
EGCG protects vascular endothelial cells from oxidative stress-induced damage by targeting the autophagy-dependent PI3K-AKT-mTOR pathway
Introduction
Oxidative stress plays a crucial role in the onset of cardiovascular damage and can affect a variety of related systems, leading to metabolic disorders, ischemic diseases and other diseases associated with cardiopulmonary function (1). Epigallocatechin gallate (EGCG) is a polyphenol natural product derived from green tea (Camellia sinensis). An expanding body of evidences indicate that EGCG has many biological activities, such as anti-inflammatory, antioxidant, anti-mutagenic and anti-cancer activities (2-5). Lots of work have been done to reveal the anti-cancer mechanism of EGCG. Studies have shown that EGCG affects multiple signaling pathways through binding to a variety of molecular targets, including transmembrane receptors, kinases and other key proteins, and ultimately leading to apoptosis, proliferation inhibition, and inhibition of invasion, angiogenesis and metastasis (6). Although it has been reported that EGCG exhibits a protective effect on the cardiovascular system (7), the protective mechanism of EGCG in oxidative stress-induced cardiovascular damage has not been fully established.
Autophagy is a defense and stress management mechanism which involves the sequestration, transport, bulk degradation and recycling of cytoplasmic components (8,9). Increasing evidence indicates that autophagy is activated in oxidative stress environment (10). Recently, a lot of studies have been done on the relationship between autophagy and apoptosis, and it is demonstrated that autophagy can act as a cell survival mechanism to protect cells from apoptosis under certain conditions (2). Under oxidative stress condition, autophagy acts primarily as a pro-survival pathway to protect cardiovascular cells from oxidative damage (7,10,11). Therefore, autophagy may play an important role in the healing effect of EGCG against oxidative stress.
It is well known that kinase mammalian target of rapamycin (mTOR) is a pivotal regulator of autophagy, and its activity can be enhanced by activating the PI3K/Akt signaling pathway (4). It has been verified that angiogenesis, as an important part of cardiac tissue regeneration and protection, can be regulated by mTOR (12). Besides, induction of autophagy through targeting mTOR pathway alleviates vascular smooth muscle cell senescence induced by Adriamycin (8). Further, inhibition of mTOR for prolonged treatment has been proved to be beneficial in reducing vasculopathy (13). In addition, through repression of the PI3K/AKT/mTOR pathway, restoring autophagy flux alleviates ox-LDL-induced human vascular endothelial cells (HUVECs) injury (14). All these evidences indicate that it is necessary to pay attention to mTOR when autophagy is investigated in cardiovascular disease.
We hypothesized that induction of autophagy may be a potential mechanism by which EGCG protects cardiovascular cells from oxidative stress damage. Herein, by examining how EGCG affects autophagy flux in endothelial cells in a dose-dependent manner, and by determining the role of mTOR in EGCG-mediated antioxidant responses, our study indicates that pretreatment with EGCG triggers autophagic survival response of endothelial cells in oxidative stress environment.
Methods
Cell culture and treatment
HUVECs were bought from ScienCell Research laboratories (USA). Cells were cultured in M199 containing 20% fetal bovine serum (FBS) (Gibco, Carlsbad, CA, USA), 200 µg/mL penicillin (Gibco), 200 µg/mL streptomycin (Gibco) and 2 mM L-glutamine (Thermo Scientific, Carlsbad, CA, USA) at 37 °C with 5% CO2 (15). EGCG powder (purity 98%) was dissolved in distilled H2O in the concentration of 10 mM, and diluted to the desired concentration. The cells were pretreated with EGCG (1, 5, 10 µmol/L) for 24 h, after which the original medium was removed and the fresh medium containing 200 µmol/L H2O2 was added. After incubated for another 4 h, the following tests were performed.
Cell viability analysis
Cell viability was measured with MTS assay. Briefly, HUVECs were seeded in 96-well tissue culture plates and pretreated with EGCG at various concentrations (1 to 10 µmol/L) or vehicle for 24 h, then treated with 200 µmol/L H2O2. After treatment, 10 µL MTS assay solution was added to each well and incubated at 37 °C for 3 h. Since MTS is light-sensitive, the procedure was performed in dark environment. Then, the optical density of each well was measured at 570 nm with a microplate reader (Molecular Device, Sunnyvale, CA, USA). The test was performed in five wells and repeated three times.
