
Bile microbiota: new insights into biliary complications in liver transplant recipients
Introduction
Biliary complications represent a major problem in patients post liver transplantation, with an incidence of 10–40% (1,2). This condition can lead to liver failure or even death and has a mortality rate ranging from 8–15% (3). Biliary infection is a frequent cause of biliary complications in liver transplant recipients (4,5). Results from previous studies have shown that bacterial and fungal pathogens can be detected in specimens of bile or bile ducts in patients with biliary complications after liver transplantation (6). Accordingly, an investigation into the microbial species and mechanisms of pathogenic microorganisms of bile in liver transplant recipients with biliary complications would provide new and important insights into the basis for these complications.
As culture-dependent methods are insensitive and inadequate to study the entire microbial community, high-throughput sequencing technology has proved to be a more effective means for investigating microbial diversity over the past decade. While variations in gut microbiota during the perioperative period in liver transplant recipients have been studied. Surgery and the application of postoperative antibiotics and immunosuppressive agents lead to intestinal microbial environment changes (7). No such information exists on bile microbiota of liver transplant recipients.
In this study, metagenomic sequencing technology was used to explore the species and abundance of microbes in the bile of patients with biliary complications after liver transplantation as compared with microbes in a matched control group of liver transplant patients without biliary complications. The findings from this study have the potential of providing new insights into the pathogenesis, as well as the diagnosis and treatment, of biliary complications after liver transplantation.
Methods
Study population
The patients enrolled in the study were liver transplant recipients treated at our center over the period from September 2015 to June 2017. The inclusion criteria consisted of post-liver transplantation patients ranging in age from 18–65 years that had signed informed consent forms. Exclusion criteria included: (I) clinical and pathological diagnosis of acute or chronic rejection, (II) imaging results showing biliary anastomotic stenosis post-liver transplantation, (III) clinical and pathological diagnosis of primary disease recurrence (virus hepatitis or tumor), (IV) invasive systemic infection and (V) antibiotic use within the prior one-month period.
This study was approved by ethics committee in Beijing Friendship Hospital. Written informed consent was obtained from participants. No organs from executed prisoners were used in this study.
Definition of the patient groups
Control group
The control group consisted of the following patients: (I) Laboratory exams showing normal liver function and ultrasound showing normal blood flow and bile duct of graft, (II) T-tube reserved during their surgery and (III) Bile duct radiography showing no stenosis or expansion.
Symptom group
The symptom group consisted of the following patients: (I) laboratory exams showing abnormal liver function, with fever and acute cholangitis, (II) imaging results showing biliary stricture and stage expansion, (III) bile duct radiography showing multiple stenosis or stage expansion with a filling defect involving the left and right hepatic ducts and (IV) T-tube or percutaneous transhepatic cholangiography drainage (PTCD) tube reserved.
Sample collection
In the symptom group, bile was collected by PTCD or T-tube after sterilization. In the control group, bile was collected aseptically from the T-tube. Samples were then stored at −80 °C until use for DNA extraction.
DNA extraction and quality control
Samples were treated using a mini-bead beater (Biospec Products, Bartlesville, OK, USA) and DNA was extracted using a QIAamp DNA Mini Kit (Qiagen, Hilden, Germany). The DNA extracts were stored at −80 °C until use. The concentration of DNA was determined spectrophotometrically while agar gel electrophoresis was used to assess DNA integrity.
PCR amplification
The V3–V4 region of the bacterial 16S rRNA gene was amplified by PCR (95 °C for 3 min, followed by 27 cycles at 95 °C for 30 s, 55 °C for 30 s and 72 °C for 45 s, and a final extension at 72 °C for 10 min) using the primers 338F 5'-barcode-ACTCCTACGGGAGGCAGCAG)-3' and 806R 5'-GGACTACHVGGGTWTCTAAT-3'. PCR was performed in triplicate in a 20-µL mixture containing 4 µL 5× FastPfu Buffer, 2 µL 2.5 mM dNTPs, 0.8 µL of each primer (5 µM), 0.4 µL FastPfu Polymerase, and 10 ng template DNA.
