ambiguous domain adaptation dataset integration reduces batch effects and improves subtype detection. Bioinformatics 2019;35:4696-706. [Crossref] [PubMed]
- Xiang S, Huang Z, Wang T, et al. Condition-specific gene co-expression network mining identifies key pathways and regulators in the brain tissue of Alzheimer's disease patients. BMC Med Genomics 2018;11:115. [Crossref] [PubMed]
- Yu CY, Xiang S, Huang Z, et al. Gene Co-expression Network and Copy Number Variation Analyses Identify Transcription Factors Associated With Multiple Myeloma Progression. Front Genet 2019;10:468. [Crossref] [PubMed]
- Zhan X, Cheng J, Huang Z, et al. Correlation Analysis of Histopathology and Proteogenomics Data for Breast Cancer. Mol Cell Proteomics 2019;18:S37-51. [Crossref] [PubMed]
- Huang Z, Han Z, Wang T, et al. TSUNAMI: Translational Bioinformatics Tool Suite For Network Analysis And Mining. BioRxiv 2019. [Crossref]
- Feng C, Huang H, Huang S, et al. Identification of potential key genes associated with severe pneumonia using mRNA-seq. Exp Ther Med 2018;16:758-66. [PubMed]
- Huang S, Feng C, Chen L, et al. Identification of Potential Key Long Non-Coding RNAs and Target Genes Associated with Pneumonia Using Long Non-Coding RNA Sequencing (lncRNA-Seq): A Preliminary Study. Med Sci Monit 2016;22:3394-408. [Crossref] [PubMed]
- Huang S, Feng C, Chen L, et al. Molecular Mechanisms of Mild and Severe Pneumonia: Insights from RNA Sequencing. Med Sci Monit 2017;23:1662-73. [Crossref] [PubMed]
- Huang S, Feng C, Zhai YZ, et al. Identification of miRNA biomarkers of pneumonia using RNA-sequencing and bioinformatics analysis. Exp Ther Med 2017;13:1235-44. [Crossref] [PubMed]
- Reilly PT, Yu Y, Hamiche A, et al. Cracking the ANP32 whips: important functions, unequal requirement, and hints at disease implications. Bioessays 2014;36:1062-71. [Crossref] [PubMed]
- Kushwaha G, Dozmorov M, Wren JD, et al. Hypomethylation coordinates antagonistically with hypermethylation in cancer development: a case study of leukemia. Hum Genomics 2016;10 Suppl 2:18. [Crossref] [PubMed]
- Kulis M, Heath S, Bibikova M, et al. Epigenomic analysis detects widespread gene-body DNA hypomethylation in chronic lymphocytic leukemia. Nat Genet 2012;44:1236-42. [Crossref] [PubMed]