TUNEL assay
Terminal deoxynucleotidyl transferase-mediated dUTP-biotin nick end labeling (TUNEL) staining was performed using a TUNEL Apoptosis Assay Kit (T2190, Solarbio, Beijing, China) and according to the manufacturer’s instructions. The drug-treated cells were fixed with paraformaldehyde (PFA) for 15 min, gently rinsed with PBS and mixed with Triton-X-100 (0.2%) (T8200, Solarbio) for 10 mins at room temperature. Add the newly prepared TUNEL assay solution and incubate at 37 °C in the dark for 1 h. Slides were rinsed with phosphate-buffer saline (PBS) three times and counterstained with 10 µg/mL PI (P4170, Sigma, MO, USA) at room temperature in the dark for 20 min. The percentage of TUNEL-positive cells in 10 randomly chosen fields (85–105 cells/field) was calculated as TUNEL positive nuclei/PI staining cells, with a fluorescence microscope (Olympus BX-51). The test was performed independently at least twice.
Transmission electron microscopy (TEM)
Cells were fixed with 2.5% glutaraldehyde overnight at 4 °C, then fixed with 1% (w/v) osmium tetroxidefor 1 h, 1% (w/v) aqueous uranyl acetate overnight, and dehydrated in 50%, 70%, and 95% pure alcohol (v/v). Samples were then infiltrated and embedded using Epon resin, resin blocks were then polymerized at 70 °C for 48 h. 0.1 µm ultrathin sections were stained with 5% lead citrate [(w/v), Sigma, MO, USA] and then observed using a Zeiss transmission electron microscope (Zeiss Inc., Thornwood, NY, USA) at an accelerating voltage of 80 kV.
Immunoblot analysis assay
Antibodies against AKT (4691S), p-AKT (4060S), mTOR (2983T), p-mTOR (5536T), LC3 (12741S), Atg5 (12994S), Atg7 (8558S), Atg12 (2011S), caspase 3 (9665S), cleaved-caspase 3 (9664S), caspase 9 (9508S) and cleaved-caspase 9 (7237S) were bought from Cell Signaling Technology (CST, Danvers, MA, USA). Antibodies against 3-methyladenine (3-MA) (M9281) and β-actin (A8481) were from Sigma. Briefly, HUVECs were lysed, then quantified proteins using the BCA assay (23225, Thermo Scientific). An equal amount of protein samples were subjected to sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and electro-transferred to a 0.45 µm polyvinylidene difluoride membranes (Millipore, HATF09025). The membranes were blocked with 5% BSA and incubated with primary antibodies at 4 °C overnight. The membranes were then incubated with secondary antibody (CST, 1:20,000) for 1 h at room temperature followed with three 10-min washes in TBS-T. bands were visualized using an enhanced chemiluminescence (ECL) system. Data within a linear range were quantified using Quantity One software (BIO-RAD, Hercules, CA, USA).
Plasmid electroporation and confocal microscopy
Briefly, resuspend the cells in serum and antibiotic-free M199 medium at a density of 1×106 cells/mL. 200 µL of cell suspension were mixed with 5 µg mCherry-EGFP-LC3 plasmid, which was a gift from Prof. Xuejun Li from Peking University Health Science Center (Addgene plasmid, 22418), then transferred to a 0.4 cm electroporation cuvette (BioRad, Hercules, USA), and electroporated using Gene Pulser Xcell™ Electroporation System (BioRad) (16). The pulse amplitude of electroporation varied between 80 and 120 V with a pulse duration of 20 ms/transfection. When the electroporation was over, the cells were incubated at room temperature for 5 min and transferred to an 8-well LAB-Tek confocal CHAMBERCVG (LAB-Tek, 155411, Hatfield, PA, USA)
After transfection, HUVECs were treated with different drugs as previously described. Immunofluorescence staining images of HUVECs transfected with mCherry-EGFP-LC3 plasmid were obtained at 40× magnification using a Leica TCS SP5 laser scanning confocal microscope. The fluorescence of EGFP (green) and mCherry (red) were excited with laser light of 488 nm and 587 nm, respectively.