Illumina MiSeq sequencing
Amplicons were extracted from 2% agarose gels, purified using the AxyPrep DNA Gel Extraction Kit (Axygen Biosciences, Union City, CA, USA) and quantified using QuantiFluor-ST (Promega, Madison, WI, USA). Purified amplicons were pooled in equimolar amounts and paired-end sequenced (2×250) on an Illumina MiSeq platform according to standard protocols. The raw reads were deposited in the NCBI Sequence Read Archive (SRA) database.
Processing of sequencing data
Raw fastq files were demultiplexed, quality-filtered by Trimmomatic and merged by FLASH with the following criteria: (I) reads were truncated at any site receiving an average quality score <20 over a 50 base pairs (bp) sliding window, (II) primers were exactly matched allowing 2 nucleotide mismatching, while reads containing ambiguous bases were removed and (III) sequences whose overlap was longer than 10 bp were merged according to their overlap sequence.
Operational taxonomy units (OTUs) were clustered with a 97% similarity cutoff using UPARSE (version 7.1, http://drive5.com/uparse/) and chimeric sequences were identified and removed using UCHIME. The taxonomy of each 16S rRNA gene sequence was analyzed by RDP Classifier (http://rdp.cme.msu.edu/) against the Silva (SSU115) 16S rRNA database using a confidence threshold of 70%.
Statistical analysis
Species diversity was evaluated with use of Shannon, Simpson, Chao, and Ace indices. Calculations of principal coordinates analysis (PCoA) were performed by Hellinger distance, Bray-curtis distance and (un) weighted UniFrac distance. LDA Effect Size (LEfse) analysis was performed to identify species in which relative abundance was significantly different among the various populations. The Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway was evaluated. A Wilcoxon rank-sum test was performed to evaluate differences between the two groups in alpha diversity, principle coordinates, and community difference analysis. A P<0.05 was required for results to be considered as statistically significant.
Results
Patients
Patients of the control group (n=13) ranged in age from 39 to 62 years (mean ± SD =51.4±8.1 years) and consisted of 10 males and 3 females. These patients included 4 cases of hepatitis B cirrhosis, 4 cases of alcoholic cirrhosis, 2 cases of hepatitis C cirrhosis, 1 case of primary biliary cirrhosis (PBC), 1 case of primary sclerosing cholangitis (PSC) and 1 case of drug-induced liver injury. One bile specimen was collected from each patient, resulting in a total of 13 bile samples for analysis.
Patients in the symptom group (n=10) ranged in age from 18 to 68 years (mean ± SD =44.7±14.6 years) and consisted of 8 males and 2 females. These patients included 3 cases of hepatitis B cirrhosis, 2 cases of alcoholic cirrhosis, 2 cases of hepatocellular carcinoma, 1 case of cryptogenic cirrhosis, 1 case of PBC, 1 case of idiopathic portal hypertension. A total of ten bile samples were collected.
No statistically significant differences were present between the two groups with regard to age, sex, body mass index (BMI), warm ischemia time, cold ischemia time, Model for End-stage Liver Disease (MELD) or trough level of FK506 (P>0.05; Table 1). Two patients received living donor liver transplantation (LDLT) while the remaining 21 patients received liver transplantation from donation after cardiac death donors (DCD). All the patients received ABO blood group compatible liver transplantation. All liver transplantation recipients received an immunosuppressive regimen of tacrolimus (FK506) + methylprednisolone + mycophenolate mofetil (MMF). Methylprednisolone was gradually withdrawn within three months after liver transplantation.

Full table
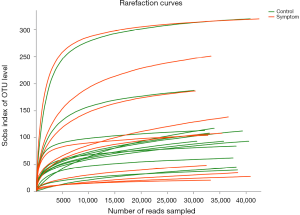
OTU cluster and Rarefaction curve
In order to reduce the impact of low-abundant OTU on subsequent statistical analysis, OTU with sequence numbers ≥5 in at least three samples and an OTU with a total number of retained sequences ≥20 were retained. The sequence data included ten bacterial phyla, subdivided into 65 families, 110 genera and 172 OTUs. According to the number of reads sampled and sobs index of OTU level, rarefaction curve of the two groups were plotted (Figure 1). In our study, the curves tended to be flat, which indicated that the reads number was relatively large enough to reflect species richness.