Small interfering RNA (siRNA) transfection
The siRNAs targeting mTOR was obtained from Santa Cruz and was a pool of three target-specific 19–25 nt siRNAs. The siRNAs were transfected into HUVECs using Lipofectamine 3000 (Invitrogen) according to the manufacturer's instructions.
Data analysis and statistics
All data were presented as the means ± SEM. Data were analyzed with GraphPad Prism 6.0 and SPSS 18.0. Statistical analysis was performed by using one-way ANOVA followed by Bonferroni correction. A value of P<0.05 was considered statistically significant.
Results
EGCG protected HUVECs from H2O2-induced apoptosis
To determine the most appropriate oxidative stress conditions, HUVECs were treated with different concentrations of H2O2 (from 100 µmol/L to 1 mmol/L). It was found that H2O2 obviously reduces cell viability of HUVECs in a dose-dependent manner (Figure 1A). Based on these results, treatment with 200 µmol/L H2O2 for 4 h was selected to provide a maximum dynamic range for quantifying protective and deleterious responses. Besides, we demonstrated that treatment with EGCG (0–40 µmol/L) alone did not significantly influence cell survival, suggesting that EGCG had no toxic effect on HUVECS (Figure 1B). To evaluate if EGCG pretreatment could protect HUVECs from oxidative stress injury, an MTS assay was carried out to assess cell viability. As illustrated, cells treated with 200 µmol/L H2O2 for 4 h showed typical characteristics of apoptosis, whereas EGCG pretreatment (5, 10 µmol/L) alleviated H2O2-induced damage and restored cell survival following the oxidative stress (Figure 1C). Further, results of TUNEL staining showed that the number of TUNEL-positive cells in the EGCG pretreatment group was significantly lower than that of the H2O2 alone treatment group (Figure 1D,E).
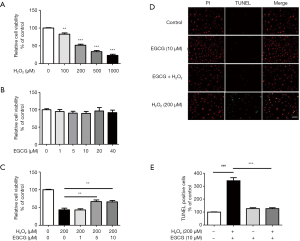
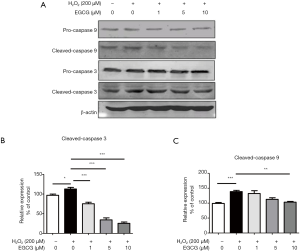
To further determine whether the protective effect of EGCG on HUVECs was related to apoptosis in vitro, we evaluated the expression of cleaved-caspase 3 and cleaved-caspase 9 by western blot analysis. The results showed that the expression levels of cleaved-caspase 3 and cleaved-caspase 9 were significantly increased after 4 h H2O2 treatment compared with the control group (Figure 2A). However, EGCG pretreatment significantly reversed this upward trend. 10 µmol/L EGCG had a significant effect on reducing cleaved-caspase 3 and cleaved-caspase 9 in HUVECs (Figure 2B,C). These results confirm that EGCG protects HUVECs from oxidative stress-induced apoptosis.
EGCG promoted autophagy in H2O2-treated HUVECs
To investigate if EGCG pretreatment affected the level of autophagy, we observed HUVECs with TEM, which is the classic method for detecting autophagy in most organisms. In the control group, the HUVECs were normal. Obvious membranolysis (white arrowhead), vacuoles, apoptosis bodies (yellow arrowhead) due to oxidative stress could be seen in H2O2 alone treatment group. While in EGCG pretreatment group, the HUVECs were protected from H2O2 -induced oxidative stress. HUVECs showed fewer features of apoptosis and more autophagosomes (red arrowhead indicating bilayer membrane structure) (Figure 3A). Besides, EGCG did not affect the morphology and ultrastructure of normal HUVECs cells. The TEM analysis is consistent with the TUNEL assay and Hoechst staining results.
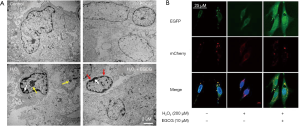
To assess the role of EGCG in autophagy in HUVECs, mCherry-EGFP-LC3 tandem tagged fluorescent protein (tfLC3) was transfected into HUVECs and then observed with a confocal microscopy. As previously described, the green LC3 puncta indicated autophagosomes, while the red LC3 puncta represented both autophagosomes and autolysosomes, the yellow spots, formed by the superposition of green and red, represented autophagosomes (9,17). In our study, the number of green, red, and yellow puncta in EGCG (10 µmol/L) pretreatment group were significantly higher (Figure 3B), which suggested that EGCG enhanced autophagy flux in HUVECs.