Distribution and abundance of bile microbiota
At the phyla level, Firmicutes was the main phylum in the control group, with its abundance being 63.08%. The other major phyla in controls were: Proteobacteria (26.07%), Actinobacteria (9.6%) and Bacteroidetes (0.43%). In the symptom group, the main phyla were: Proteobacteria (55.19%), Firmicutes (32.36%), Actinobacteria (10.24%) and Bacteroidetes (1.23%) (Figure 2).
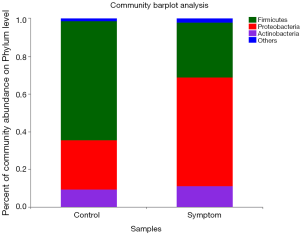
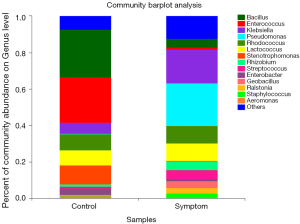
At the genera level, Bacillus was the most common genus of the control group (26.15%). The other major genera in controls were: Enterococcus (24.88%), Stenotrophomonas (10.74%), Rhodococcus (8.58%), Lactobacillus (8.29%), Klebsiella (6.17%) and Enterobacter (3.48%). The most common genus of the symptom group was Pseudomonas (23.31%), with other major genera being: Klebsiella (18.42%), Lactococcus (9.61%), Rhodococcus (9.59%), Rhizobium (5.08%), Bacillus (4.77%) and Streptococcus (4.76%) (Figure 3).
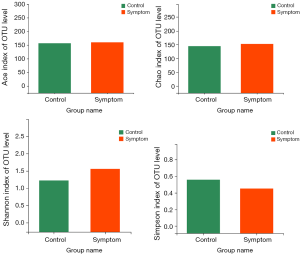
Alpha diversity analysis
Alpha diversity provides a measure of species diversity within a sample. Shannon’s and Simpson’s are common indices for measures of species diversity including species richness and evenness. The greater the Shannon index, the higher the diversity, while an opposite relationship comprises the Simpson index. Chao and Ace are indices used to access species richness. The greater the value in these indices, the higher the diversity and richness of the microbial community. No statistically significant differences were obtained among all cohorts (P>0.05) as measured with these commonly used Shannon, Simpson, Chao and Ace indices (Figure 4).
Analysis of diversity differences: PCoA
PCoA is based on the OTU classified taxon data and involves analyzing the intrinsic composition of the microbial structure between the two groups. In our study, a number of methods were used to calculate the distance matrix, including the Hellinger, Bray-curtis and (un) weighted UniFrac. PCoA analysis of the four distances revealed that the two groups could not be completely discriminated (Figure 5).
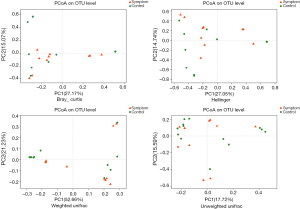
Results of a PCoA based on OTU composition. Each point represents a sample. Groups are color coded and distance between the two points indicates differences in community composition, with shorter distances denoting smaller differences. PCoA, principal coordinates analysis; OTU, operational taxonomy unit.
Community difference analysis: bacterial differences at phyla and genera levels
At the phyla level, statistically significant differences were obtained between the control and symptom groups for Firmicutes and Proteobacteria. Firmicutes was found to be enriched in the control group (P=0.03775) and Proteobacteria in the symptom group (P=0.0143) (Figure 6).
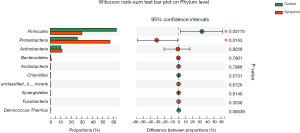
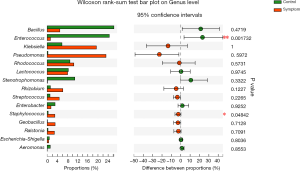
At the genera level, statistically significant differences were present between the control and symptom groups for Enterococcus and Staphylococcus. While Enterococcus was found to be enriched in the control group (P=0.001732), Staphylococcus was enriched in the symptom group (P=0.04842) (Figure 7).