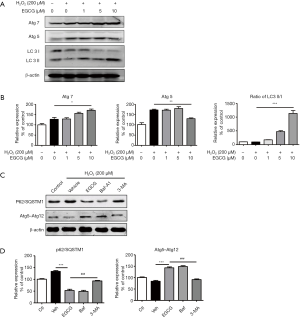
Next, the expression levels of microtubule associated protein 1 light chain 3-II (LC3-II), which is an LC3-conjugate and an autophagosomal marker, was assessed (17). Western blot analysis indicated that the ratio of LC3-II/I significantly elevated in a dose-dependent manner in EGCG (1, 5, 10 µmol/L) pretreatment cells compared with H2O2 alone treated cells. EGCG also increased the expression of Atg5 and Atg7, which have been believed to be molecules essential for autophagy (Figure 4A,B). Consistently, when pretreated with EGCG before H2O2 stimulation in HUVECs, the expression of Atg5–Atg12 complex increased, and the level of P62/SQSTM1 significantly decreased. Whereas, when given 10 mM 3-MA, an autophagy inhibitor, for 1 h before exposed to H2O2, could weaken the effect of EGCG (Figure 4C,D).
EGCG downregulated the PI3K-AKT-mTOR signaling pathway in HUVECs exposed to oxidative stress
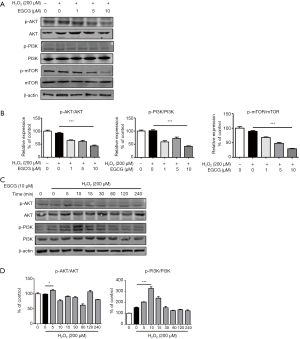
To further determine the mechanism of EGCG-induced autophagy, we examined Ser473 phosphorylation of AKT, which measured both AKT-mTORC1 and mTORC2 activity (18). As illustrated, the levels of p-AKT, p-PI3K and p-mTOR were markedly decreased in a dose-dependent manner due to EGCG pretreatment compared with these in H2O2 alone treatment group (Figure 5A,B). Moreover, within 4 h, by observing 200 µmol/L H2O2 combined 10 µmol/L EGCG treatment group in terms of time, we found that p-Akt and p-PI3K were markedly reduced after 60 min in HUVECs (Figure 5C,D).
Knockdown of mTOR partially promoted EGCG-induced autophagy
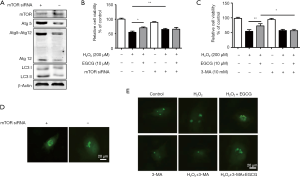
We used a siRNA interference vector to knock down human mTOR gene expression in HUVECs. Western blot analysis showed that the amount of mTOR protein expression was reduced by 60–70% after 48 h transfection; as expected, the levels of Atg5, Atg12, and Atg5–Atg12 complex were increased compared with control group transfected with nontargeting scramble siRNA transiently (Figure 6A). We detected the subcellular localization of GFP-LC3, a green fluorescent autophagy reporter protein, and the results showed that mTOR-siRNA promoted GFP-LC3 puncta formation (Figure 6B). Besides, we found that knockdown of mTOR could promoted the protective effect of EGCG (Figure 6C). Next, we evaluated if inhibition of autophagy by 3-MA affected the EGCG-induced protection in HUVECs. We pretreated the cells with EGCG for 24 h, one sample of which was given 10 mmol/L of 3-MA, and finally injured the cells by exposed to 200 µmol/L H2O2 for 4 h. Compared with the EGCG + H2O2 groups, cell viability in EGCG + 3-MA + H2O2 treatment groups were reduced (Figure 6D). Consistently, GFP-LC3 in HUVECs treated 3-MA showed a significant decrease (Figure 6E). Together, these data suggest that knockdown of mTOR partially promote EGCG-induced autophagy, whereas inhibiting autophagy in HUVECs can eliminate the cytoprotective effects of EGCG.