LEfse analysis
LEfSe analysis was used to detect differential abundances of species between the control and symptom groups. We identified differential distributions of species using the default cut-offs (LDA score >2.0; P value <0.05). The results showed that the relative abundances of Bacilli, Firmicutes, Enterococcaceae, Enterococcus, Planctomycetacia, Brevibacillus and Micrococcus were significantly decreased, while those of Proteobacteria, Bosea, Staphylococcus, Trichococcus, Ruminococcaceae-UCG-002, Epilithonimonas, Fusicatenibacter, Subdoligranulum, Lachnospiraceae-UCG-004, Pelomonas, Blautia, Ruminococcus-1, Roseburia and Empedobacter significantly increased in the symptom group as compared with the control group (Figures 8,9).
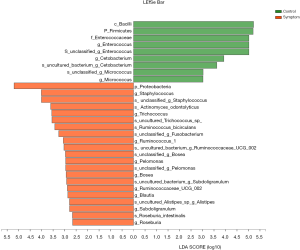
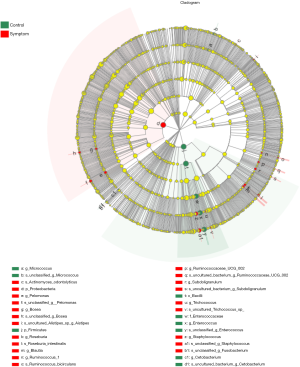
LEfSe analysis of differential species abundance between the control and symptom groups. The LDA score is represented on the abscissa with higher scores indicating greater differences. The green and red horizontal bars represent species enrichment in the control and symptom groups, respectively.
A cladogram plot of differential species abundance between the control and symptom groups. The green and red areas represent species enrichment in the control and symptom groups, respectively.
Differences in KEGG pathway levels
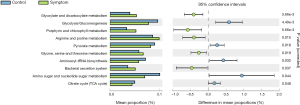
The composition of the OTU sequence was transformed into KEGG orthology to analyze differences in metabolic pathways between the control and symptom groups. We identified 5 significantly enriched and 5 depleted pathways in bile samples in the symptom versus control group.
Bile samples in the symptom group were significantly enriched in pathways related to glyoxylate and dicarboxylate metabolism, porphyrin and chlorophyll metabolism, arginine and proline metabolism, glycine, serine and threonine metabolism and bacterial secretion system. Bile samples depleted in pathways were related to glycolysis/gluconeogenesis, pyruvate metabolism, aminoacyl-tRNA biosynthesis, amino sugar and nucleotide sugar metabolism and citrate cycle (TCA cycle) (Figure 10).
Discussion
To the best of our knowledge, this study represents the first to apply high-throughput sequencing technology for the study of biliary microbiota in patients with biliary complications after liver transplantation. With use of this technique, it was possible to examine the role of biliary microbes associated with the onset, occurrence and development of biliary complications following liver transplantation.
As bile acids present in the bile contain a certain amount of antimicrobial activity (8), the biliary tract of healthy people is generally considered to be a relatively sterile environment (9). However, in certain diseases or infectious conditions, bile acid secretion can be reduced (10). A few studies have used modern molecular methods to characterize the microbes present in bile, mainly in relation to various disease conditions (11,12). In our study, the main bacterial phyla in bile samples of the symptom group were Proteobacteria, Firmicutes, Actinobacteria and Bacteroidetes, while the main bacterial genera in this group were Pseudomonas, Klebsiella, Lactococcus and Rhodococcus. In a study on bile microbiota in patients with PSC the most common phyla were Bacteroidetes, Firmicutes, Proteobacteria, Fusobacteria and Actinobacteria, and the most common genera were Prevotella, Streptococcus, Veillonella, Fusobacterium and Haemophilus. Streptococcus abundance was shown to be positively correlated with an increase in disease severity and it may play a pathogenic role in the progression of this condition (8).
Biliary complications, which are characterized by abnormal liver function, biliary stricture and obstruction, represent one of the major problems associated with liver transplantation (13,14). If treatments such as medications, endoscopy, interventional therapy are ineffective, these complications can eventually lead to liver failure and even death (15,16). Biliary stricture can be characterized as either anastomotic or non-anastomotic. The former is mainly related to surgical techniques or local ischemia. Therefore, in this study, patients with biliary anastomotic stricture after liver transplantation were not included. In a prospective study reported by Gotthardt (6), 213 patients with biliary complications after liver transplantation underwent endoscopic retrograde cholangio-pancreatography (ERCP), and bile samples were collected during the first endoscopic treatment. Bacteria or fungi were found in 180 patients, with 57% being Gram-positive and 24% Gram-negative bacterial isolates, and the main species obtained were Enterococcus and Escherichia coli. The results of this study suggest that infections of Enterobacteria and fungi are significantly associated with the clinical signs of cholangitis, a condition which significantly lowers survival rates in patients with biliary tract injury.
In our study, patients in the symptom group included those whose imaging results indicated the presence of the non-anastomotic biliary stricture, and the patients displayed clinical signs such as fever and cholangitis. At the phyla level, bile samples in the symptom group predominantly consisted of Proteobacteria, with this content being significantly greater than that observed in controls. Pseudomonas and Klebsiella were the dominant genus in the symptom group, but were not significantly different from that of the control group. Bile samples from patients in the symptom group were also assessed in bacterial cultures, with these cultures showing the presence of Acinetobacter baumannii, radiation Rhizobium, Enterobacter cloacae, Klebsiella pneumoniae, Pseudomonas aeruginosa, and Chryseobacterium indologenes. Among these, five species belong to Proteobacteria, suggesting that Proteobacteria may play a significant role in the occurrence and development of biliary tract injury after liver transplantation.
Enterococcus, a bacterium that can grow in a bile-rich environment, is a normal flora of the human intestine and one of its probiotics. This bacterium can regulate the balance of intestinal flora and also possesses certain immune regulatory and anti-allergic effects (17). In our study, Enterococcus was found to be enriched in bile samples from the control group. The absence of enrichment in symptom patients may be due to changes in the bile microenvironment associated with biliary complications and a disruption of the inter-dependent balance of bile flora. In this way, the reduction in abundance of Enterococcus in biliary complication patients suggests that Enterococcus may play an important role in maintaining the stability and balance of bile microecology.
Staphylococcus, a conditional pathogen, belongs to the phylum Firmicutes, class Bacilli, order Bacillales and family Staphylococcaceae. Some strains of Staphylococcus can produce a variety of pathogenic factors such as exotoxin, enterotoxin, agglutination factor and bio film, which represent common bacterium of biliary infection. We found that Staphylococcus was enriched in the bile of patients with biliary complications from the symptom group. These findings suggest that this bacterium may contribute to the biliary injury in immunodeficient patients after liver transplantation.
There exist some salient differences in the metabolic pathways of bile samples between the two groups. However, the 16s rRNA sequencing analysis is limited and predictive in the study of KEGG pathways. Whether these pathways are involved in the development and progression of biliary complications after liver transplantation will require further studies which can incorporate a combination of metagenomics and metabolomics.
There remain some limitations in this study. Notably, an increase in sample size and multicenter experiences will be required to corroborate these findings. In addition, combining genomics, metabolomics and transcriptomics will be necessary to achieve a more comprehensive and deep understanding of the pathogenesis of biliary complications after liver transplantation.
In summary, this study represents the first to apply high-throughput sequencing to study bile microbiota of patients with biliary complications after liver transplantation. Our results reveal that significant differences are present in species composition, diversity and differential species within bile samples from the control versus symptom groups. The species and pathways enriched in bile samples of the symptom group may be involved in the pathogenesis of biliary complications after liver transplantation. These findings provide new and important information for understanding the mechanism of biliary injury after liver transplantation.