Discussion
It is well known that oxidative stress exhibits a critical role in many physiological and pathological processes in humans, including cancer, ischemic injury (19), chronic inflammation (6), type II diabetes (20) and arteriosclerosis (10,14,21-23). Thus, eliminating excess intracellular reactive oxygen species and preventing oxidative stress injury might be powerful interventions for the prevention and treatment of these diseases. EGCG, a polyphenol with antioxidant property belonging to the catechin family, is beneficial to human vascular endothelial, including myocyte hypertrophy, cardiac hypertrophy and fibrosis developing from aorta constriction (3). In this study, our data showed that EGCG could protect HUVECs from oxidative stress injury by inducing a beneficial autophagy.
Autophagy is an essential cellular mechanism which plays “housekeeping” role in normal physiological processes and defects in autophagy regulation play a central role in number of diseases. Autophagy can adapt cells to intracellular stress environmental, including oxidative stress, endoplasmic reticulum stress, starvation, hypoxia. Numerous studies have shown that autophagy acts primarily as a pro-survival process to protect cells from intracellular stress environmental, but sometimes, autophagy can also lead to cell death (24,25). Besides, the relationship between apoptosis and autophagy has also been extensively studied. Apoptosis and autophagy can be initiated with similar stimuli, while in some case, they are initiated in a mutually exclusive manner (26). In our study, H2O2-induced apoptosis in HUVECs was reduced and autophagy was increased when pretreated with EGCG.
In recent studies, EGCG in high doses (>40 µmol/L) suppress cancer cells by inducing apoptosis and by inhibiting autophagic processes (27,28). However, several studies also illustrate that EGCG in low doses (<20 µmol/L) play a preventive role in cancer developing via promoting autophagic processes during the early phase of cancer (29). Based on these studies, we observed that EGCG in low doses (<20 µmol/L) showed an autophagy-inductive effect on both cancer cells and normal cells. In our study results, we found that EGCG inhibited the apoptosis process at a concentration of 10 µmol/L, but the effect was not strong enough to completely protect HUVECs from oxidative stress-caused cell death. Since EGCG is a multi-target compound, we can confirm that the protective effect of EGCG on H2O2-caused HUVECs cell death is not only based a single mechanism.
In the following studies, we analyzed the underlying molecular mechanisms involved in EGCG-mediated autophagy induction. We know that the PI3K-AKT-mTOR signaling pathway plays an essential role in many biological and pathological processes in cancer cells and normal cells (30-32). mTOR is one essential regulator of autophagy because of its ability to sense stress, growth factor and nutrient states (33). We all know that mTOR signaling is very vital in regulating lysosomal function (34). It has been confirmed that EGCG competitively inhibits PI3K and mTOR with Ki values of 380 and 320 nM, respectively (35). We found that EGCG down-regulated phosphorylation of mTOR in a dose dependent manner, which is consistent with the opinion that mTOR mediates autophagy by regulating phosphorylation (36). Knockdown of mTOR partially promoted the protective effect of EGCG on oxidative stress. This result provided solid evidence on our hypothesis that EGCG reverses H2O2-caused damage on HUVECs via targeting mTOR.
Conclusions
In this work, we illustrated EGCG in low doses can protect vascular endothelial cells from oxidative stress-induced damage by inducing the autophagy and inhibiting apoptosis. In particular, pretreatment with EGCG significantly improved the survival of HUVECs from H2O2-induced cell death, upregulated the levels of Atg 5, Atg 7, LC3 II/I, and the Atg5–Atg12 complex and downregulated apoptosis-related proteins expression. In addition, we verified that EGCG induced autophagy by targeting the PI3K-AKT-mTOR pathway. All of these results indicate a novel role for EGCG in the treatment of oxidative stress-related cardiovascular and cerebrovascular diseases.
Acknowledgments
Funding: This work was supported by the Shaanxi Fourth People’s Hospital (grant number: 2017SY-003). We are grateful to Prof. Li for kindly donated the mCherry-EGFP-LC3 plasmid.
Footnote
Conflicts of Interest: The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. All animal experiments were performed according to NIH Guide for the Care and Use of Laboratory Animals and approved by the Institutional Animal Care and Use Committee of the Fourth People’s Hospital of Shaanxi (China).
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Maiese K, Chong ZZ, Hou J, et al. Oxidative stress: Biomarkers and novel therapeutic pathways. Exp Gerontol 2010;45:217-34. [Crossref] [PubMed]
- Johnson R, Bryant S, Huntley AL. Green tea and green tea catechin extracts: an overview of the clinical evidence. Maturitas 2012;73:280-7. [Crossref] [PubMed]
- Khurana S, Venkataraman K, Hollingsworth A, et al. Polyphenols: benefits to the cardiovascular system in health and in aging. Nutrients 2013;5:3779-827. [Crossref] [PubMed]
- Meydani M, Hasan ST. Dietary polyphenols and obesity. Nutrients 2010;2:737-51. [Crossref] [PubMed]
- Shankar S, Ganapathy S, Srivastava RK. Green tea polyphenols: biology and therapeutic implications in cancer. Front Biosci 2007;12:4881-99. [Crossref] [PubMed]
- Fürst R, Zundorf I. Plant-derived anti-inflammatory compounds: hopes and disappointments regarding the translation of preclinical knowledge into clinical progress. Mediators Inflamm 2014;2014:146832. [Crossref] [PubMed]
- Ge D, Jing Q, Meng N, et al. Regulation of apoptosis and autophagy by sphingosylphosphorylcholine in vascular endothelial cells. J Cell Physiol 2011;226:2827-33. [Crossref] [PubMed]
- Mizushima N, Levine B, Cuervo AM, et al. Autophagy fights disease through cellular self-digestion. Nature 2008;451:1069-75. [Crossref] [PubMed]
- Kimura S, Noda T, Yoshimori T. Dissection of the autophagosome maturation process by a novel reporter protein, tandem fluorescent-tagged LC3. Autophagy 2007;3:452-60. [Crossref] [PubMed]
- Jain SK, McVie R, Duett J, et al. Erythrocyte membrane lipid peroxidation and glycosylated hemoglobin in diabetes. Diabetes 1989;38:1539-43. [Crossref] [PubMed]
- Xie Y, You SJ, Zhang YL, et al. Protective role of autophagy in AGE-induced early injury of human vascular endothelial cells. Mol Med Rep 2011;4:459-64. [PubMed]
- Lemaître V, Dabo AJ, D'Armiento J. Cigarette smoke components induce matrix metalloproteinase-1 in aortic endothelial cells through inhibition of mTOR signaling. Toxicol Sci 2011;123:542-9. [Crossref] [PubMed]
- Mancini D, Pinney S, Burkhoff D, et al. Use of rapamycin slows progression of cardiac transplantation vasculopathy. Circulation 2003;108:48-53. [Crossref] [PubMed]
- Mueller CF, Laude K, McNally JS, et al. ATVB in focus: redox mechanisms in blood vessels. Arterioscler Thromb Vasc Biol 2005;25:274-8. [Crossref] [PubMed]
- Jaffe EA, Nachman RL, Becker CG, et al. Culture of human endothelial cells derived from umbilical veins. Identification by morphologic and immunologic criteria. J Clin Invest 1973;52:2745-56. [Crossref] [PubMed]
- Pan Y, Zhong LJ, Zhou H, et al. Roles of vimentin and 14-3-3 zeta/delta in the inhibitory effects of heparin on PC-3M cell proliferation and B16-F10-luc-G5 cells metastasis. Acta Pharmacol Sin 2012;33:798-808. [Crossref] [PubMed]
- Klionsky DJ, Abeliovich H, Agostinis P, et al. Guidelines for the use and interpretation of assays for monitoring autophagy in higher eukaryotes. Autophagy 2008;4:151-75. [Crossref] [PubMed]
- Pankiv S, Clausen TH, Lamark T, et al. p62/SQSTM1 binds directly to Atg8/LC3 to facilitate degradation of ubiquitinated protein aggregates by autophagy. J Biol Chem 2007;282:24131-45. [Crossref] [PubMed]