Acknowledgments
Funding: This study was supported by the National Natural Science Foundation of China (Grant No. 81570586).
Footnote
Conflicts of Interest: The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. This study was approved by ethics committee in Beijing Friendship Hospital (No. 2017-P2-060-02). Written informed consent was obtained from participants.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Dubbeld J, van Hoek B, Ringers J, et al. Biliary complications after liver transplantation from donation after cardiac death donors: an analysis of risk factors and long-term outcome from a single center. Ann Surg 2015;261:e64. [Crossref] [PubMed]
- Nemes B, Gámán G, Doros A. Biliary complications after liver transplantation. Expert Rev Gastroenterol Hepatol 2015;9:447-66. [Crossref] [PubMed]
- Op den Dries S, Sutton ME, Lisman T, et al. Protection of bile ducts in liver transplantation: looking beyond ischemia. Transplantation 2011;92:373-9. [Crossref] [PubMed]
- Yoshizumi T, Shirabe K, Ikegami T, et al. Decreased immunoglobulin G levels after living-donor liver transplantation is a risk factor for bacterial infection and sepsis. Transpl Infect Dis 2014;16:225-31. [Crossref] [PubMed]
- Moy BT, Birk JW. A Review on the Management of Biliary Complications after Orthotopic Liver Transplantation. J Clin Transl Hepatol 2019;7:61-71. [Crossref] [PubMed]
- Gotthardt DN, Weiss KH, Rupp C, et al. Bacteriobilia and fungibilia are associated with outcome in patients with endoscopic treatment of biliary complications after liver transplantation. Endoscopy 2013;45:890-6. [Crossref] [PubMed]
- Doycheva I, Leise MD, Watt KD. The Intestinal Microbiome and the Liver Transplant Recipient: What We Know and What We Need to Know. Transplantation 2016;100:61-8. [Crossref] [PubMed]
- Pereira P, Aho V, Arola J, et al. Bile microbiota in primary sclerosing cholangitis: Impact on disease progression and development of biliary dysplasia. PLoS One 2017;12:e0182924. [Crossref] [PubMed]
- Verdier J, Luedde T, Sellge G. Biliary Mucosal Barrier and Microbiome. Viszeralmedizin 2015;31:156-61. [PubMed]
- Bauer TM, Steinbrückner B, Brinkmann FE, et al. Small intestinal bacterial overgrowth in patients with cirrhosis: prevalence and relation with spontaneous bacterial peritonitis. Am J Gastroenterol 2001;96:2962-7. [Crossref] [PubMed]
- Shen H, Ye F, Xie L, et al. Metagenomic sequencing of bile from gallstone patients to identify different microbial community patterns and novel biliary bacteria. Sci Rep 2015;5:17450. [Crossref] [PubMed]
- Ye F, Shen H, Li Z, et al. Influence of the Biliary System on Biliary Bacteria Revealed by Bacterial Communities of the Human Biliary and Upper Digestive Tracts. PLoS One 2016;11:e0150519. [Crossref] [PubMed]
- Axelrod DA, Dzebisashvili N, Lentine KL, et al. Variation in biliary complication rates following liver transplantation: implications for cost and outcome. Am J Transplant 2015;15:170-9. [Crossref] [PubMed]
- Palanisamy AP, Taber DJ, Sutter AG, et al. Clinical outcomes and costs associated with in-hospital biliary complications after liver transplantation: a cross-sectional analysis. J Gastrointest Surg 2015;19:282-9. [Crossref] [PubMed]
- Liu Y, Jia JD, Sun LY, et al. Characteristics of bile microbiota in liver transplant recipients with biliary injury. Int J Clin Exp Pathol 2018;11:481-9. [PubMed]
- Arain MA, Attam R, Freeman ML. Advances in endoscopic management of biliary tract complications after liver transplantation. Liver Transpl 2013;19:482-98. [Crossref] [PubMed]
- Bodera P, Chcialowski A. Immunomodulatory effect of probiotic bacteria. Recent Pat Inflamm Allergy Drug Discov 2009;3:58-64. [Crossref] [PubMed]