- Zhu S, Li W, Ward MF, et al. High mobility group box 1 protein as a potential drug target for infection- and injury-elicited inflammation. Inflamm Allergy Drug Targets 2010;9:60-72. [Crossref] [PubMed]
- Munir KM, Chandrasekaran S, Gao F, et al. Mechanisms for food polyphenols to ameliorate insulin resistance and endothelial dysfunction: therapeutic implications for diabetes and its cardiovascular complications. Am J Physiol Endocrinol Metab 2013;305:E679-86. [Crossref] [PubMed]
- Brownlee M. Biochemistry and molecular cell biology of diabetic complications. Nature 2001;414:813-20. [Crossref] [PubMed]
- Fouraux MA, Deneka M, Ivan V, et al. Rabip4' is an effector of rab5 and rab4 and regulates transport through early endosomes. Mol Biol Cell 2004;15:611-24. [Crossref] [PubMed]
- Vaziri ND, Rodriguez-Iturbe B. Mechanisms of disease: oxidative stress and inflammation in the pathogenesis of hypertension. Nat Clin Pract Nephrol 2006;2:582-93. [Crossref] [PubMed]
- Levine B, Yuan J. Autophagy in cell death: an innocent convict? J Clin Invest 2005;115:2679-88. [Crossref] [PubMed]
- Galluzzi L, Bravo-San Pedro JM, Levine B, et al. Pharmacological modulation of autophagy: therapeutic potential and persisting obstacles. Nat Rev Drug Discov 2017;16:487-511. [Crossref] [PubMed]
- Lum JJ, DeBerardinis RJ, Thompson CB. Autophagy in metazoans: cell survival in the land of plenty. Nat Rev Mol Cell Biol 2005;6:439-48. [Crossref] [PubMed]
- Chen L, Ye HL, Zhang G, et al. Autophagy inhibition contributes to the synergistic interaction between EGCG and doxorubicin to kill the hepatoma Hep3B cells. PLoS One 2014;9:e85771. [Crossref] [PubMed]
- Lecumberri E, Dupertuis YM, Miralbell R, et al. Green tea polyphenol epigallocatechin-3-gallate (EGCG) as adjuvant in cancer therapy. Clin Nutr 2013;32:894-903. [Crossref] [PubMed]
- Irimie AI, Braicu C, Zanoaga O, et al. Epigallocatechin-3-gallate suppresses cell proliferation and promotes apoptosis and autophagy in oral cancer SSC-4 cells. Onco Targets Ther 2015;8:461-70. [PubMed]
- Cui H, Wu S, Shang Y, et al. Pleurotus nebrodensis polysaccharide(PN50G) evokes A549 cell apoptosis by the ROS/AMPK/PI3K/AKT/mTOR pathway to suppress tumor growth. Food Funct 2016;7:1616-27. [Crossref] [PubMed]
- Xiaokaiti Y, Wu H, Chen Y, et al. EGCG reverses human neutrophil elastase-induced migration in A549 cells by directly binding to HNE and by regulating alpha1-AT. Sci Rep 2015;5:11494. [Crossref] [PubMed]
- Yang Y, Cong H, Han C, et al. 12-Deoxyphorbol 13-palmitate inhibits the expression of VEGF and HIF-1alpha in MCF-7 cells by blocking the PI3K/Akt/mTOR signaling pathway. Oncol Rep 2015;34:1755-60. [Crossref] [PubMed]
- Jung CH, Ro SH, Cao J, et al. mTOR regulation of autophagy. FEBS Lett 2010;584:1287-95. [Crossref] [PubMed]
- Sinha D, Valapala M, Shang P, et al. Lysosomes: Regulators of autophagy in the retinal pigmented epithelium. Exp Eye Res 2016;144:46-53. [Crossref] [PubMed]
- Van Aller GS, Carson JD, Tang W, et al. Epigallocatechin gallate (EGCG), a major component of green tea, is a dual phosphoinositide-3-kinase/mTOR inhibitor. Biochem Biophys Res Commun 2011;406:194-9. [Crossref] [PubMed]
- Alers S, Loffler AS, Wesselborg S, et al. Role of AMPK-mTOR-Ulk1/2 in the regulation of autophagy: cross talk, shortcuts, and feedbacks. Mol Cell Biol 2012;32:2-11. [Crossref] [PubMed]


